Abstract
The phosphorylation-dependent anchorage of retinoblastoma protein Rb in the nucleus is essential for its function. We show that its pocket C domain is both necessary and sufficient for nuclear anchorage by transiently expressing green fluorescent protein (GFP) chimeras of Rb fragments in tissue culture cells and by extracting the cells with hypotonic solutions. Solid phase binding assays using glutathione S-transferase-fusion of Rb pockets A, B, and C revealed a direct association of lamin C exclusively to pocket C. Lamina-associated polypeptide (LAP) 2α, a binding partner of lamins A/C, bound strongly to pocket C and weakly to pocket B. When LAP2α was immunoprecipitated from soluble nuclear fractions, lamins A/C and hypophosphorylated Rb were coprecipitated efficiently. Similarly, immunoprecipitation of expressed GFP-Rb fragments by using anti-GFP antibodies coprecipitated LAP2α, provided that pocket C was present in the GFP chimeras. On redistribution of endogenous lamin A/C and LAP2α into nuclear aggregates by overexpressing dominant negative lamin mutants in tissue culture cells, Rb was also sequestered into these aggregates. In primary skin fibroblasts, LAP2α is expressed in a growth-dependent manner. Anchorage of hypophosphorylated Rb in the nucleus was weakened significantly in the absence of LAP2α. Together, these data suggest that hypophosphorylated Rb is anchored in the nucleus by the interaction of pocket C with LAP2α–lamin A/C complexes.
INTRODUCTION
Vertebrate nuclei are highly organized structures in which chromosomes occupy discrete territories (Croft et al., 1999), and activities such as DNA replication, transcription, and RNA processing occur within discrete nuclear bodies (Lamond and Earnshaw, 1998). This level of organization implies that architectural proteins link chromatin to the nuclear envelope (NE) or to a nucleoskeleton, and similar proteins also anchor regulators of transcription and DNA replication to nuclear bodies. The recent description of human genetic diseases that arise as a result of mutations in nuclear architectural proteins (Hutchison et al., 2001; Wilson, 2000) highlights the importance of the structure of the nucleus in relation to its function.
The major structural framework in the nucleus is the nuclear lamina, which determines both the size and shape of the nucleus and its mechanical stability (reviewed by Moir and Goldman, 1995; Vaughan et al., 2000). The major components of the lamina are the nuclear lamins, which are members of the intermediate filament family, and lamina-associated polypeptides (LAPs). Lamins have reported functions in DNA replication (Meier et al., 1991; Jenkins et al., 1993; Ellis et al., 1997; Spann et al., 1997; Moir et al., 2000) and nuclear pore organization (Lenz-Bohme et al., 1997; Liu et al., 2000; Smythe et al., 2000). In addition, one member of the LAP family, LAP2β, has also been reported to influence NE growth (Yang et al., 1997) and DNA replication (Gant et al., 1999). A second member of the LAP family, emerin, when mutated gives rise to Emery-Dreifuss muscular dystrophy, implying that it also has important functions, possibly in tissue-specific transcription regulation (reviewed by Morris and Manilal, 1999; Cohen et al., 2001).
A recently described member of the LAP family is LAP2α. The LAP2 protein originally described in rat nuclear envelopes (Foisner and Gerace, 1993) has now been shown to be one member of a family of nuclear proteins derived from a single gene by alternative splicing (Harris et al., 1994; Berger et al., 1996). Three abundant proteins are expressed from the human LAP2 gene, namely, LAP2α (75 kDa), LAP2β (51 kDa), and LAP2γ (39 kDa) (Harris et al., 1995). Of these proteins, LAP2β and γ are both type II transmembrane proteins that differ only by the insertion of a β-specific domain of 109 amino acids in LAP2β. In contrast, LAP2α shares a 187-amino acid N-terminal domain with LAP2β and γ, but this is followed by a 506-amino acid α-specific domain lacking transmembrane regions. LAP2α has been shown to be distributed throughout the nucleus, rather than at the NE (Dechat et al., 1998). Complexes of LAP2α and A-type lamins form architectural, interchromosomal structures (Dechat et al., 1998, 2000). The interaction of LAP2α with chromatin seems to require the α-specific domain (Vlcek et al., 1999) and is likely regulated by cell cycle-dependent phosphorylation (Dechat et al., 1998). These important findings imply that LAP2α may have a number of functions in higher order chromatin interactions. This includes functions in the mitotic assembly/disassembly of the nucleus and/or as an anchorage protein for transcription regulators.
The retinoblastoma protein p110Rb (Rb) controls progression through the cell cycle by negatively regulating the E2F transcription factor in a phosphorylation-dependent manner (Chellappan et al., 1991). Rb has a well-characterized domain structure consisting of an N-terminal domain followed by three C-terminal pocket domains termed A, B, and C (Figure 1). The N-terminal domain is capable of oligomerization (Hensey et al., 1994) and binds to an 84-kDa protein that colocalizes with centers of RNA processing (Durfee et al., 1994). The large A/B pocket binds to the E2F transcription factor (Cao et al., 1992; Lees et al., 1993) and D-type cyclins (Dowdy et al., 1993; Ewen et al., 1993). It also forms a complex with histone deacetylase (Brehm et al., 1998; Luo et al., 1998; Magnaghi-Jaulin et al., 1998), presumably leading to long-range silencing of genes required for cell division (Zhang et al., 2000). Pocket C has been shown to contain a nuclear localization signal sequence (NLS; Zacksenhaus et al., 1993) and binds both the c-Abl tyrosine kinase (Welch and Wang, 1993, 1995) and MDM2 (Xiao et al., 1995).
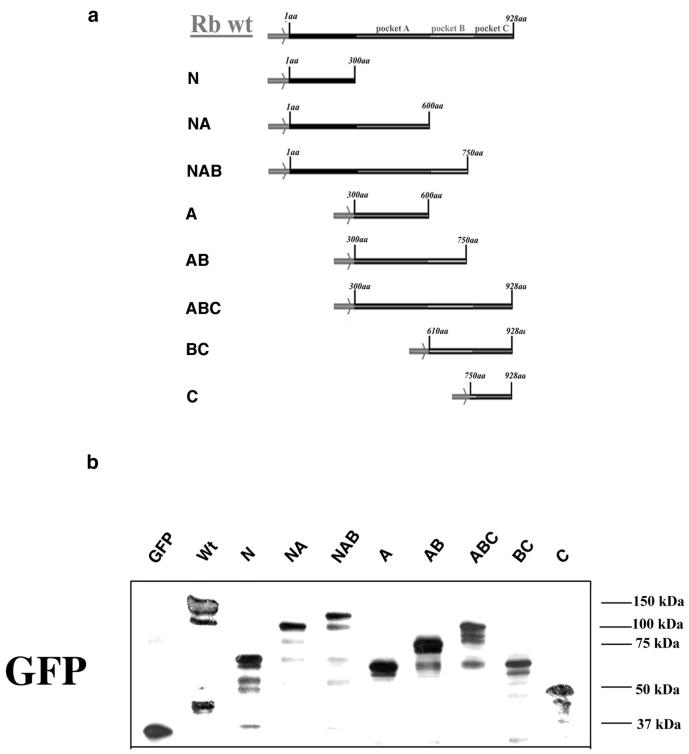
Figure 1.
