Abstract
The small GTPase Ran has been found to play pivotal roles in several aspects of cell function. We have investigated the role of the Ran GTPase cycle in spindle formation and nuclear envelope assembly in dividing Caenorhabditis elegans embryos in real time. We found that Ran and its cofactors RanBP2, RanGAP, and RCC1 are all essential for reformation of the nuclear envelope after cell division. Reducing the expression of any of these components of the Ran GTPase cycle by RNAi leads to strong extranuclear clustering of integral nuclear envelope proteins and nucleoporins. Ran, RanBP2, and RanGAP are also required for building a mitotic spindle, whereas astral microtubules are normal in the absence of these proteins. RCC1(RNAi) embryos have similar abnormalities in the initial phase of spindle formation but eventually recover to form a bipolar spindle. Irregular chromatin structures and chromatin bridges due to spindle failure were frequently observed in embryos where the Ran cycle was perturbed. In addition, connection between the centrosomes and the male pronucleus, and thus centrosome positioning, depends upon the Ran cycle components. Finally, we have demonstrated that both IMA-2 and IMB-1, the homologues of vertebrate importin α and β, are essential for both spindle assembly and nuclear formation in early embryos.
INTRODUCTION
The small GTPase Ran has been extensively studied due to its central role in major cellular processes such as nucleocytoplasmic transport, spindle microtubule dynamics, and nuclear envelope (NE) assembly (see recent reviews by Dasso, 2001; Kuersten et al., 2001; Macara, 2001; Moore, 2001; Vasu and Forbes, 2001). Although very diverse, these processes seem to all depend on the GTP- and GDP-bound forms of Ran interacting differentially with downstream targets. When in the GTP conformation Ran binds to a class of importin β-like proteins and thereby regulates their affinity for various interaction partners (Kuersten et al., 2001). Ran on its own has weak GTPase activity, which can be stimulated ∼105-fold by the Ran GTPase-activating protein RanGAP (Klebe at al., 1995). However, in complex with importin β-like proteins Ran is usually inaccessible to RanGAP. Proteins harboring a certain type of Ran binding domain, such as Ran binding proteins (RanBPs) 1 and 2, can overcome this inhibition, making RanGTP accessible to RanGAP (Bischoff and Görlich, 1997; Floer et al., 1997; Askjaer et al., 1999). A single guanine nucleotide exchange factor for Ran, termed RCC1, has been described previously (Bischoff and Ponstingl, 1991; Klebe et al., 1995). Ran is thought to be mainly soluble, although it binds to mitotic chromatin, whereas RCC1 also binds to chromatin, and a proportion of RanGAP and RanBP1/2 is tethered to the cytoplasmic filaments of nuclear pore complexes (Kuersten et al., 2001; Macara, 2001, Bilbao-Cortés et al., 2002).
When cells enter mitosis, the stability of cytoplasmic microtubules is dramatically reduced and spindle microtubules are nucleated by centrosomes and chromatin (Compton, 2000; Wittmann et al., 2001). This process can be reproduced in meiotic Xenopus egg extracts in the presence of an exogenous source of chromatin. Strikingly, addition of RanGTP or mutant forms of Ran locked in a GTP-like conformation to such extracts stimulates microtubule assembly and can even induce the formation of centrosome-independent bipolar spindles in the presence or absence of chromatin (Carazo-Salas et al., 1999; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999; Zhang et al., 1999). RCC1 also stimulates spindle formation in experiments with wild-type Ran, whereas a mutant form of Ran with low affinity for guanine nucleotides inhibits spindle formation, presumably by inhibiting RCC1 (Carazo-Salas et al., 1999; Kalab et al., 1999). From these studies a model emerged in which chromatin-bound RCC1 generates RanGTP in the vicinity of the chromosomes in mitosis. This high local concentration of RanGTP, which has been visualized experimentally (Kalab et al., 2002), is then believed to stabilize mitotic microtubules that are assembled into a spindle structure by motor proteins (reviewed by Dasso, 2001; Moore, 2001).
In yeast, the Ran system has also been proposed to regulate mitotic spindle formation independently of nuclear transport (Ouspenski, 1998; Fleig et al., 2000; Quimby et al., 2000). However, yeast have a closed mitosis where the spindle is formed within the nucleus, suggesting the mechanisms underlying spindle assembly are likely to differ from those proposed on the basis of the in vitro data described above (Dasso, 2001). In mammalian cells, only the role of RanBP1 in mitosis has been investigated. On overexpression of RanBP1, ∼50% of mitotic cells had abnormal spindles, typically monopolar or multipolar (Guarguaglini et al., 2000). On the other hand, reducing RanBP1 activity by microinjection of anti-RanBP1 antibodies delayed progression through mitosis by slowing down metaphase and anaphase (Guarguaglini et al., 2000). Finally, a recent report demonstrated that the Ran system is required for proper positioning of chromosomes in Caenorhabditis elegans embryos, presumably reflecting a role of Ran in spindle formation (Bamba et al., 2002). Two potential downstream targets for Ran in spindle formation have been identified, namely, TPX2 (Gruss et al., 2001) and NuMA (Nachury et al., 2001; Wiese et al., 2001). Interestingly, the effect of Ran on spindle assembly via TPX2 and NuMA is mediated by regulating their interaction with importin α and importin β, and is thus analogous to how Ran controls nucleocytoplasmic transport.
An additional function of Ran has been reported. NE assembly can be studied in vitro by adding chromatin to Xenopus egg extracts (Gant and Wilson, 1997; Vasu and Forbes, 2001). Membrane vesicles dock to the chromatin and fuse to form a closed envelope. Nuclear pore complexes form and are inserted into the NE. The reconstituted nucleus starts to grow in a nuclear import-dependent manner. It was found that either RanGTP or RanGDP together with RCC1 is necessary for early membrane fusion steps in NE assembly (Hetzer et al., 2000). Similarly, beads coated with wild-type Ran formed pseudonuclei surrounded by an NE, whereas beads attached to mutant forms of Ran incapable of cycling between GTP- and GDP-bound conformations failed to do so (Zhang and Clarke, 2000). In fission yeast, the RCC1 homologue pim1p is required for maintenance of a normal NE after mitosis (Demeter et al., 1995). However, because the NE does not break down during mitosis in yeast, it is difficult to rule out the possibility that the observed fragmentation of the NE is not caused by a protein import defect. More recently, it was reported that perturbing the Ran system in C. elegans by double-stranded RNA-mediated interference (RNAi) gives rise to embryos where the chromatin no longer associated with nucleoporins, indicative either of a defect in NE assembly or in nuclear pore complex assembly or insertion into the NE (Bamba et al., 2002). Importin β has recently been proposed to be a downstream effector of Ran in NE assembly on the basis of in vitro experiments, including the ability of importin β to attract nuclear pore complex-containing membranes when attached to the surface of inert beads (Zhang et al., 2002).
We have analyzed the effects of perturbation of the Ran system in vivo by using C. elegans embryos and time-lapse microscopy. C. elegans embryos can be efficiently depleted of single proteins by RNAi. When combined with live cell imaging RNAi can provide detailed insight into the function of individual genes in morphogenetic processes. We demonstrate that Ran itself is absolutely required for formation of a mitotic spindle but not for nucleation of astral microtubules. Depletion of RanBP2 or RanGAP gives a similar phenotype, demonstrating that the GTPase activity of Ran is also required for spindle formation. Targeting RCC1 by RNAi results in a strong defect early in spindle formation, however, this defect is temporary and a spindle is eventually assembled. In contrast, we show that all components of the Ran GTPase cycle are required for formation of a closed NE after cell division and for centrosome-pronuclear attachment. Finally, we demonstrate that IMA-2 and IMB-1, C. elegans homologues of importin α and β, are essential for both spindle formation and NE assembly.
MATERIALS AND METHODS
Worm Strains
The C. elegans Bristol strain N2 was used for analysis of RNAi effects on brood size and viability. Strain N2 was also used to create transgenic nematodes by microinjection. DP38 unc-119(ed3) (Maduro and Pilgrim, 1995) and AZ212 (Praitis et al., 2001) were provided by the Caenorhabditis Genetic Center. A heterogeneous population containing nematodes carrying the deletion allele ok256 of the ima-2 gene was obtained from The Caenorhabditis elegans Gene Knockout Consortium. WH204, which expresses a green fluorescent protein (GFP)::β-tubulin fusion protein in the germline, and strains expressing GFP::histone H2B and GFP::α-tubulin fusion proteins from extrachromosomal arrays have been described previously (Oegema et al., 2001; Strome et al., 2001).
Plasmid Constructs
Plasmid pJH4.52 contains the promoter and noncoding sequences from the C. elegans pie-1 gene as well as the coding sequence for GFP-tagged C. elegans histone H2B (Strome et al., 2001). To generate pPAG20 a multiple cloning sequence was polymerase chain reaction (PCR) amplified and used to replace the histone H2B sequence of pJH4.52. Using sequence information from ACeDB (http://www.wormbase.org), we PCR amplified the genomic DNA sequence of the C. elegans emerin gene (emr-1, M01D7.6) and the C. elegans RCC1 gene (ran-3). The emr-1 PCR product was inserted downstream of the gfp sequence in pPAG20 to generate plasmid pPAG7, whereas the ran-3 PCR product was used to replace the histone H2B sequence of pJH4.52, generating plasmid pPAG38.