Schematic representation of GFP-Rb fusion constructs (a). Transient expression of GFP-Rb fusion constructs in HEK293 cells. GFP-Rb fusion proteins were transiently expressed in HEK293 cells for 72 h. Cells were solublized in sample buffer and proteins analyzed by SDS-PAGE and immunoblotting by using monoclonal antibodies to GFP (b).
Binding of Rb to its targets is regulated by phosphorylation in a cell cycle-dependent manner (Lees et al., 1991; reviewed by Wang et al., 1994). Anchorage of Rb in the nucleus is also regulated by phosphorylation in that hypophosphorylated Rb, capable of binding E2F, is anchored in the nucleus during G1 phase of the cell cycle (Mittnacht and Weinberg, 1991; Mancini et al., 1994). Internal deletions or mutations of Rb within a region spanning pockets B and C promote tumorigenesis and prevent nuclear anchorage of the protein (Mittnacht and Weinberg, 1991), giving rise to the hypothesis that nuclear anchorage of Rb as well as binding to E2F are both essential for its function. Clearly, if the identity of the protein (or proteins) that anchor Rb to the nucleoskeleton were known, a test of this hypothesis would be possible. Previous reports have revealed that Rb binds to A-type lamins in blot overlay assays through its large pocket domain (Ozaki et al., 1994), suggesting that lamin A might anchor Rb within the nucleus. Herein, we report that LAP2α, which we have previously identified as an interaction partner for nucleoskeletal lamin A/C, binds to Rb within a region spanning pockets B and C. Disruption of the normal distribution of lamins A/C and LAP2α leads to a similar disruption of Rb distribution, and hypophosphorylated Rb is not retained in the nucleus of cells expressing low levels of LAP2α. Taken together, these data suggest that LAP2α–lamin A/C complexes have an important role in the nuclear anchorage of Rb.
MATERIALS AND METHODS
Cell Cultures
Cells were cultured in DMEM (Invitrogen, Paisley, United Kingdom). Human embryonic kidney (HEK) 293 cells were supplemented with 10% fetal calf serum (FCS). Human dermal fibroblasts (HDF) were supplemented with 10% newborn calf serum (NCS) as recommended. The osteosarcoma cell line SA0S-2 was supplemented with 10% FCS. Cultures were maintained at 60–70% confluence. To induce quiescence in HDF, cultures were transferred to medium containing 0.5% NCS for 5 d. To stimulate HDF to reenter the cell cycle, quiescent cultures were transferred to medium containing 10% NCS.
Transient Expression of Fusion Constructs in Cells
HEK293 cells were grown on DMEM supplemented with 10% fetal calf serum (FCS). Cell cultures were maintained in an incubator with 5% CO2 at 37°C. For transfections, cells were grown to 25–30% confluence in 45-mm dishes. A mixture of 5 μg of plasmid DNA, 0.12 M CaCl2, and HBS (70 mM NaCl, 0.5 mM Na2HPO4, HEPES, pH 7.0) was added to 2 ml of culture medium. The medium was replaced after 24 h, and fusion proteins were transiently expressed in cells for an additional 24 h.
Immunofluorescence and Confocal Microscopy
Cells grown on glass coverslips were either fixed with 4% formaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature and permeabilized in PBS/0.1% Triton X-100 for 5 min or extracted with hypotonic buffer (10 mM HEPES-KOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 0.5 mM dithiothreitol [DTT]) followed by fixation. Primary and secondary antibodies were applied in PBS/1% NCS (PBS/NCS) for 1 h at room temperature. Primary antibodies used were as described in Table 1 and diluted in PBS/NCS. Secondary antibodies were donkey anti-mouse, goat, or rabbit IgG conjugated to tetramethylrhodamine B isothiocyanate (TRITC) or Cy5 (Jackson Immunoresearch, West Grove, PA) and diluted 1:100 in PBS/NCS. After several washes in PBS, samples were mounted in Mowiol/4,6-diamidino-2-phenylindole (DAPI)/DABCO and viewed with an inverted fluorescent microscope Axiovert 10 (Zeiss, Oberkochen, Germany), fitted with a 12-bit charge-couple device camera controlled with IPLab software or with a Radiance 2000 confocal microscope imaging system with LaserSharp software (Bio-Rad, Hercules, CA).
Table 1.
Antibodies used in this study
| Antibody | Target | Antibody type | Dilution | Source |
|---|---|---|---|---|
| Lamin C | Lamin C | Rabbit polyclonal | 1:50 for fluorescence | Venables et al., 2001 |
| LN43 | Lamin B2 | Mouse monoclonal | 1:100 for blotting | Dyer et al., 1999 |
| Lamin B1 | Lamin B1 | Goat polyclonal | 1:50 for fluorescence | Vaughan et al., 2001 |
| Jo12 | Lamin A/C aa 319–572 | Mouse monoclonal | 1:10 for blotting and fluorescence | Venables et al., 1997 |
| IF8 | Pocket A of Rb | Mouse monoclonal | 1:1000 for blotting 1:50 for fluorescence | Bartek et al., 1992 |
| Ab-2 | Pocket C of RB | Rabbit polyclonal | 1:50 for fluorescence | Calbiochem |
| Phospho-Rb (Ser780) | Phospho-Ser780 of Rb | Rabbit polyclonal | 1:1000 for blotting and fluorescence | Sigma/RBI |
| 204–41 | NuMa | Mouse monoclonal | 1:20 for fluorescence | Calbiochem |
| LAP15 | Lap2-α specific αα 187–693 | Mouse monoclonal | 1:100 for blotting and 1:10 for fluorescence | Dechat et al., 1998 |
GFP-Rb deletion constructs were expressed in HEK293 cells and their distribution between the nucleus and the cytoplasm was observed before and after extraction with hypotonic buffer. +, the presence of a fusion protein within a compartment; −, the absence of fusion protein within a compartment.