For generation of RNAi constructs the following sequences were obtained by either PCR or reverse-transcription (RT)-PCR: full length C. elegans Ran (ran-1, nucleotides [nt] 1–648 of predicted open reading frame [ORF]); C. elegans RanGAP (ran-2, nt 1789–2533 of predicted ORF); three different C. elegans RCC1 fragments (ran-3, nt 1–732, 396-1013, and 1033–1600 of predicted ORF); C. elegans RanBP2/Nup358 (npp-10, nt 1555–2238 of predicted ORF); three C. elegans importin α homologues (ima-1, nt 364-1158 of predicted ORF; ima-2, nt 814-1389 of predicted ORF; ima-3, nt 745-1542 of predicted ORF); and two different C. elegans importin β fragments (imb-1, nt 916-1425 and 1659–2340 of predicted ORF). In addition, fragments of three C. elegans genes (F07C3.4, Y48G8AL.1, and W09G3.3) with similarity to human RCC1, but more distantly related than ran-3, were cloned. In all cases except for the 3′ imb-1 fragment, only intron-less sequences were amplified. The DNA fragments were inserted into either pPD129.36 L4440 (Timmons and Fire, 1998) or pCRII TOPO (Invitrogen, Carlsbad, CA) and then subcloned into pPD129.36. As negative control in RNAi experiments, the empty pPD129.36 vector was used. Detailed cloning information is available on request.
Generation of Transgenic Worms
To generate transgenic worms expressing GFP::emerin fusion proteins, we initially performed microinjections into the gonads of wild-type adult hermaphrodites (Strome et al. 2001). Worms were injected with a mixture containing 50 μg/ml genomic DNA (linearized with PvuII), 1 μg/ml pPAG7 (linearized with SacII), and 1 μg/ml pRF4 rol-6(su1006) (linearized with EcoRI). Several independent lines were obtained of which two (XA3510 and XA3511) showed relatively stable GFP::emerin expression over multiple generations. XA3510, N2 qaEx3510[rol-6(su1006) pie-1::gfp::emr-1] was used in this study and maintained at ∼24.5°C. Eventually, GFP::emerin expression was silenced in XA3510 and XA3511. To overcome this problem we exploited the gold particle bombardment method (Praitis et al. 2001). The rescuing gene (unc-119) and the gfp-fusion gene were, however, on separate plasmids: a 1:1 mixture of plasmids pDP#MM051 (Maduro and Pilgrim, 1995) and pPAG7 was precipitated on 1.5- to 3-μm gold particles (Sigma-Aldrich). Roughly 10,000 DP38 unc119(ed3) worms were used per bombardment and from six bombardments we obtained two independent transformed lines with wild-type behavior. One line was positive for GFP expression, giving rise to strain XA3504, unc-119(ed3) qaEx3504[unc-119 (+) pie-1::gfp::emr-1]. XA3504 is maintained at ∼20°C and has so far been stable for >60 generations. Two independent strains expressing GFP::RCC1 (XA3515 and XA3516) were generated similarly to XA3504 by using plasmid pPAG38. XA3515 showed germline expression for approximately eight generations before being silenced, whereas XA3516 still is GFP positive. To obtain a strain that expresses both GFP::histone H2B and GFP::β-tubulin we crossed the strains AZ212 and WH204, giving rise to strain XA3501.
From The Caenorhabditis elegans Gene Knockout Consortium we obtained ima-2(ok256) worms carrying a ∼1.8-kb deletion in the 3′ region of the ima-2 gene. To map precisely the mutation we cloned and sequenced the ok256 allele. Sequence information has been submitted to Wormbase. The ok256 allele was kept in the population by PCR screening for heterozygous individuals carrying the deletion: single worms were incubated in 2.5 μl of lysis buffer (10 mM Tris-HCl, pH 8.5, 50 mM KCl; 2.5 mM MgCl2, 0.45% Nonidet-40, 0.45% Tween 20, 0.01% gelatin, 0.2 mg/ml proteinase K) at −80°C for 15 min, and then at 60°C for 60 min and finally at 95°C for 15 min. Standard AmpliTaq (22.5 μl; Applied Biosystems, Foster City, CA) PCR mix containing primers 5′-GAA GAG GGA AAG GAT GAG GG and 5′-GTT TGA TGT TTT CAC CGC CT was added to the worm lysate followed by heating to 94°C for 3 min before PCR for 35 cycles (94°C, 40 s; 58°C, 40 s; and 72°C, 90 s). After outcrossing ima-2(ok256) worms with N2 wild-type males for seven generations, we named the strain XA3513 ima-2(ok256). Strain XA3503 was then created by crossing XA3513 with WH204 followed by selecting worms heterozygous for the ima-2(ok256) allele and homozygous for gfp::β-tubulin.
RNAi
RNA-mediated interference was performed by feeding the worms with bacteria that express double-stranded RNA (dsRNA) (Fraser et al., 2000). RNAi plates were generated as follows: Escherichia coli strain HT115(DE3) transformed with plasmid pPD129.36 harboring the relevant RNAi fragment was grown for 8 h at 37°C in the presence of ampicillin. Isopropyl β-d-thiogalactoside was added to a concentration of 1 mM, and the bacteria were seeded onto NGM plates containing 50 μg/ml ampicillin and 1 mM isopropyl β-d-thiogalactoside. Plates were left to dry at room temperature over night and then used immediately or stored at 16°C. To analyze the effect of RNAi on brood size and viability L3 larvae were incubated on the RNAi plates at 16°C for 70 h. Young adults from these plates were then transferred to individual RNAi plates and incubated at 20°C for 20 h. The adult hermaphrodites were removed from the plates. After further incubation of the plates the number of oocytes, embryos, and larvae on the plates was determined. To obtain RNAi embryos for imaging, L3 and L4 larvae from the GFP strains were incubated on the RNAi plates at 20°C or 25°C for 16 to 48 h, depending on the gene.
Live Embryo Imaging
Embryos were mounted in M9 buffer on 2% agarose pads and covered with a coverslip. Observation and recording of epifluorescence and transmitted light was with 1) Leica confocal microscope TCS SP2 with an HCX PL APO 100×/1.40-0.7 objective. Images were captured using integrated Leica software (Leica Microsystems, Wetzlar, Germany) and processed with NIH Image. 2) Nikon Eclipse TE200 (Nikon Instech, Kanagawa, Japan) with a Plan Fluor 100×/1.30 objective and a PerkinElmer Life Sciences Ultraview real-time confocal spinning disk unit (Perkin Elmer Life Sciences, Boston, MA). Data were processed with IDL (Research Systems, Boulder, CO) and NIH Image. 3) Zeiss Axioplan 2 Imaging (Carl Zeiss Microimaging, Thornwood, NY), with a Plan Apochromat 63×/1.40 objective equipped either with a Hamamatsu CA742-95 camera (Hamamatsu Photonics, Hamamatsu, Japan) or a Visitech International QLC100 spinning disk confocal (Visitech International, Sunderland, United Kingdom). MetaMorph software (Universal Imaging, West Chester, PA) was used to control hardware and acquire and process images. With all systems, laser intensities were adjusted so that no effect on development was seen. Depending on the system and on the GFP strain being analyzed images were collected at 6- to 20-s intervals for a total of 20–40 min.
Immunofluorescence
AZ212 worms from RNAi plates were dissected directly on poly-l-lysine–coated glass slides. The eggshell was opened by freeze cracking and the embryos were fixed in methanol for 20–30 min at −20°C. After rehydration in phosphate-buffered saline (PBS) with 0.1% Tween 20 and blocking with 3% milk the embryos were incubated with either a 1:500 dilution of monoclonal antibody (mAb) 414 against nucleoporins (Jackson Immunoresearch Laboratories, West Grove, PA), a 1:500 dilution of mAb DM1A against α-tubulin (Sigma-Aldrich, St. Louis, MO), a 1:250 dilution of polyclonal antibodies against phosho-histone H3 (Upstate Biotechnology, Lake Placid, NY), and/or a 1:100 dilution of polyclonal antibodies against EMR-1 (Gruenbaum et al., 2002). After incubation for 45–120 min at room temperature embryos were washed for 5 min in PBS with 0.05% Tween 20 followed by 5 min in PBS or for 60 min in PBS with 0.1% Tween 20. Alexa Fluor 546 and Alexa Fluor 633 secondary antibodies (Molecular Probes, Eugene, OR) were used at 1:2000 dilution in PBS for 45–120 min. The embryos were washed as described above and mounted. Confocal images were obtained on a Leica TCS SP2 microscope as described above.
RESULTS
Identification of Essential Ran and Ran-related Genes
To study the role of Ran in mitotic spindle formation and NE assembly, we identified C. elegans homologues for Ran and Ran cofactors (RanGAP, RanBP2, and RCC1) based on published data (Gönczy et al., 2000) and database searches (Table 1; Bamba et al., 2002). Because importin α and β have been demonstrated to play critical roles in spindle formation and/or NE assembly in Xenopus extracts (Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001; Zhang et al., 2002) we included the C. elegans homologues in our studies. Geles and Adam (2001) have described three C. elegans homologues of importin α, IMA-1, -2, and -3, whereas importin β is homologous to C. elegans IMB-1 (Geles and Adam, 2001; Table 1). Because RNAi against IMA-1 gives no observable phenotype (Geles and Adam, 2001; our unpublished data) IMA-1 was not analyzed further.
Table 1.
RNAi effects on brood size and viability
| C. elegans genea | Sequence | Human gene | PNb | nc | Brood sized | Lethality (%)e | |
|---|---|---|---|---|---|---|---|
| Controlf | 18 | 132 ± 37 | 0 ± 0 | ||||
| Ran | ran-1 | K01G5.4 | Ran | 1.6e−102 | 12 | 14 ± 16 | 100 ± 0 |
| RanGAP | ran-2 | C29E4.3 | RanGAP | 4.7e−73 | 7 | 140 ± 29 | 99 ± 1 |
| RanBP2 | npp-10 | F59A2.1 | RanBP2 | 2.6e−65 | 7 | 122 ± 60 | 100 ± 0 |
| RCC1 | ran-3 | C26D10.1 | RCC1 | 6.0e−34 | 10 | 151 ± 45 | 91 ± 13 |
| IMA-2 | ima-2 | F26B1.3 | Importin α | 5.3e−90 | 10 | 149 ± 43 | 99 ± 2 |
| IMA-3 | ima-3 | F32E10.4 | Importin α | 2.8e−116 | 9 | 35 ± 25 | 99 ± 2 |
| IMB-1 | imb-1 | F28B3.8 | Importin β | 9.8e−217 | 14 | 74 ± 60 | 100 ± 1 |
Left column shows gene product names used in this study; right column shows systematic gene name.
Probability values obtained in blast searches with human protein sequences against the WormPep database at http://www.wormbase.org.
Number of adult hermaphrodites analyzed for RNAi effects on brood size and embryonic lethality.
Average number of embryos being laid per hermaphrodite in 20 h ± SD. See MATERIALS AND METHODS for details.
Average percentage of embryos that do not hatch ± SD.
Control experiments were done by feeding the worms bacteria containing empty plasmid.
As an initial characterization of the seven genes (Table 1), we evaluated the effect of RNAi on brood size and embryo viability. We found that all seven genes are required for embryonic development because 91–100% of the embryos failed to hatch (Table 1). For some of the genes this is in agreement with previous observations (IMA-2, Fraser et al., 2000; Ran, RanGAP, and RanBP2, Gönczy et al., 2000; and IMA-3, Geles and Adam, 2001), whereas others had not been tested before. RNAi of Ran significantly reduced the brood size, indicating that Ran is also required at some step during oogenesis, whereas RNAi of any of the regulators of Ran's nucleotide state did not affect brood size (Table 1). Thus, it is possible that Ran might have a function in oogenesis different from its GTPase activity, although it is also possible that RNAi fails to efficiently deplete Ran's regulators from the gonads.
Ran Is Required for Spindle Formation
Previous RNAi experiments have shown that Ran, RanGAP, and RanBP2 are involved in pronuclear and nuclear appearance and possibly also spindle formation (Gönczy et al., 2000). IMA-2 has similarly been proposed to play a role in pronuclear formation (Zipperlen et al., 2001). However, differential interference contrast (DIC) microscopy, the method of choice for those high-throughput screens, does not give detailed insight to the subcellular structures that are affected or the defects caused. We therefore retested these genes, as well as RCC1, IMB-1, and IMA-3, with a range of fluorescent markers and antibodies.
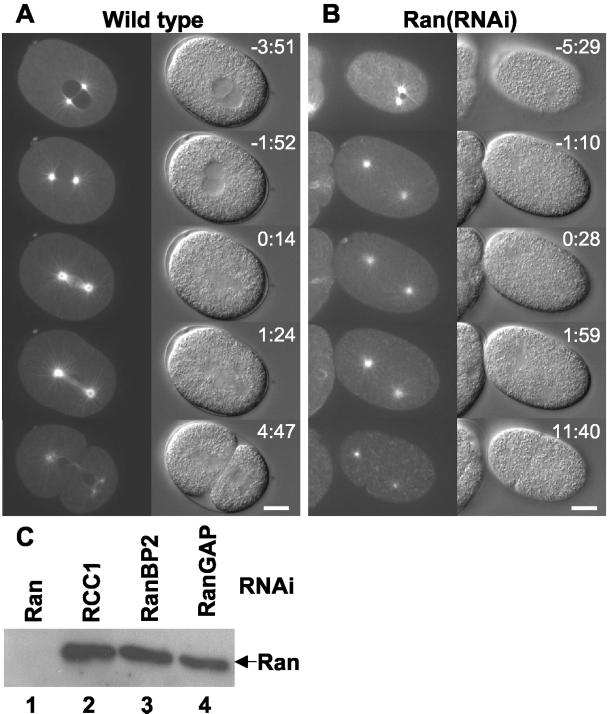
To investigate spindle formation in time-lapse microscopy we initially used a strain expressing a fusion protein of GFP and α-tubulin (Oegema et al., 2001). In normal one-cell embryos the two centrosomes are associated with the sperm pronucleus until pronuclear meeting when the centrosomes border the junction between the two pronuclei (Figure 1A, −3:51. Note that in all time-lapse experiments time is indicated relative to anaphase onset; Video 1). The pronuclei then migrate together to the center of the embryo and the pronuclear envelopes break down as visualized by entry of soluble GFP::tubulin into the nuclear space (Figure 1A, −1:52). A spindle is rapidly formed, followed soon by anaphase (Figure 1A, 0:14), telophase, and cytokinesis (Figure 1A, 4:47). When Ran is depleted by RNAi (Figure 1C), several features are striking (4 of 5 embryos). First, the pronuclei are very small (Figure 1B, −5:29; Video 2) as reported previously (Gönczy et al., 2000). Second, the embryos fail to set up a spindle, although the centrosomes are still capable of nucleating astral microtubules (Figure 1B, −1:10–1:59). Third, from being closely associated with the male pronucleus the centrosomes immediately move far apart and after several minutes undergo strong sideward movements, as in normal anaphase (Video 2). This process (“spindle rocking”) together with cleavage furrow ingression was used throughout this study to define anaphase in embryos where no spindle was formed. Fourth, in some embryos, the centrosomes are not properly positioned so that the first cleavage leads to an abnormal bisection of the embryo as seen in Figure 1B (11:40) where the posterior cell P1 is larger than the anterior AB. Taken together, these data indicate that Ran is required for spindle formation, but not for astral microtubule nucleation.
Figure 1.
Ran is required for spindle formation. Still images from time-lapse microscopy are shown from wild-type (A) and Ran(RNAi) (B) embryos expressing GFP::α-tubulin. Anterior of the embryos is on the left and time (minutes:seconds) relative to anaphase onset is indicated. For the wild-type embryo the following events are depicted: pronuclear meeting (−3:51), pronuclear envelope breakdown (−1:52), metaphase-to-anaphase transition (0:14), anaphase (1:24), and telophase and cytokinesis (4:47). In the Ran(RNAi) embryo no spindle is formed, nevertheless the embryo eventually divides (11:40). Bar, 10 μm. (C) Western blot analysis of Ran in embryonic lysates from worms depleted of Ran (lane 1), RCC1 (lane 2), RanBP2 (lane 3), or RanGAP (lane 4). Expression of Ran is efficiently and specifically inhibited by Ran dsRNA. Equal loading of protein was verified with antibodies against LEM-2 (our unpublished data).
Figure 4.
Depletion of RCC1 leads to defects in chromosome segregation. Embryos expressing GFP::β-tubulin and GFP::histone H2B from either control worms (A) or worms depleted of RCC1 (B) were observed by time-lapse microscopy. Still images are shown with indication of time (minutes:seconds) relative to anaphase onset. Anterior of the embryos is on the left. RCC1 depletion causes problems in the initial phase of spindle formation and in DNA segregation. Bar, 10 μm. (C) Comparison of one-cell embryos from GFP::RCC1 worms grown on either control bacteria (left), or bacteria expressing dsRNA corresponding to RCC1 (middle). RNAi against RCC1 decreases the level of fluorescence to that of N2 embryos with no GFP gene (right). Dotted white lines indicate pronuclei and plasma membranes. Bar, 10 μm. (D) Distance between the two centrosomes was measured relative to the total length of the embryo. Shown is the average of several wild-type (diamonds, n = 9), RCC1(RNAi) (squares, n = 10), and RanGAP(RNAi) (triangles, n = 5) embryos as a function of time. Each embryo was aligned relative to anaphase onset, which defines t0. Vertical lines represent the SD at each time point. Also shown is a single Ran(RNAi) embryo (crosses). For wild-type embryos, approximate timing of pronuclear meeting and pronuclear envelope breakdown (NEBD) is indicated as is DNA segregation and spindle rocking.
GTPase Activity of Ran Is Essential for Assembly of Spindle Microtubules
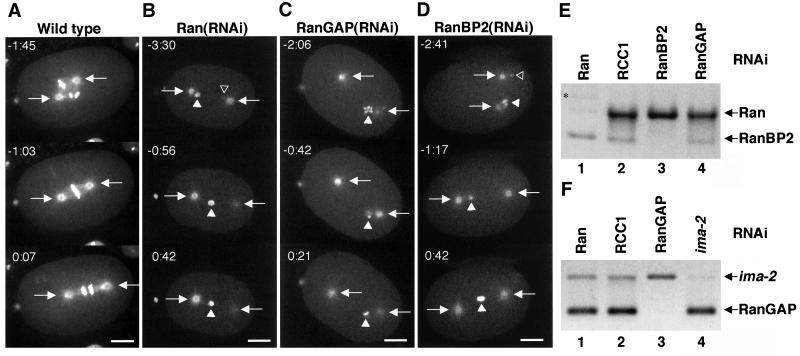
Chromosomes are proposed to play an active and important role in spindle formation (Compton, 2000; Wittmann et al., 2001). Depletion of Ran results in small pronuclei that are difficult to follow in DIC or GFP::α-tubulin recordings. The lack of a mitotic spindle in Ran(RNAi) embryos could therefore be a consequence of aberrant chromatin localization which in turn would cause a lack of chromatin-mediated microtubule-nucleating activity between the two centrosomes. To address this possibility, we created a transgenic strain that expresses GFP::histone H2B and GFP::β-tubulin by crossing the strains AZ212 (Praitis et al., 2001) and WH204 (Strome et al., 2001). This strain allowed us to precisely follow the localization and morphology of the chromatin concomitantly with centrosome position and microtubule dynamics. An example is shown in Figure 2A where three still pictures of a time-lapse recording represent prometaphase (−1:45), metaphase (−1:03), and anaphase (0:07) (Video 3).