Solid Phase Overlay Assay
Lamin C cDNA in pBlueScript KS+ (a gift from G. Krohne, University of Würzburg, Würzburg, Germany) and LAP2α cDNA in pET23a plasmid (Dechat et al., 1998) were transcribed in vitro by using T7 polymerase (Promega, Madison, WI), and RNAs were translated in vitro by using rabbit reticulocyte lysate (Promega) and [35S]methionine (PerkinElmer Life Sciences, Boston, MA), according to the manufacturers' instructions. Glutathione S-transferase (GST)-fusion constructs of Rb corresponding to pocket A (aa 379–612), pocket B (aa 612–792), and pocket C (aa 792–928), and GST fusion of lamin C covering its C-terminal domain (aa 319–572) were separated by SDS-PAGE (Laemmli, 1970). Gels were stained with Coomassie or transblotted onto nitrocellulose (0.2 μm; Schleicher & Schuell, Dassel, Germany) in 48 mM Tris-HCl, pH 9.4, 39 mM glycine by using the Mini Transblot system (Bio-Rad). Nitrocellulose membranes were incubated in overlay buffer (10 mM HEPES, pH 7.4, 100 mM NaCl, 5 mM MgCl2, 2 mM EGTA, 0.1% Triton X-100, 1 mM DTT) for 1 h. Filters were then blocked with 2% bovine serum albumin (wt/vol) in overlay buffer for 1 h and probed with reticulocyte lysate containing in vitro translated 35S-labeled proteins, diluted 1:50 in overlay buffer plus 1% bovine serum albumin (wt/vol) and 1 mM phenylmethylsulfonyl fluoride, for 3 h at room temperature. After extensive washing in overlay buffer, nitrocellulose was air dried, and bound proteins were detected by autoradiography.
Preparation of Cell Extracts
Cells were grown in 90-mm petri dishes. Medium was aspirated from the petri dishes and the cultures were washed twice with PBS. Cultures were extracted by incubation with hypotonic buffer (10 mM HEPES-KOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 0.5 mM DTT) for 30 min at 4°C. The buffer was removed and the cells were washed twice with fresh hypotonic buffer and then scraped directly into radioimmunoprecipitation assay buffer for SDS-PAGE.
Immunoprecipitation
Mouse IgG Dynabeads (Dynal Biotech, Oslo, Norway) were coupled to lamin A/C or LAP2α-specific antibody by incubation for 12 h at 4°C in the presence of 1% bovine serum albumin. Asynchronously growing cells were extracted with hypotonic solution containing 10 mM KCl, 10 mM HEPES-KOH, pH 7.4, 1.5 mM MgCl2, 0.1% Triton X-100, 0.5 mM DTT, and protease inhibitors. After a 10-min incubation at 4°C, nuclei were isolated with homogenization and samples were centrifuged for 5 min in Eppendorf microcentrifuge. Nuclei were extracted with buffer containing 0.5 M NaCl and samples were centrifuged for 5 min at 13,000 rpm. Soluble fractions after dialysis to PBS/0.1% Triton X-100 were processed for immunoprecipitation by using LAP2α and lamin A/C-specific antibody coupled to 100 μl of mouse IgG Dynabeads. After 2-h incubation at 4°C, beads were washed with PBS/0.1%Triton X-100 (3 × 5 volumes) and prepared for gel electrophoresis and immunoblotting.
Gel Electrophoresis and Immunoblotting
One-dimensional SDS-PAGE was performed according to Laemmli (1970). For immunoblotting, proteins separated on gels were electrophoretically transferred onto nitrocellulose (0.2 mm; Schleicher & Schuell) in 48 mM Tris-HCl, pH 9.4, 39 mM glycine by using the Mini Transblot system (Bio-Rad). For the immunological detection of proteins the enhanced chemiluminescence system was used. Primary antibodies were as described in Table 1. Secondary antibodies were goat anti-mouse IgG conjugated to horseradish peroxidase (Jackson Immunoresearch).
Cloning of GFP-Rb Fusion Constructs
A series of N- and C-terminal deletion constructs of Rb were created by restriction digestion of full-length wild-type Rb cloned into BlueScript SK vector followed by subcloning into the pEGFP vector. Constructs that expressed green fluorescent protein (GFP)-tagged polypeptides were made in GFP plasmid (CLONTECH, Palo Alto, CA) that contains a multiple cloning site downstream from the cytomegalovirus early promoter. All cloning procedures were performed according to standard methods (Sambrook et al., 1989).
For construct GFP-Rb Δ 628C, Rb cDNA was digested/repaired with BsaHI/Klenow-EcoRI, and the resulting 0.9-kb fragment was cloned into GFPc1 between BglII/Klenow-EcoRI sites into GFPc 1.
For construction of GFP-Rb Δ 328C, Rb cDNA was digested with EcoRI-PstI, and the 0.9-kb fragment was subcloned into GFP-Rb Δ 628C between EcoRI-PstI sites.
For construct GFP-Rb Δ 180C, Rb cDNA was digested with EcoRI-SspI, and the resulting 1.4-kb fragment was subcloned into GFP-Rb 628C between EcoRI-SmaI.
For construct GFP-Rb Δ 300N328C, Rb cDNA was digested with EcoRi-PstI, and the 0.9-kb fragment was cloned into GFPc1 between EcoRI-PstI sites.
For construct GFP-Rb Δ 300N180C, Rb cDNA was digested with EcoRI-SspI, and the resulting 1.4-kb fragment was cloned into GFPc-1 between EcoRI-SmaI.
For construction of GFP-Rb Δ 300N, a 3.1-kb Rb cDNA fragment was cloned into GFPc1 between EcoRI-KpnI sites.
For construct GFP-Rb Δ610N, Rb cDNA was digested with BglII-BglII, and the resulting 1.8-kb fragment cloned into GFPc1 between BglII-BamHI sites.
For construction of GFP-Rb Δ 750N, Rb cDNA was digested with SspI-BglII, and the 1.3-kb fragment was cloned into GFPc3 between EcoRI/Klenow-BamHI.
For GFP-Rbwt, a 3.1-kb EcoRI-KpnI fragment was subcloned into GFP-Rb Δ 628C between EcoRI-KpnI sites.