Figure 2.
Depletion of Ran, RanGAP, or RanBP2 causes similar defects on spindle formation. Embryos expressing GFP::β-tubulin and GFP::histone H2B from either control worms (A) or worms depleted of Ran (B), RanGAP (C), or RanBP2 (D) were observed by time-lapse microscopy. Still images are shown with indication of time (minutes:seconds) relative to anaphase onset. Arrows point to the centrosomes, whereas open and closed triangles indicate the pronucleus originating from the sperm and the oocyte, respectively. The sperm pronucleus is in the focal plane only in certain images. Note that the chromatin aligns on a metaphase plate in the wild-type embryo, whereas the pronuclei fail to meet in RNAi embryos and no spindle is formed. Bar, 10 μm. (E) RNA prepared from embryos depleted of Ran (lane 1), RCC1 (lane 2), RanBP2 (lane 3), or RanGAP (lane 4) was analyzed by RT-PCR with primers specific for Ran and RanBP2. Ran and RanBP2 mRNA are efficiently and specifically depleted by dsRNA corresponding to Ran and RanBP2, respectively. Star in lane 1 indicates a RanBP2 PCR product from contaminating genomic DNA. (F) RNA prepared from embryos depleted of Ran (lane 1), RCC1 (lane 2), RanGAP (lane 3), or IMA-2 (lane 4) was analyzed by RT-PCR with primers specific for RanGAP and ima-2. RanGAP and ima-2 mRNA are efficiently and specifically depleted by dsRNA corresponding to RanGAP and ima-2, respectively. Similarly, RCC1 mRNA was inhibited by RCC1 dsRNA (our unpublished data; Figure 4C).
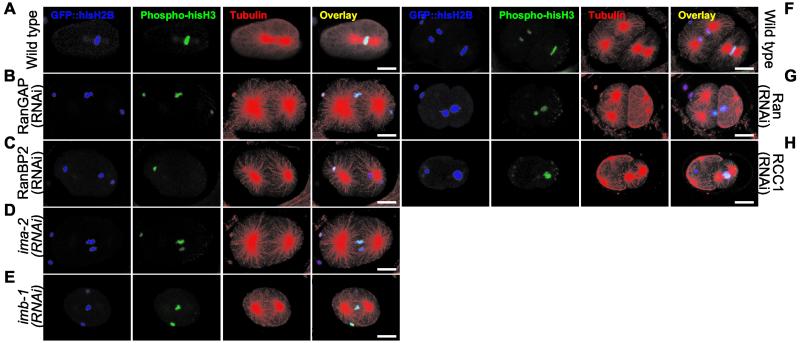
When we targeted Ran, RanGAP, or RanBP2 with RNAi (Figure 2, E–F), joining of the two pronuclei was never observed, although oocyte pronuclear migration still took place (10 of 10 embryos where both pronuclei could be followed almost continuously; Videos 4–6). In approximately half of the cases (6 of 10) one of the pronuclei was eventually positioned in line between the two centrosomes, either in the middle (Figure 2B, closed triangle) or toward one of the centrosomes (Figure 2C, closed triangle). Most often (5 of 6) the pronucleus found between the centrosomes was from the oocyte (Figure 2, B–D, closed triangle), whereas the sperm pronucleus was more or less closely attached to one centrosome (Figure 2, B and D, open triangle). In other cases (4 of 10), neither of the pronuclei aligned between the centrosomes (our unpublished data). Importantly, spindles were never assembled in embryos where Ran, RanGAP, or RanBP2 was depleted, regardless of chromatin position. Analysis of fixed embryos was also carried out because the combination of live cell imaging and immunofluorescence provides resolution in time together with high sensitivity. Staining of microtubules with anti-α-tubulin antibodies confirmed our live cell recordings because we could not detect spindles in embryos depleted of Ran, RanGAP, or RanBP2, even in cases where mitotic chromatin was positioned between two centrosomes (Figure 3, A–C, F and G; 28 of 28 embryos). Immunofluorescence with antibody against phosphorylated histone H3 was used to confirm that the embryos were in prometaphase or metaphase. Thus, our data strongly indicate that the GTPase activity of Ran is essential for the assembly of microtubules that connect chromatin to the centrosomes.
Figure 3.
RNAi embryos fail to assemble microtubules into spindles around mitotic chromatin. Embryos from GFP::histone H2B worms grown on either control bacteria (A and F), or bacteria expressing dsRNA corresponding to RanGAP (B), RanBP2 (C), ima-2 (D), imb-1 (E), Ran (G), or RCC1 (H) were fixed and analyzed by immunofluorescence. GFP::histone H2B (depicted in blue) was used to visualize total chromatin, whereas mitotic chromatin and microtubules were recognized with anti-phospho-histone H3 (green) and anti-α-tubulin (red) antibodies. An overlay of the three stains is shown on the right. Bar, 10 μm.
RNAi of RCC1 Causes DNA Segregation Defects
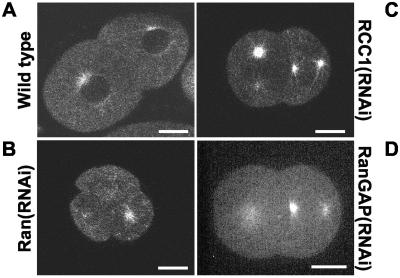
Inhibition of RCC1 activity by the Ran mutant T24N has been shown to interfere with spindle microtubule assembly in Xenopus egg extracts (Carazo-Salas et al., 1999; Kalab et al., 1999). Based on this and on our observation that Ran GTPase activity is necessary for spindle formation we predicted that inhibiting RCC1 expression would also block spindle formation by preventing formation of RanGTP. As with Ran depletions RCC1(RNAi) embryos are generally reduced in size (∼15% shorter than wild type) and have smaller pronuclei (Figure 4; our unpublished data). However, in contrast to the RNAi experiments described above the centrosomes in RCC1(RNAi) embryos stay associated with the sperm pronucleus. Instead of migrating to join the oocyte pronucleus at the normal position approximately one-third of the embryo length from the posterior pole the sperm pronucleus either remains at the cortex or moves only slightly away from it (14 of 14 embryos; our unpublished data). On joining, the two pronuclei migrate together toward the center of the embryo (Figure 4, −4:00; Video 7). Note that diffuse nucleoplasmic GFP::histone H2B staining is seen around the condensed chromatin before NE breakdown in wild-type embryos (Figure 4A, −3:12) but not in RNAi embryos (Figure 4B, −4:00; see Video 7 for earlier time points), suggestive of a defect in retention of GFP::histone H2B in the pronuclei. At this stage the centrosomes in the RCC1(RNAi) embryos moved away from the pronuclei (Figure 4, compare A and B at −1:48), creating a situation similar to Ran(RNAi) embryos in terms of distance between the centrosomes and the chromatin (Figure 2B). However, in the RCC1(RNAi) embryos this situation is rescued, giving rise to spindles that are indistinguishable from those in wild-type embryos (Figure 4, compare A and B at −0:12). Although the chromosomes are aligned between spindle microtubules in a seemingly normal metaphase (Figure 4B, −0:12) they clearly fail to segregate properly in anaphase (Figure 4B, 0:48). At later time points chromatin bridges are still observed across the cleavage furrow whereas wild-type embryos have completed cytokinesis and formed growing, spherical nuclei (Figure 4, A and B, 3:12). Consistent with the normal appearance of the mitotic spindle most (18 of 21) embryos divided into a larger AB and a smaller P1 daughter cell (Figure 4B).
Immunofluorescence analysis of RCC1(RNAi) embryos confirmed that DNA segregation is affected, yet mitotic spindles can assemble in embryos depleted of RCC1 (Figure 3H, compare missegregation of chromatin at the division of AB on the left with spindle formation in P1 on the right).
The observed pattern of an early spindle defect and later rescue could have been due to inefficient depletion of RCC1. To address this several approaches were taken: 1) In addition to RNAi-by-feeding we also tried to prevent RCC1 expression by injection of dsRNA into the gonads. 2) We targeted the RCC1 mRNA with three different dsRNAs covering in total the whole open reading frame. 3) Incubations on RNAi bacteria plates were prolonged up to 48 h. 4) RNAi was performed at either 20 or 25°C. None of these measures led to more severe effects on the spindle (our unpublished data). 5) Finally, we quantified the efficiency of RNAi by targeting RCC1 in a transgenic strain that expresses GFP::RCC1 in the germline. RCC1(RNAi) embryos from this strain had <5% GFP::RCC1 signal left when quantified by confocal microscopy (Figure 4C).
Centrosome Position Is Regulated by Ran System via Spindle Formation
To have another and more quantitative way of analyzing spindle formation, we measured the distance between the centrosomes relative to the total length of the embryo in time-lapse experiments. Aligning the data from nine different wild-type embryos relative to anaphase showed a very reproducible pattern (Figure 4D, Wild-type). Joining of the two pronuclei and setting up the spindle at NE breakdown do not change the distance between the centrosomes dramatically. Approximately 60 s before DNA segregation the spindle poles start to move apart, first slowly and later with higher velocity until maximum distance is reached and the embryo divides.