RESULTS
C-Terminal Domain of Rb Is Both Necessary and Sufficient for Nuclear Anchorage
A series of N- and C-terminal deletion constructs of Rb fused to GFP (Figure 1a) were created and transiently expressed in HEK293 cells. The molecular weights and the expression levels of GFP-Rb fusion proteins were investigated by Western blotting of whole cell lysates by using either a monoclonal anti-GFP antibody (Figure 1b) or anti-Rb antibodies. As expected all constructs were expressed at high levels and corresponded to the expected mobility of the GFP chimeras. To investigate the cellular localization of the transiently expressed constructs we performed immunofluorescence microscopy by using cells fixed 72 h after transfection. Wild-type GFP-Rb was distributed throughout the nucleoplasm (Figure 2a and Table 2) identical to endogenous Rb (Figure 2c). In contrast, GFP-N terminus was restricted to the cytoplasm and was excluded from the nucleus (Figure 2a and Table 2). Identical patterns were observed with the constructs GFP-NA, GFP-A, and GFP-AB (our unpublished data; but see Table 2). Because the NLS has been mapped to the C terminus of Rb (Zacksenhaus et al., 1993) the cytoplasmic localization of these constructs was expected. Unexpectedly, the construct GFP-NAB was distributed between the cytoplasm and the nucleus (Figure 2a and Table 2). Finally, the constructs GFP-ABC, GFP-BC (Table 2), and GFP-C (Figure 2a and Table 2) were restricted to the nucleus, consistent with the presence of a functional NLS in pocket C. In summary, it can be concluded that all constructs containing the C-terminal domain (and hence the NLS) localize to the nucleus where their distribution is indistinguishable from that of wild-type GFP-Rb. Domains lacking the C-terminal domain do not localize to the nucleus, except for GFP-NAB, which localized to the nucleus and cytoplasm. Transfection experiments were also carried out on primary HDF and HeLa cells, revealing identical results (our unpublished data).
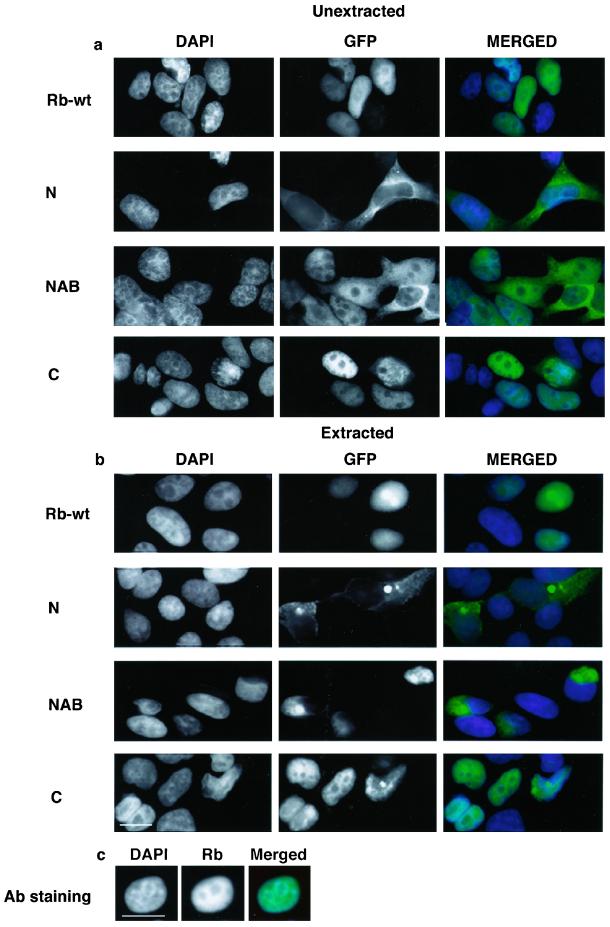
Figure 2.
Cellular localization of GFP-Rb chimeras. HEK293 were transiently transfected with GFP-Rb fusion constructs and fixed with 4% formaldehyde with or without prior extraction with hypotonic buffer. Cells were processed for immunofluorescence microscopy. The DNA was stained with DAPI. Each micrograph shows the distribution of DNA (left) or GFP (middle) in black and white or merged images (right) in color. Rb-wt, wild-type Rb fused to GFP; N, N-terminal domain fused to GFP; NAB, N terminus + pockets A and B fused to GFP; C, pocket C fused to GFP. (a) Distribution of GFP chimeras in fixed cells. (b) Distribution of GFP chimeras in cells fixed after extraction with hypotonic buffers. (c) Distribution of endogenous Rb in cells fixed after hypotonic extraction. Bar, 10 μm.
Table 2.
Cellular distribution of GFP-Rb fusion constructs
| Construct | Distribution before extraction
|
Distribution after extraction
|
||
|---|---|---|---|---|
| Nuclear | Cytoplasmic | Nuclear | Cytoplasmic | |
| GFP-wt | + | − | + | − |
| GFP-N | − | + | − | + |
| GFP-NA | − | + | − | + |
| GFP-NAB | + | + | − | + |
| GFP-A | − | + | − | + |
| GFP-AB | − | + | − | + |
| GFP-ABC | + | − | + | − |
| GFP-BC | + | − | + | − |
| GFP-C | + | − | + | − |
GFP-Rb deletion constructs were expressed in HEK293 cells and their distribution between the nucleus, and the cytoplasm was observed before and after extraction with hypotonic buffer. + indicates the presence of a fusion protein within a compartment; − indicates the absence of fusion protein within a compartment.
In previous reports, Rb was retained in the nucleus after extraction with hypotonic solutions in a phosphorylation-dependent manner (Mittnacht and Weinberg, 1991). In the model cell lines used in the current investigation (HDF or HEK293) treatment of cells with hypotonic solutions before fixation and staining also retained a significant fraction of Rb in the nucleus (our unpublished data). Because Rb mutants with internal deletions in a region spanning pockets B and C expressed in some tumor cells were not retained in the nucleus (Mittnacht and Weinberg, 1991), nuclear anchorage is likely mediated by these subdomains. To determine whether the pocket domains B and C are indeed involved in nuclear anchorage, HDF (our unpublished data) and HEK293 cells that had been transfected with GFP-Rb chimeras were extracted with hypotonic solutions. As expected wild-type GFP-Rb was retained in the nuclei of numerous cells, similar to endogenous protein (Figure 2b and Table 2). For those cells transfected with constructs that localized to the cytoplasm (e.g., GFP-N terminus; Figure 2b and Table 2) cytoplasmic staining was retained in some but not all cells. Importantly, for the construct GFP-NAB, which localized to the cytoplasm and the nucleoplasm, only the cytoplasmic staining was retained after extraction with hypotonic solutions and the nucleoplasmic staining was abolished completely (Figure 2b and Table 2). All chimeras containing pocket C, including GFP-ABC, GFP-BC (Table 2), and GFP-C (Figure 2b and Table 2) were mostly retained in nuclei, after extraction with hypotonic solutions. Thus, all constructs possessing pocket C were retained in the nucleus, whereas the construct GFP-NAB, which was partially localized within the nucleus, was not retained. These data suggest that pocket C contains a motif that is both necessary and sufficient for anchorage of Rb in the nucleus.