Although no spindle is formed in RanGAP(RNAi) embryos the centrosomes still rock from side to side, which was used as an anaphase indicator in aligning the curves from five independent embryos [Figure 4D, RanGAP(RNAi)]. The two centrosomes move rapidly apart ∼3 min before mitosis to a final separation (Figure 4D) and position (Figure 2C) identical to normal embryos. Ran(RNAi) embryos followed a pattern very similar to RanGAP depletions (3 of 3 embryos). Due to the more dramatic movements of centrosomes and pronuclei seen when targeting Ran several frames in the time-lapse recordings were out of focus with respect to the centrosomes, making averaging more difficult. Instead, a representative example of a single Ran(RNAi) embryo is presented in Figure 4D, showing a similar pattern as RanGAP depletions. As described above, RCC1(RNAi) embryos also show premature separation of the centrosomes but are able to reverse this and build a functional spindle. When averaging 10 independent embryos the phenotype seems highly reproducible [Figure 4D, RCC1(RNAi)]. Initially, the centrosomes are associated with the sperm pronucleus but as microtubules connecting chromatin with the centrosomes appear, the centrosomes start to move prematurely apart to a maximum separation comparable with Ran(RNAi) and RanGAP(RNAi) embryos (Figure 4D, approximately −180 s; Figure 4B, −1:48). Later, the centrosomes move closer to each other again and reach an almost normal distance at metaphase (Figure 4D, approximately −30–0 s; Figure 4B, −0:18). During anaphase and telophase the centrosomes in RCC1(RNAi) embryos behave as in wild-type embryos (Figure 4D, 0–240 s), although chromatin segregation is clearly affected (Figure 4B, 0:48–3:12).
In summary, these data demonstrate that although spindle assembly is defective in the absence of Ran or RanGTP hydrolysis, the cues that control final centrosome position are functional. We suggest that the premature centrosome separation may be caused by a lack of Ran-dependent attachment of the centrosomes to the pronuclei.
IMA-2 and IMB-1, Homologues of Vertebrate Importin α and β, Are Essential for Spindle Formation
We initially analyzed IMA-2 and IMA-3 requirement by RNAi in embryos expressing GFP::histone H2B. ima-3(RNAi) embryos were significantly reduced in size (our unpublished data) in agreement with a role of IMA-3 in oogenesis (Geles and Adam, 2001; Table 1). However, spindle formation seemed relative normal as judged from the presence of a metaphase plate and DIC images (our unpublished data) and IMA-3 was therefore not studied further. ima-2(RNAi) embryos, on the other hand, had clear spindle defects (Figure 3D) and were examined more closely.
An ima-2 deletion allele was generated and isolated by the C. elegans Gene Knockout Consortium. We obtained the ima-2(ok256) allele and sequenced it to characterize the deletion. The ima-2(ok256) allele lacks 1782 base pairs, from nt 325 downstream of the start codon to nt 283 downstream of the stop codon. This leads to a shortening of the protein from 532 to 76 amino acid residues from IMA-2 plus another 36 unrelated amino acid residues encoded by sequences downstream of the deletion. We therefore expect ima-2(ok256) to be a null allele. In the wild-type genome the distance from the ima-2 stop codon to the start codon of the downstream gene F26B1.2 is only 647 base pairs. RNAi against F26B1.2 produced no detectable effects arguing against any role of F26B1.2 in our study (Fraser et al., 2000).
By outcrossing the ima-2(ok256) worms for several generations we obtained strain XA3513. Analysis of XA3513 by single worm PCR showed that ima-2(ok256) homozygotes are viable but produce only inviable embryos. This demonstrates that IMA-2 is required either for proper oogenesis or early embryogenesis or both, consistent with its expression pattern (Geles and Adam, 2001).
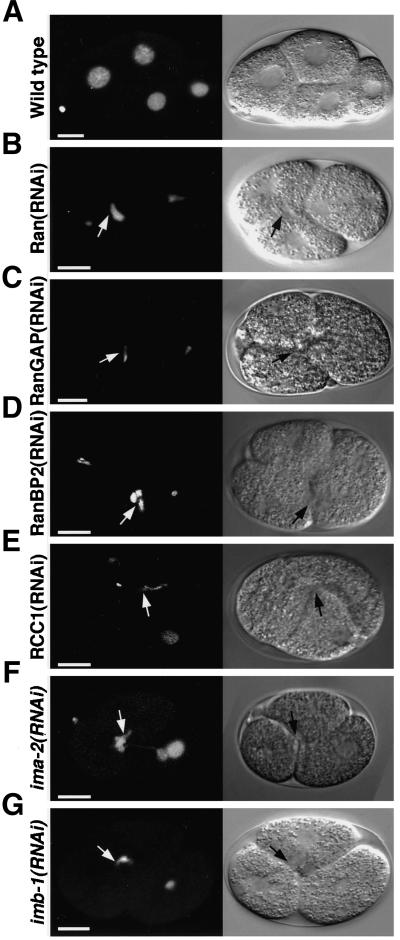
To study microtubule dynamics in the context of the ima-2(ok256) allele we crossed XA3513 with WH204 (Strome et al., 2001), giving rise to strain XA3503. Observation of embryos from ima-2(ok256) worms revealed that the centrosomes initially stay relatively close to the sperm pronucleus and that pronuclear migration was normal (compare Figure 5B, first panel and Video 8 with Figure 5A and Video 1). However, the centrosomes then move apart to an intermediate degree of separation (Figure 5C, which displays the average of 5 embryos). The centrosomes remain separated, but no mitotic spindle was assembled and eventually the embryos enter mitosis as visualized by further centrosome separation, “spindle” rocking, and cytokinesis (Figure 5, B, 0:48–9:28, and C). In addition to the failure of spindle formation, the timing from pronuclear joining to rocking of the centrosomes was abnormally long in embryos from ima-2(ok256) worms (504 ± 36 s [n = 5] compared with 332 ± 45 s in wild-type embryos [n = 3]). This suggests that IMA-2 could be involved in determining the onset or timing of mitosis.
Figure 5.
Embryos devoid of IMA-2 or IMB-1 fail to form mitotic spindles. Embryos expressing GFP::β-tubulin from either a wild-type hermaphrodite (A) or from an ima-2(ok256) homozygous worm (B) were observed by time-lapse microscopy. Still images are shown with indication of time (minutes:seconds) relative to anaphase onset. Bar, 10 μm. (C) Distance between the two centrosomes were measured relative to the total length of the embryo. Shown is the average of several wild-type (diamonds, n = 9; same as in Figure 4C) and ima-2(ok256) (crosses, n = 5) embryos as function of time. Each embryo was aligned relative to anaphase onset, which defines t0. Vertical lines represent the SD at each time point. For wild-type embryos approximate timing of pronuclear meeting and pronuclear envelope breakdown (NEBD) is indicated as is DNA segregation and spindle rocking. (D) An embryo expressing GFP::β-tubulin and GFP::histone H2B from worms depleted of IMB-1 was observed by time-lapse microscopy. Still images are shown with indication of time (minutes:seconds) relative to anaphase onset. Bar, 10 μm. (E) RNA prepared from embryos depleted of RanGAP (lanes 1 and 3) or IMB-1 (lanes 2 and 4) was analyzed by RT-PCR with primers specific for imb-1 (lanes 1 and 2) and Ran (lanes 3 and 4). imb-1 mRNA is efficiently and specifically depleted by dsRNA corresponding to imb-1 (lane 2).
Depletion of IMB-1 from embryos by RNAi give rise to a strong spindle defect like those seen in embryos from ima-2(ok256) worms and in Ran(RNAi) embryos. The centrosomes lose their association with the sperm pronucleus early (Figure 5D, −6:40; 3 of 3 embryos) and move far apart (Figure 5D, −3:20). The pronuclei never meet. The oocyte pronucleus partly aligns between the centrosomes but spindle assembly does not occur (Figure 5D, −3:20–0:40; see also Figure 3E and Video 15).
Disrupting the Ran Cycle Leads to Aneuploidy and Abnormal Chromatin Morphology
Following embryos that express GFP::histone H2B beyond the first division allowed us to investigate the fate of chromatin when the Ran cycle is abrogated. Measured from the completion of P0 division, AB in RNAi embryos divides at the same time as AB in control embryos. In contrast, P1 division in RNAi embryos is significantly delayed compared with wild-type embryos, resulting in prolonged three-cell embryo stages (Figure 6; Videos 9–11; n > 50). Consistent with the finding that a functional Ran system as well as IMA-2 and IMB-1 are necessary for proper spindle function the DNA content of the cells is not equally distributed between the daughter cells and chromatin is often seen trapped at cell junctions (Figure 6, arrows). When any of the Ran-related genes was targeted by RNAi highly unstructured chromatin was present after the first division and throughout the next cell cycle (Figure 6; our unpublished data; >90% for each gene), suggesting severe defects in NE reformation and perhaps DNA decondensation. In some cases, we observed that the chromatin eventually had time to round up in P1 as a consequence of the delayed division (Figure 6, E and F).
Figure 6.
The Ran GTPase cycle, IMA-2, and IMB-1 are essential for normal chromatin appearance. Embryos from GFP::histone H2B worms grown on either control bacteria (A), or bacteria expressing dsRNA corresponding to Ran (B), RanGAP (C), RanBP2 (D), RCC1 (E), ima-2 (F), or imb-1 (G) were observed by time-lapse microscopy. Single still images are shown illustrating that division of P1 is often delayed in RNAi embryos, which leads to prolonged appearance of three-cell-stage embryos. Arrows indicate examples of abnormal chromatin structures that often are trapped at the cleavage furrow. Bar, 10 μm.
NE Formation Is Dependent on RanGTP Production and Hydrolysis
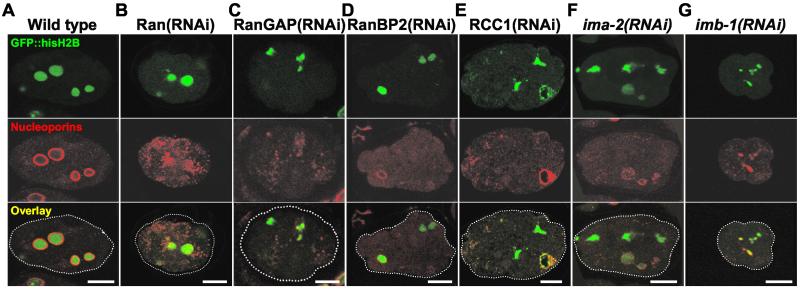
Although RNAi against any component of the Ran cycle affected spindle formation, the embryos always went through several rounds of cytokinesis. This allowed us to investigate the role of the Ran GTPase cycle in NE formation after mitosis. Initially, we examined the distribution of nucleoporins by immunofluorescence with mAb414 (Davis and Blobel, 1987). In wild-type embryos a bright and continuous mAb414 signal surrounds each nucleus (Lee et al., 2000; Figure 7A). In contrast, when we disrupted the Ran system by targeting any of its components mAb414 stained aggregates, many of which were not associated with chromatin. In cases where association was detected the mAb414 signal was not uniform. We observed both situations within the same embryo for each of the genes analyzed (Ran, RanGAP, RanBP2, and RCC1, n > 10 for each gene). A single example for each gene is depicted in Figure 7, B–E. Similar results on depletion of Ran were reported by Bamba et al. (2002).
Figure 7.
RNAi against Ran and its cofactors affects targeting of nuclear pore proteins. Embryos from GFP::histone H2B worms grown on either control bacteria (A), or bacteria expressing dsRNA corresponding to Ran (B), RanGAP (C), RanBP2 (D), RCC1 (E), ima-2 (F), or imb-1 (G) were fixed and analyzed with the anti-nucleoporin mAb mAb414. Top row shows GFP::histone H2B in green, middle row shows mAb414 staining in red, and bottom row shows an overlay of the two signals. Bar, 10 μm.
Because NEs can assemble in the absence of nuclear pore complex insertion (Macaulay and Forbes, 1996), we wished to determine whether the disrupted nucleoporin localization was a consequence of a lack of NE assembly. For this purpose, transgenic strains that express GFP fused to the inner nuclear membrane protein emerin (EMR-1; Lee et al., 2000) were generated. GFP::EMR-1 can be used as marker for the NE. Embryos from these strains show a distinct NE signal as well as cytoplasmic staining like that seen with endoplasmic reticulum proteins (Figure 8A, Video 12). In older embryos the ratio of NE to total signal increases (our unpublished data), suggesting that early embryos may store EMR-1 protein in the endoplasmic reticulum for later use. Antibodies raised against EMR-1 give rise to similar staining (see below).
Figure 8.
Disruption of the Ran GTPase cycle prevents nuclear envelope formation. Embryos from GFP::EMR-1 worms grown on either control bacteria (A), or bacteria expressing dsRNA corresponding to Ran (B and C), RanGAP (D), RanBP2 (E), RCC1 (F), ima-2 (G), or imb-1 (H) were observed by time-lapse microscopy. Single still images are shown demonstrating that no nuclear structure surrounded by a uniform EMR-1 staining is seen in embryos where the Ran cycle (B and D–F), IMA-2 (G) or IMB-1 (H) is targeted by RNAi. An example of a less severely affected embryo is shown (C). Bar, 10 μm.
In embryos depleted of Ran or any of its cofactors, RanBP2, RanGAP, or RCC1, no NE-like GFP::EMR-1 signal was detected after the first cell division in the majority of cases (67%) (Figure 8, B and D–F, Video 13). In other embryos, presumably where the RNAi had been less efficient, aberrant nuclei with a strong aggregation of GFP::EMR-1 next to the NE became apparent after a prolonged time (Figure 8C). Thus, the distribution of GFP::EMR-1 in RNAi embryos was similar to the mislocalization of nucleoporins described above. When endogenous EMR-1 was visualized by immunofluorescence with polyclonal antibodies (a gift of Y. Gruenbaum and K. Wilson), identical results were obtained (supplementary Figure 1).
As a further proof that a closed NE is not assembled after mitosis when the Ran cycle is inactivated, we determined whether soluble GFP::β-tubulin was uniformly distributed in the embryos. In normal embryos tubulin is excluded from the pronuclei and the nuclei and can therefore be used as marker for NE breakdown and reformation (Figure 5A and 9). In contrast, Ran(RNAi) embryos never showed such exclusion after mitosis (Figure 9B; see also Figure 1, 4 of 4 embryos) and targeting the Ran cofactors also produced uniform GFP::β-tubulin staining (Figure 9, C and D, 5 of 6 embryos). We conclude that not only Ran but also its ability to cycle between RanGDP and RanGTP under the influence of RanGAP, RanBP2, and RCC1, is essential for an early step of NE assembly in vivo in these embryos.
Figure 9.
Nuclear exclusion of soluble GFP::β-tubulin depends on the Ran GTPase cycle. Embryos from GFP::β-tubulin worms grown on either control bacteria (A), or bacteria expressing dsRNA corresponding to Ran (B), RCC1 (C), or RanGAP (D) were observed by time-lapse microscopy combined with confocal z-scanning. Single still images are shown illustrating that nuclei in a control embryo exclude GFP::β-tubulin (A), whereas no area of exclusion is detected when the Ran cycle is affected (B–D). Bar, 10 μm.
Depletion of IMA-2 or IMB-1 Affects NE Formation
We next wished to investigate whether IMA-2 and IMB-1 also play a role in regeneration of the NE after mitosis in vivo. Interestingly, multiple small structures surrounded by GFP::EMR-1 were detectable in ima-2(RNAi) embryos early (∼1 min) after anaphase onset, when nuclei are formed in wild-type embryos (3 of 3 embryos, compare Videos 12 and 14). However, in ima-2(RNAi) embryos (Figure 2F) these structures grew only slowly in size and a significant amount of GFP::EMR-1 accumulated at the centrosomes (Figure 8, compare A and G). Presumably the appearance of multiple micronuclei after division is a consequence of the lack of spindle formation described above. The fact that the structures only grow slowly in size combined with the observation that ima-2(ok256) embryos do not show nuclear exclusion of soluble GFP::β-tubulin even late after mitosis (compare last panels in Figure 5, A and B, 4 of 4 embryos) suggests that IMA-2 is required either for fusion of nuclear membranes or for assembly of nuclear pore complexes.
RNAi against IMA-3 was clearly effective as judged from embryonic lethality (Table 1) and alterations in embryo size and organization (our unpublished data). Despite this, nuclei were formed in ima-3(RNAi) embryos upon cell division and grew to nearly normal size (our unpublished data), illustrating that IMA-3 is not required for nuclear formation in early embryos. Embryos depleted of IMB-1 showed a strong defect in GFP::EMR-1 recruitment to the NE-like structures after mitosis (Figure 7H and Video16, 3 of 3 embryos). As with embryos where Ran or its cofactors had been targeted, imb-1(RNAi) embryos instead showed clustering of GFP::EMR-1 at the centrosomes. In addition, nuclear exclusion of soluble GFP::β-tubulin was not observed in embryos depleted of IMB-1 (our unpublished data; 3 of 3 embryos). Furthermore, immunofluorescence analysis demonstrated that chromatin in ima-2(RNAi) and imb-1(RNAi) embryos is most often not surrounded by a smooth and continuous nucleoporin staining (Figure 7, compare F and G with A; >80% for each gene).
DISCUSSION
RanGTP production in the vicinity of chromosomes has recently been shown to mediate chromosomal nucleation of spindle microtubules in vitro (see INTRODUCTION). In this article, we describe the first use of live cell imaging to investigate the function of Ran and its regulators in mitotic spindle assembly and NE formation in vivo. In agreement with biochemical data, largely obtained using extracts of Xenopus eggs and with suggestive genetic evidence from yeasts (see INTRODUCTION and below), we provide evidence that the Ran system is essential for both spindle and NE assembly in C. elegans embryos. We have, however, found unexpected differences in the requirement for Ran cofactors compared with those predicted from in vitro studies. In addition, a role for the importin α and β homologues IMA-2 and IMB-1 in spindle and NE formation was demonstrated. Finally, the components of the Ran system are shown to be required for normal comigration of centrosomes with the male pronucleus.
In Vivo, Ran Is Essential for Spindle Formation …
When Ran was depleted from C. elegans embryos before the first zygotic mitosis, the centrosomes did not remain associated with the sperm pronucleus as in wild-type embryos but instead moved far apart early in mitosis. The mechanism by which centrosomes are anchored to the (pro-)nuclear membrane in interphase and prophase is largely unknown. However, because NE formation is dependent on Ran (see below) a likely explanation for the premature release of the centrosomes from the sperm pronucleus is the lack of a membrane-associated anchoring protein. The association of centrosomal microtubules with the NE has recently been shown to accelerate the process of NE breakdown in vertebrate cells (Beaudouin et al., 2002; Salina et al., 2002) and this requires an as yet undefined connection between the NE and microtubules. This connection could be related to the C. elegans Ran-dependent centrosome-anchoring activity.