LAP2α and Lamin C Both Bind to C Terminus of Rb
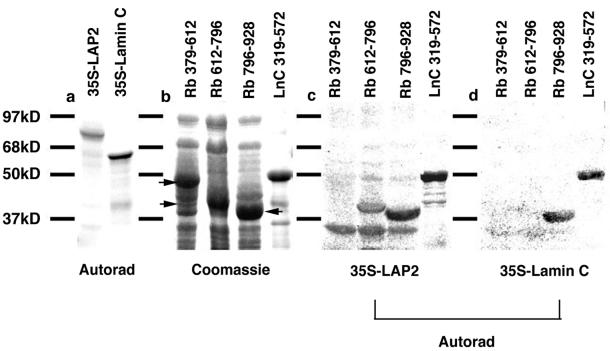
In a previous investigation, lamin A was shown to interact with the pocket domain of Rb in blot overlay assays (Ozaki et al., 1994). To test whether A-type lamins and their nucleoskeletal interaction partner LAP2α interact directly with Rb pocket C, and might thus be responsible for the nuclear anchorage of Rb, we used similar assays. Furthermore, because binding of pRb to lamin A has been demonstrated previously using this assay (Ozaki et al., 1994), herein we concentrated on lamin C. Three different GST-fusion constructs of Rb corresponding to pocket A (aa 379–612), pocket B (aa 612–792), and pocket C (aa 792–928) were resolved on SDS-PAGE (Figure 3b). The resolved proteins were then transferred to nitrocellulose and overlayed with [35S]methionine-labeled LAP2α or lamin C (Figure 3a). Neither LAP2α (Figure 3b) nor lamin C (Figure 3c) interacted with pocket A of Rb. Although lamin C did not interact with pocket B of Rb (Figure 3c), LAP2α showed a weak interaction (Figure 3b). Both lamin C (Figure 3c) and LAP2α (Figure 3b) interacted strongly with pocket C of Rb. A GST fusion protein containing the tail domain of lamin C (aa 319–572; Figure 3d) represented a positive control in this experiment, because we have shown previously that LAP2α and lamin C both interact with this domain (Dechat et al., 2000). Taken together, these data reveal that LAP2α and lamin C are both capable of interacting with the nuclear anchorage domain in Rb and suggest that these proteins are involved in the nuclear anchorage of Rb.
Figure 3.
LAP2α binds to pocket B and C, and lamin C to pocket C of Rb. Recombinant GST fusions of Rb domains corresponding to pockets A (379–612), B (612–792), and C (792–928), and GST fusions of lamin C C-terminal domain (319–572) were separated by SDS-PAGE and stained with Coomassie, or transferred to nitrocellulose, overlaid with vitro translated 35S-labeled recombinant LAP2α or lamin C. Bound proteins were detected by autoradiography. (a) Translated 35S-labeled LAP2α (LAP2) and lamin C. (b) Recombinant GST-protein fragments resolved on SDS-PAGE and stained with Coomassie Blue. (c) Filter overlayed with 35S-LAP2. (d) Filter overlayed with 35S-lamin C. Note, lower bands in 35S-LAP2α autoradiogramm are unspecific interactions with bacterial protein.
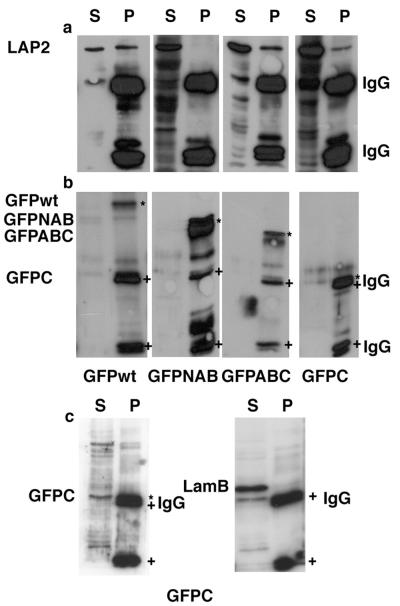
To confirm that these interactions also occur under more physiological conditions in cell lysates, GFP-Rb chimeras were expressed in HEK293 cells. Cell extracts were then prepared and the GFP-Rb chimeras immunoprecipitated with anti-GFP antibodies. Immunoprecipitates were resolved on SDS-PAGE, transferred to nitrocellulose, and blotted with a GFP antibody to detect the fusion proteins (Figure 4, b and c) and with a LAP2α-specific antibody (Figure 4a). As expected, LAP2α coimmunoprecipitated with GFP-Rb, GFP-ABC, and GFP-C. In contrast, LAP2α did not coimmunoprecipitate with GFP-NAB. In contrast, lamin B2, the major lamin in HEK293 cells, did not coprecipitate with GFP-C, excluding the possibility that pocket C is a sticky domain (Figure 4d).
Figure 4.
LAP2α coimmunoprecipitates with Rb fusion proteins from nuclear extracts of cells transfected with GFP-Rb fusion constructs. HEK293 cells were transiently transfected with GFP-Rb fusion constructs for 72 h. Nuclei were isolated from cells and extracted with solutions containing 500 mM NaCl. Soluble fractions were dialyzed to 100 mM NaCl and processed for immunoprecipitation by using GFP-specific monoclonal antibody coupled to mouse IgG Dynabeads. Immunoprecipitates were analyzed by immunoblotting with monoclonal antibody against LAP2α (a) and GFP (b) or as a control with monoclonal antibody against GFP and lamin B2 (c). wt, wild-type Rb fused to GFP; NAB, N terminus plus pockets A and B of Rb fused to GFP; ABC, pockets A, B, and C fused to GFP; C, pocket C fused to GFP. IgG indicates the positions of immunoglobulin light and heavy chains. LAP2 indicates the position of LAP2α. S indicates protein remaining in the cell extract supernatant after immunoprecipitation. P indicates material recovered in the immunoprecipitate. The positions of GFP-chimeras and IgG heavy and light chains are also indicated in b and c with * and + symbols, respectively. NB GFPC has a mobility that is almost identical to IgG heavy chain.
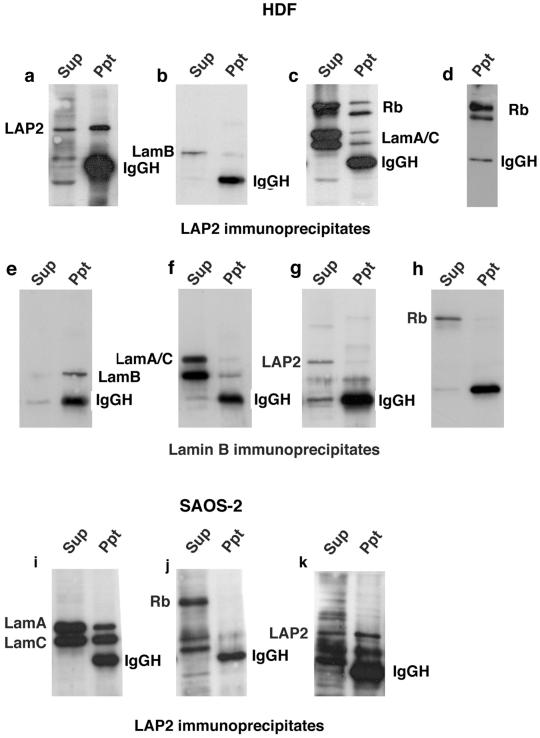
To determine whether endogenous Rb interacts with either LAP2α or lamin C in cells, we prepared cell extracts from HDF. LAP2α was immunoprecipitated from the extracts by using an LAP2α-specific antibody (Figure 5a). Fractions of lamins A/C and Rb both coimmunoprecipitated with LAP2α (Figure 5c). Lamin B2, however, did not coimmunoprecipitate with LAP2α (Figure 5b). Two forms of Rb with slightly different mobility were detected in cell extracts (Figure 5c). A faster migrating underphosphorylated form and a slower migrating, more heavily phosphorylated form. The faster migrating form coimmunoprecipitated more efficiently with LAP2α, suggesting that LAP2α binds preferentially the underphosphorylated form of Rb. To confirm that the two Rb isoforms were differentially phosphorylated, we blotted LAP2α immunoprecipitates with phosphospecific Rb antibodies. Even though the faster migrating form was quantitatively the largest component of the immunoprecipitate (Figure 5c), it was detected less efficiently with the phospho-antibodies (Figure 5d). Thus, the Rb isoform that coprecipitated preferentially with LAP2α was indeed hypophosphorylated.
Figure 5.
Coimmunoprecipitation of RB and lamin A/C with LAP2α. Nuclei were isolated from asynchronously dividing HDF and extracted with solutions containing 500 mM NaCl. Soluble fractions were dialyzed to 100 mM NaCl and processed for immunoprecipitation by using LAP15 to immunoprecipitate LAP2α (a–d) or LN42 to immunoprecipitate lamin B2 (e–f). The supernatant (sup) and immunoprecipitate (Ppt) were both resolved on SDS-PAGE and immunoblotted. LAP15 was used to detect LAP2α (a and e), LN43 to detect lamin B2 (b and f), and JoL2 and IF8 to detect lamin A/C and Rb on the same filter (c), or IF8 to detect Rb alone (h). Alternatively, filters were probed with phospho-Rb (Ser780) to detect phosphorylated forms of Rb (d). Nuclei were also isolated from the osteosarcomma cell line SAOS-2 and processed for immunoprecipitation by using LAP15 to immunoprecipitate LAP2α (i–k). Sup and Ppt were both resolved on SDS-PAGE and immunoblotted with JoL2 to detect lamins A/C (i), IF8 to detect Rb (j), or LAP15 to detect LAP2α (k). In all panels, LamA indicates the position of lamin A, LamC indicates the position of lamin C, lamB indicates the position of lamin B2, LAP2 indicates the position of LAP2α, and Rb indicates the position of RB.
As a control, immunoprecipitation reactions were performed in parallel with an anti-lamin B2 antibody. Lamin B2 was efficiently precipitated from the extract (Figure 5e). Although some lamin A/C did coimmunoprecipitate with lamin B2 (Figure 5f), neither LAP2α (Figure 5g) nor Rb (Figure 5h) was detected in lamin B2 immunoprecipitates.
Because lamin C and LAP2α both seemed to bind to pocket C of Rb we predicted that Rb would not associate with LAP2α–lamin A/C complexes in cell lines containing certain Rb deletion mutants. SAOS-2 is an osteosarcoma cell line expressing an Rb mutant containing a C-terminal deletion, covering pocket C (Mittnacht and Weinberg, 1991). Cell extracts were prepared from SAOS-2 cells and immunoprecipitated with the LAP2α antibody. LAP2α and lamins A/C were both recovered efficiently in LAP15 immunoprecipitates (Figure 5, i and k). However, the Rb deletion mutant was absent (Figure 5j), suggesting that pocket C is essential for the interaction between Rb and LAP2α–lamin A/C complexes.
Dominant Negative Lamin Mutants Sequester Rb and LAP2α within Nuclear Aggregates
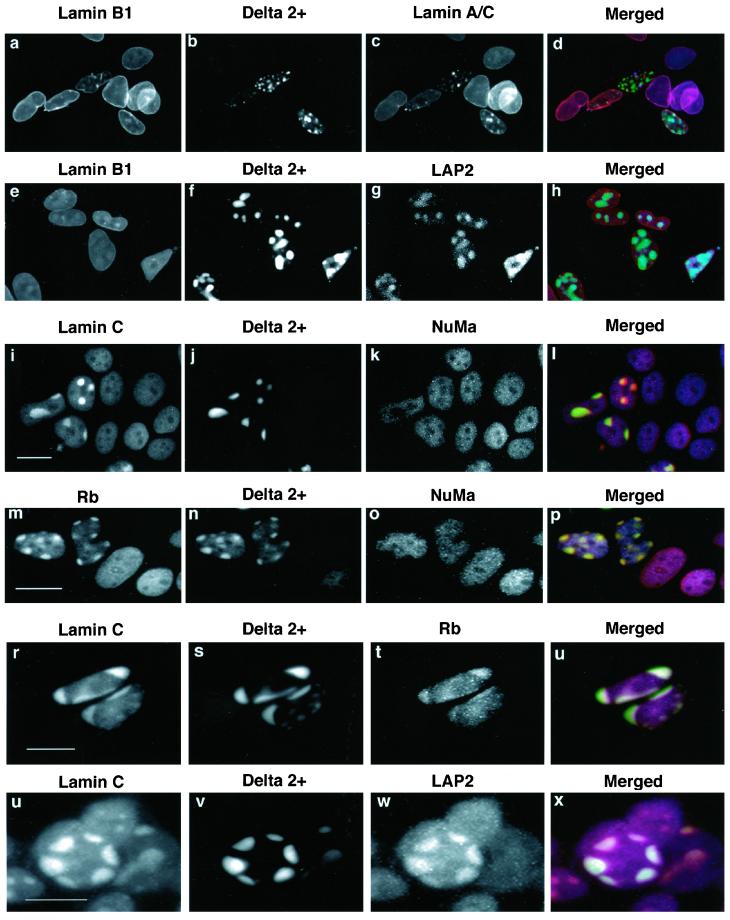
Our data suggest that LAP2α and lamins A/C interact with the nuclear anchorage domain of Rb in blot overlay assays and in cell extracts. To obtain evidence for an interaction of the proteins in vivo, we investigated whether induced LAP2α/lamin A/C redistribution influences the distribution of Rb in the nucleus. In recent publications, we have shown that dominant negative lamin mutants that sequester endogenous A-type lamins into nuclear aggregates (Izumi et al., 2000; Vaughan et al., 2001) also sequester LAP2α into the same aggregates (Dechat et al., 2000). If LAP2α and lamins A/C do influence the distribution of Rb, sequestration of LAP2α and lamins A/C into nuclear aggregates should result in a similar sequestration of Rb but not other nuclear matrix proteins. HEK293 cells were transfected with the dominant negative lamin mutant GFP-delta 2+. Transfected cells were costained with different combinations of antibodies to detect lamins A/C, LAP2α, Rb, and lamin C or as controls with lamin