A few microtubules were often seen to connect the centrosomes and the pronuclei at this stage even in Ran(RNAi) embryos. We believe these are astral microtubules from the centrosomes that inefficiently associate with the pronuclei, because the capacity of centrosomes to nucleate microtubules seemed unaffected in these embryos. In strong contrast, no spindle microtubules were seen between centrosomes and chromatin. Neither could we detect bipolar spindles in embryos where one of the pronuclei was aligned between the two centrosomes nor half-spindles between single centrosomes and isolated pronuclei. This is not because a haploid DNA complement provides insufficient stimulus to induce spindle formation because when oocytes are fertilized with anucleate sperm, they develop into embryos with normal spindles (Sadler and Shakes, 2000). Thus, we conclude that the ability of embryonic chromatin to stimulate the nucleation of spindle microtubules is dependent on Ran.
Our data show in addition that the cues for positioning of the centrosomes along the anterior-posterior axis are independent of spindle formation or of attachment of the centrosomes to the pronuclei. In some cases, meiotic defects can affect downstream mitotic division (Oegema et al., 2001). However, polar bodies were always produced in Ran(RNAi) embryos and we did not detect abnormal chromatin content in the oocyte pronucleus as measured by GFP::histone H2B levels. This suggests that the meiotic spindle in C. elegans may be formed by an alternative, Ran-independent mechanism.
Recently, RNAi-mediated depletions of Ran from C. elegans embryos were reported to cause chromosome positioning and segregation defects (Bamba et al., 2002; discussed further below). A role for Ran in mitotic spindle formation has also been demonstrated in fission yeast where a point mutation in the Ran gene (spi1) caused the appearance of monopolar and tiny spindles in a significant proportion of the cells (Fleig et al., 2000). In addition, the yeast mutant showed altered interphase microtubule arrays in the cytoskeleton, implying that Ran in this organism may be generally involved in chromatin-independent microtubule nucleation (Fleig et al., 2000).
… and for Nuclear Envelope Formation
Recently, it has been demonstrated that GTP hydrolysis by Ran is essential for membrane fusion events early during NE assembly in vitro (Hetzer et al., 2000; Zhang and Clarke, 2000; see INTRODUCTION). Our results extend this demonstration in vivo. In contrast to the smooth NE staining of the inner nuclear membrane protein EMR-1 in wild-type embryos (Lee et al., 2000) we observed that EMR-1 in Ran(RNAi) embryos was found in large clusters that most likely represent membrane aggregates. These structures were often not associated with chromatin, suggesting that aggregation was caused by a block to reassociation of NE membrane material with chromatin after mitosis. In some embryos, chromatin was actually surrounded by EMR-1. However, strong perinuclear aggregation was always seen in these embryos. This is most easily explained as the result of lower RNAi efficiency in these embryos that would allow some reassociation of EMR-1–containing membranes with chromatin but not allow NE assembly to proceed to completion.
Importantly, we observed very similar defects in NE assembly upon Ran depletion when we compared living embryos expressing GFP::EMR-1 with immunostaining of endogenous EMR-1. The two methods also provide reciprocal assurance against fixation artifacts or GFP::EMR-1 overexpression and/or mislocalization behavior. Localization of nucleoporins confirmed the results of Bamba et al. (2002) and showed that the observed defects were not restricted to proteins of the inner nuclear membrane. Consistent with the absence of a NE, Ran(RNAi) embryos failed to establish nuclei that could exclude cytoplasmic GFP::β-tubulin. Finally, it should be noted that the defects in NE assembly are most likely not attributable to spindle failure. When ICP-1 is targeted by RNAi, spindle formation and DNA segregation are affected without preventing subsequent NE formation (Galy, Askjaer, and Mattaj, unpublished observations). Our data therefore leads us to conclude that Ran is essential for an early step in NE reassembly after mitosis.
A recent study came to the same conclusion based on the use of a nucleoporin antibody and fixed specimens (Bamba et al., 2002). For dynamic cellular processes, especially in fast dividing embryos, time-lapse analysis provide much more information because it enables a detailed kinetic description (see next section). Second, because NE assembly can take place without detectable nucleoporin insertion (Macaulay and Forbes, 1996), analysis of a marker of the inner nuclear membrane is a significantly more informative and reliable method of evaluation of the presence or absence of a NE.
Differential Requirement for RanBP2, RanGAP, and RCC1 in Mitosis
When we depleted embryos of the Ran regulators RanBP2, RanGAP, and RCC1, defects in NE formation were observed that were very similar to the defects seen in Ran(RNAi) embryos. Thus, we can conclude that a functional Ran cycle, including GTP hydrolysis, is a necessity for NE assembly after mitosis in living cells, as in vitro (Hetzer et al., 2000; Zhang and Clarke, 2000).
The effects of targeting RanBP2 and RanGAP were also very similar to the Ran RNAi phenotype in spindle formation, indicating either that hydrolysis of GTP by Ran is essential for spindle formation or that RanGTP concentrations need to be kept low away from the chromatin. In mammalian cells, RanBP1 has been implicated in regulation of the mitotic spindle since injection of antibodies against RanBP1 leads to mitosis defects, including delayed metaphase and cytokinesis failure (Guarguaglini et al., 2000). Also in budding yeast RanBP1 has been suggested to be required for proper spindle formation (Ouspenski, 1998). However, in this case the activity of RanBP1 is independent of RanGAP, suggesting that the effects observed in this organism, where spindle formation takes place within a closed NE, may have a different basis from those observed in C. elegans. Parenthetically, it is interesting to note that although many metazoans have both RanBP1 and RanBP2 genes, C. elegans does not encode a RanBP1 homologue. Our data strongly suggest that RanBP2 takes over all of RanBP1's functions in the nematode.
Because Ran mutants locked in the GTP conformation stimulate delocalized microtubule nucleation in vitro (Carazo-Salas et al., 1999; Kalab et al., 1999; Wilde and Zheng, 1999), we expected that an accumulation of cytoplasmic RanGTP in RanGAP(RNAi) and RanBP2(RNAi) embryos would lead to an increase in microtubule formation. We did not detect such an increase, perhaps because delocalized RanGTP only gives rise to short or unstable microtubules in the embryos that would be indistinguishable from the soluble tubulin staining. Regardless, our data suggest that both spindle and NE formation in vivo requires that RanGTP can be reconverted to RanGDP.
In contrast to embryos from which other components of the RanGTPase cycle are depleted the centrosomes in RCC1(RNAi) embryos stay associated with the sperm pronucleus. This suggests that anchoring of the centrosomes to the sperm pronucleus could be RanGDP dependent. The centrosomes then separate prematurely and at the same time microtubules are seen connecting the centrosomes to the chromatin. Later, the centrosomes move closer again and a seemingly normal spindle is assembled that however fails to segregate the chromatin properly to the two daughter cells. We can envisage several different explanations for the specific lack of effect of RCC1 depletion on spindle formation compared with the requirement for Ran, RanGAP, and RanBP2 in this process and for all four proteins in NE assembly.
A formal possibility is that RanGDP rather than RanGTP is required for spindle formation in C. elegans. This must be considered unlikely, because all known functions of Ran studied to date, including spindle formation in vitro, require RanGTP. Alternatively, it might be that the reduction of RCC1 in RNAi animals is incomplete, and leaves sufficient activity to support spindle formation. Both RT-PCR analysis of RCC1 mRNA and quantitation of the reduction of GFP::RCC1 fusion expression in embryos from dsRNA-treated worms suggest that RCC1 expression is reduced to <5% of wild-type. Furthermore, the degree of embryonic lethality in these animals (>90%) is similar to that achieved for the other genes studied here. Analysis of vertebrate systems shows that different processes require different levels of RCC1 activity. For example, tsBN2 cells, which are temperature sensitive for RCC1 activity, support both spindle formation (Nishitani et al., 1991; Compton and Cleveland, 1993) and RanGTP-dependent nuclear export (Richards et al., 1997) but not nuclear import (Tachibana et al., 1994; Richards et al., 1997). Spindle formation and NE assembly in Xenopus extracts are differentially sensitive to the RCC1 inhibitor RanT24N (Carazo-Salas et al., 1999; Hetzer et al., 2000), with spindle formation being more sensitive than NE assembly. It is therefore possible, but in our view unlikely, that residual RCC1 provides the explanation for our observations. Finally, it is possible that there is an activity in C. elegans that is redundant with RCC1 for nucleotide exchange on Ran and that functions in spindle, but not NE, assembly. Although combined RNAi against RCC1 and the three genes most closely related in sequence to RCC1 in the C. elegans genome did not block spindle formation (our unpublished data), it is difficult to definitively rule out either this explanation, or the others listed above.
It has been reported by others that targeting either Ran, RanGAP, or RCC1 expression in C. elegans embryos by RNAi gives similar spindle assembly defects (Bamba et al., 2002). However, the penetrance of RNAi against RCC1 in that study was reported to be low (∼15% of embryos), whereas we consistently see the phenotype described above in >80% of the embryos and RNAi against RCC1 resulted in >90% lethality in our hands (Table 1). Because Bamba et al. (2002) analyzed fixed embryos we believe that the defect they reported most likely reflects the temporary delay that we observed in the initial phase of spindle formation, which was subsequently rescued as mitosis progressed.
IMA-2 and IMB-1 Are Required for Spindle Assembly and NE Formation in Embryonic Cells
We have characterized a deletion allele of ima-2 and found that embryos produced by ima-2(ok256) homozygous worms as well as imb-1(RNAi) embryos fail to assemble spindles while their centrosomes nucleate normal levels of astral microtubules. As in the case of RNAi against RanGAP and RanBP2, these results would not be predicted by simple application of the model for Ran function on spindle assembly derived from experiments in Xenopus egg extracts. There, importin α and β have been shown to act as inhibitors of microtubule nucleation by sequestering TPX2 or NuMA (Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001), and one might therefore expect to see an increase in microtubule abundance upon depletion of IMA-2 or IMB-1. Moreover, injection into mammalian cells of an importin β fragment, which cannot interact with Ran, has a dominant negative effect on spindle formation (Nachury et al., 2001). There is no obvious C. elegans homologue of TPX2. This could imply that IMA-2/IMB-1 and importin α/β function differently in spindle formation. However, more data on downstream mediators of the IMA-2 and IMB-1 effect is required before strong conclusions can be reached.