B1 or NuMa. GFP-delta 2+ formed aggregates in transfected cells (Figure 6b, f, j, n, s, and v). In the cells containing aggregates, lamin A/C (Figure 6c), lamin C (Figure 6, i and r), LAP2α (Figure 6, g and w), and a fraction of Rb (Figure 6, m and t) were all sequestered to the aggregates. Other nuclear matrix proteins such as NuMa (Figure 6, k and o) and lamin B1 (Figure 6, a and e), however, remained normally distributed. The use of triple fluorescence in these experiments clearly demonstrated that Rb but not other nuclear matrix proteins (NuMa and lamin B1) were sequestered (Figure 6, a–p). Triple fluorescence also demonstrated that LAP2α, lamin C, and Rb were all present in the same aggregates (Figure 6, r–x). Taken together, these experiments show that, when the distribution of lamins A/C is perturbed, the distribution of LAP2α and a fraction of Rb are also specifically disturbed, supporting the existence of a complex of these three proteins in the nucleus.
Figure 6.
Redistribution of LAP2α and Rb by expression of dominant negative mutants. The dominant negative mutant GFPΔ2+ (GFP-delta2+) was transiently expressed in HEK293 cells. Cells were processed for triple immunofluorescence microscopy by using the following combinations of antibody. Goat anti-lamin B1 followed by TRITC donkey anti-goat to detect lamin B1 (a and e). JoL2 followed by Cy5 donkey anti-mouse to detect lamin A/C (c). LAP15 followed by Cy5 donkey anti-mouse to detect LAP2α (g and w). Ab5 followed by Cy5 donkey anti-mouse to detect Rb (m and k). Anti-lamin C followed by TRITC donkey anti-rabbit to detect lamin C (i, r, and u). Anti-NuMa followed by Cy5 donkey anti-mouse to detect number (k and o). GFP-Δ2+ was detected using fluorescein isothiocyanate filters (b, f, j, n, s, and v). Individual staining patterns are presented in black and white. Merged images (d, h, l, p, u, and x) are displayed in color with TRITC staining presented in red the GFP signal in green and the Cy5 signal in blue. Yellow indicates areas of spectral overlap between red and green. Cyan indicates areas of spectral overlap between red blue and green. Bars, 10 μm.
Hypophosphorylated Rb Is Not Anchored in Nucleus of Cells Lacking LAP2α
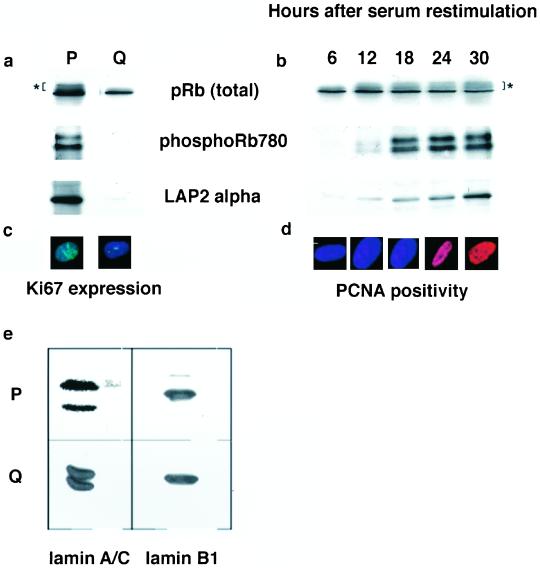
LAP2α is expressed in a growth-dependent manner in primary skin fibroblasts. By using immunoblotting, LAP2α is readily detected in exponentially dividing cultures of HDF along with phosphorylated forms of Rb (Figure 7a). When HDF were induced to enter quiescence by serum starvation (judged by absence of expression of Ki67; Figure 6c), neither LAP2α nor phospho-isoforms of Rb were detected (Figure 7a). In contrast, expression of hypophosphorylated Rb, lamins A/C, and lamin B1 did not change as cells progressed from a proliferating to a quiescent state (Figure 7, a and e). When HDF were induced to reenter the cell cycle by serum restimulation, LAP2α expression was slowly increasing between 12 and 30 h after serum restimulation, whereas total Rb protein remained constant within 30 h of restimulation (Figure 7b). Phospho-isoforms of Rb were first detected 18 h after serum restimulation (Figure 7b). Cells entered S phase between 24 and 30 h after serum restimulation as judged by staining with proliferating cell nuclear antigen, a marker for DNA replication (Figure 7d). Consequently, there is a 12-h period after serum restimulation when hypophosphorylated Rb is present in HDF in the absence of LAP2α.
Figure 7.
Changes in the expression of LAP2α and phospho-Rb in proliferating and quiescent HDF. Exponentially dividing HDF where induced to enter a quiescent state by withdrawal of serum (a and e). Cells were stained with Ki67 before and after serum withdrawal to confirm that cells had entered a quiescent state (c). Cell extracts were prepared from proliferating (P) and quiescent cells (Q) and prepared for immunoblotting. Immunoblots were probed with IF8 to detect total Rb, Rb780 to detect phospho-Rb, LAP15 to detect LAP2α (all in a), and JoL2 to detect lamins A/C or anti-lamin B1 (both in e). Quiescent cells were induced to reenter the cell cycle by readdition of serum to growth media (b and d). Reentry into the cell cycle was monitored by immunostaining with anti-proliferating cell nuclear antigen antibodies (d). Cell extracts were prepared at 6-h intervals after serum restimulation and used for immunoblotting (b). Immunoblots were probed with IF8 to detect total Rb, Rb780 to detect phospho-Rb, and LAP15 to detect LAP2α. Asterisk (*) indicates slowly migrating phosphorylated forms of Rb.