The chromatin in ima-2(RNAi) and imb-1(RNAi) embryos was often fragmented as a consequence of spindle failure but was nevertheless associated with EMR-1 early after division. The micronuclei seen at this stage did not fuse or grow normally, suggesting either that a closed, functional NE was not formed or that protein import was defective. Because no nuclear exclusion of soluble GFP::β-tubulin was observed in ima-2(ok256) embryos or imb-1(RNAi) embryos we favor the former possibility, implying a role for IMA-2 and IMB-1 in NE assembly before its function in nuclear protein import.
In summary, our data demonstrates that spindle assembly and NE formation in C. elegans embryos depend on the Ran GTPase cycle functioning, at least in part, via IMA-2 and IMB-1. However, there are aspects of our data that are unexpected in the light of the models derived from previous in vitro studies. These suggest either that the detailed mechanisms by which Ran affects mitotic events in the nematode and in vertebrates may be significantly different or that our incomplete understanding of both systems masks their intrinsic similarity. Further analysis of these processes in C. elegans and other in vivo systems is required to provide the answer.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to A. Desai, M. Kirkham, and T. Zimmermann for practical assistance and especially to A.A. Hyman for sharing unpublished data and materials. We thank J. Ahringer, J. Austin, D. Bilbao-Cortés, Y. Gruenbaum, C. Malone, R. Saffrich, G. Seydoux, J. White, and K. Wilson for providing materials and technical advice. W. Antonin, A. Becskei, D. Bilbao-Cortés, C. Franz, V. Hachet, M. Hetzer, A.A. Hyman, A. Segref, C.A. Schatz, and T.C. Walther are acknowledged for discussions and critical reading of the manuscript. Some strains used in this work were provided by the Caenorhabditis Genetic Center, which is funded by the National Institutes of Health National Center for Research Resources. This work was supported by the Louis-Jeantet Foundation. P.A. and V.G. were funded by The Carlsberg Foundation and European Molecular Biology Oraganization, respectively.
Abbreviations used:
- DIC
differential interference contrast
- dsRNA
double-stranded RNA
- GFP
green fluorescent protein
- NE
nuclear envelope
- ORF
open reading frame
- RNAi
dsRNA-mediated interference
Footnotes
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–06–0346. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–06–0346.
REFERENCES
- Askjaer P, et al. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol. 1999;19:6276–6285. doi: 10.1128/mcb.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamba C, Bobinnec Y, Fukuda M, Nishida E. The GTPase Ran regulates chromosome positioning and nuclear envelope assembly in vivo. Curr Biol. 2002;12:503–507. doi: 10.1016/s0960-9822(02)00741-8. [DOI] [PubMed] [Google Scholar]
- Bilbao-Cortés D, Hetzer M, Laengst G, Becker PB, Mattaj IW. Ran binds to chromatin by two distinct mechanisms. Curr Biol. 2002;12:1151–1156. doi: 10.1016/s0960-9822(02)00927-2. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Görlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. doi: 10.1016/s0092-8674(01)00627-4. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Compton DA. Spindle assembly in animal cells. Annu Rev Biochem. 2000;69:95–114. doi: 10.1146/annurev.biochem.69.1.95. [DOI] [PubMed] [Google Scholar]
- Compton DA, Cleveland DW. NuMA is required for proper completion of mitosis. J Cell Biol. 1993;120:947–957. doi: 10.1083/jcb.120.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M. Running on Ran. nuclear transport and the mitotic spindle. Cell. 2001;104:321–324. doi: 10.1016/s0092-8674(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc Natl Acad Sci USA. 1987;84:7552–7556. doi: 10.1073/pnas.84.21.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter J, Morphew M, Sazer S. A mutation in the RCC1-related protein pim-1 results in nuclear envelope fragmentation in fission yeast. Proc Natl Acad Sci USA. 1995;92:1436–1440. doi: 10.1073/pnas.92.5.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleig U, Salus SS, Karig I, Sazer S. The fission yeast Ran GTPase is required for microtubule integrity. J Cell Biol. 2000;151:1101–1111. doi: 10.1083/jcb.151.5.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Blobel G, Rexach M. Disassembly of RanGTP-karyopherin beta complex, an intermediate in nuclear protein import. J Biol Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Gant TM, Wilson KL. Nuclear assembly. Annu Rev Cell Dev Biol. 1997;13:669–695. doi: 10.1146/annurev.cellbio.13.1.669. [DOI] [PubMed] [Google Scholar]
- Geles KG, Adam SA. Germline and developmental roles of the nuclear transport factor importin alpha3 in C. elegans. Development. 2001;128:1817–1830. doi: 10.1242/dev.128.10.1817. [DOI] [PubMed] [Google Scholar]
- Gönczy P, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Lee KK, Liu J, Cohen M, Wilson KL. The expression, lamin-dependent localization and RNAi depletion phenotype for emerin in C. elegans. J Cell Sci. 2002;115:923–929. doi: 10.1242/jcs.115.5.923. [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Guarguaglini G, Renzi L, D'Ottavio F, Di Fiore B, Casenghi M, Cundari E, Lavia P. Regulated Ran-binding protein 1 activity is required for organization, and function of the mitotic spindle in mammalian cells in vivo. Cell Growth Differ. 2000;11:455–465. [PubMed] [Google Scholar]
- Hetzer M, Bilbao-Cortes D, Walther TC, Gruss OJ, Mattaj IW. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol Cell. 2000;5:1013–1024. doi: 10.1016/s1097-2765(00)80266-x. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pu RT, Dasso M. The Ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2956. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 1995;34:639–647. doi: 10.1021/bi00002a031. [DOI] [PubMed] [Google Scholar]
- Kuersten S, Ohno M, Mattaj IW. Nucleocytoplasmic transport. Ran, beta and beyond. Trends Cell Biol. 2001;11:497–503. doi: 10.1016/s0962-8924(01)02144-4. [DOI] [PubMed] [Google Scholar]
- Lee KK, Gruenbaum Y, Spann P, Liu J, Wilson KL. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol Biol Cell. 2000;11:3089–3099. doi: 10.1091/mbc.11.9.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:5705–5794. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay C, Forbes DJ. Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTPγS, and BAPTA. J Cell Biol. 1996;132:5–20. doi: 10.1083/jcb.132.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD. The Ran-GTPase and cell-cycle control. Bioessays. 2001;23:77–85. doi: 10.1002/1521-1878(200101)23:1<77::AID-BIES1010>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Ohtsubo M, Yamashita K, Iida H, Pines J, Yasudo H, Shibata Y, Hunter T, Nishimoto T. Loss of RCC1, a nuclear DNA-binding protein, uncouples the completion of DNA replication from the activation of cdc2 protein kinase and mitosis. EMBO J. 1991;10:1555–1564. doi: 10.1002/j.1460-2075.1991.tb07675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Ouspenski II. A RanBP1 mutation which does not visibly affect nuclear import may reveal additional functions of the Ran GTPase system. Exp Cell Res. 1998;244:171–183. doi: 10.1006/excr.1998.4174. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimby BB, Wilson CA, Corbett AH. The interaction between Ran and NTF2 is required for cell cycle progression. Mol Biol Cell. 2000;11:2617–2629. doi: 10.1091/mbc.11.8.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards SA, Carey KL, Macara IG. Requirement of guanosine triphosphate-bound ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- Sadler PL, Shakes DC. Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development. 2000;127:355–366. doi: 10.1242/dev.127.2.355. [DOI] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- Strome S, Powers J, Dunn M, Reese K, Malone CJ, White J, Seydoux G, Saxton W. Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol Biol Cell. 2001;12:1751–1764. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana T, Imamoto N, Seino H, Nishimoto T, Yoneda Y. Loss of RCC1 leads to suppression of nuclear protein import in living cells. J Biol Chem. 1994;269:24542–24545. [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Vasu SK, Forbes DJ. Nuclear pores and nuclear assembly. Curr Opin Cell Biol. 2001;13:363–375. doi: 10.1016/s0955-0674(00)00221-0. [DOI] [PubMed] [Google Scholar]
- Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Hyman A, Desai A. The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol. 2001;3:28–34. doi: 10.1038/35050669. [DOI] [PubMed] [Google Scholar]
- Zhang C, Clarke PR. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science. 2000;288:1429–1432. doi: 10.1126/science.288.5470.1429. [DOI] [PubMed] [Google Scholar]
- Zhang C, Hughes M, Clarke PR. Ran-GTP stabilizes microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J Cell Sci. 1999;112:2453–2461. doi: 10.1242/jcs.112.14.2453. [DOI] [PubMed] [Google Scholar]
- Zhang C, Hutchins JRA, Mühlhaüsser P, Kutay U, Clarke PR. Role of importin-β in the control of nuclear envelope assembly by Ran. Curr Biol. 2002;12:498–502. doi: 10.1016/s0960-9822(02)00714-5. [DOI] [PubMed] [Google Scholar]
- Zipperlen P, Fraser AG, Kamath RS, Martinez-Campos M, Ahringer J. Roles for 147 embryonic lethal genes on C. elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 2001;20:3984–3992. doi: 10.1093/emboj/20.15.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.