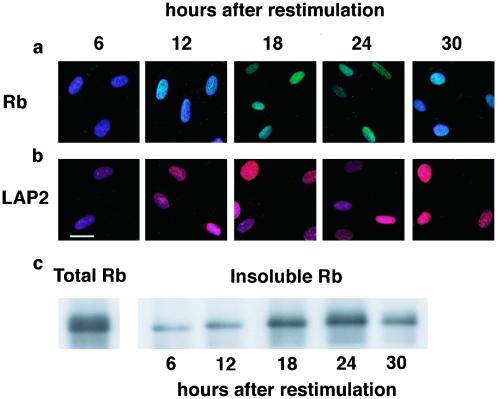
We predicted that hypophoshorylated Rb would be susceptible to extraction between 6 and 12 h after serum restimulation, if LAP2α is required for its nuclear anchorage. To test this prediction, HDF were extracted to remove nonanchored Rb and then costained with DAPI (to detect chromatin) and either anti-Rb or anti-LAP2α antibodies (Figure 8, a and b). Six hours after serum restimulation, little or no LAP2α was detected in the nuclei of HDF, and Rb was no longer detected after extraction. LAP2α was more readily detected 12 h after serum restimulation, and at this time Rb could be detected after extraction. Between 18 and 24 h after serum restimulation, LAP2α was readily detected and Rb was retained throughout the nucleus of extracted cells (Figure 8, a and b). Thirty hours after serum restimulation as cells entered S phase and Rb became hypophosphorylated (Figure 7, b and d), Rb was no longer detected in the nucleus after extraction, although LAP2α was present in the nucleus (Figure 8, a and b). To confirm and extend these findings, HDF were extracted at the same time intervals after serum restimulation but were prepared for immunoblotting rather than immunofluorescence. Six and 12 h after serum restimulation, only a fraction of Rb was insoluble after hypotonic extraction (Figure 8, c and d). Significantly more Rb was insoluble between 18 and 24 h after serum restimulation (Figure 8d) when levels of LAP2α had increased (Figure 7b). Thirty hours after serum restimulation, the amount of insoluble Rb declined (Figure 8d) as cells entered S phase (Figure 7d). These data support the hypothesis that expression of LAP2α is essential for the nuclear anchorage of hypophosphorylated Rb before cells enter S phase.
Figure 8.
Rb is not anchored in the nucleus in the absence of LAP2α. Quiescent HDF were stimulated to reenter the cell cycle by refeeding with serum. The cells were extracted with hypotonic solution at 6-h intervals after restimulation and either stained with IF8 (a) to detect Rb, LAP15 to detect LAP2α (b), or immunoblotted with IF8 (c). The micrographs in a and b are two color-merged images in which DAPI (blue) is used to stain DNA, IF8 staining is shown in green, and LAP15 staining is shown in red. Where IF8 (a) or LAP15 (b) staining is absent the predominant color is blue. In c, total Rb in unextracted cells is shown in the left-hand panel (18 h after restimulation). Bar (b), 10 μm.
DISCUSSION
LAP2α and Lamins A/C Anchor Rb in Nucleus
In this investigation, we showed the C pocket to be necessary and sufficient for anchorage of GFP-Rb chimeras in the nucleus. Two nuclear architectural proteins, lamin C and LAP2α, bind across this domain in blot overlay assays. LAP2α binds efficiently to hypophosphorylated Rb and lamins A/C in cell extracts, provided that pocket C is present. When LAP2α and lamins A/C are forced to accumulate into aggregates formed by dominant negative lamin mutants, Rb also accumulates in those aggregates. Finally, when LAP2α is not present in cells entering G1 phase from a quiescent state, hypophosphorylated Rb is not anchored in the nucleus. Taken together, these data suggest that LAP2α and lamins A/C have an important role in the nuclear anchorage of Rb.
In a previous study, hypophosphorylation of Rb was correlated with its anchorage (defined by resistance to extraction with hypotonic buffers) in the nucleus (Mittnacht and Weinberg, 1991). The N terminus of Rb had been shown to be involved in its oligomerization (Hensey et al., 1994) and binding to proteins located at sites of RNA processing (Durfee et al., 1994). These findings gave rise to the suggestion that this domain of Rb may facilitate binding to nuclear bodies. However, loss of nuclear anchorage of Rb occurs in mutant proteins carrying deletions spanning pockets B and C (amino acids 702–767; Mittnacht and Weinberg, 1991). The behavior of GFP-Rb chimeras in our study suggests that sequences located in pocket C (amino acids 750–928) are both necessary and sufficient for nuclear anchorage. In our studies, GFP-chimeras expressing pocket C are resistant to extraction with hypotonic solution consistent with the Mittnacht and Weinberg data. Although we cannot exclude the possibility that Rb is anchored in the nucleus through both its C-terminal and N-terminal domains, pocket C does have an essential role.
LAP2α binds to GST-Rb fragments corresponding to pockets B and C; therefore, it seems likely that binding of LAP2α occurs across these two pocket domains. Moreover, LAP2α binds preferentially (but not exclusively) to hypophosphorylated Rb. In a previous report, lamin A was identified as an Rb binding protein that also associated with the large pocket domain (Ozaki et al., 1994). We can confirm that A-type lamins associate with Rb in blot overlay assays, through pocket C. Based upon this evidence LAP2α and lamins A/C both seem to be involved in nuclear anchorage of Rb. Because lamins A/C and LAP2α have overlapping binding sites but also seem to be present in the same Rb complexes, we propose that the two proteins cooperate in anchoring Rb within the nucleus.
Form and Function of Lamin–LAP2α–Rb Complexes
Specific nuclear processes occur within organizational centers referred to as nuclear bodies (reviewed by Lamond and Earnshaw, 1999). Some authors have favored the view that nuclear metabolism occurs on a nucleoskeleton with properties resembling the intermediate filament cytoskeleton (Hozak et al., 1993). Others have suggested that a more local organization is probable in which architectural proteins such as NuMa may assemble into mini-platforms, providing surfaces for transcription or splicing (Harborth and Osborn 1999). If the proposal of Harborth and Osborn is true, proteins capable of forming oligomeric complexes on chromosome surfaces might be sufficient to tether regulatory proteins at those sites. LAP2α and lamins A/C both posses the properties necessary for formation of oligomeric complexes. LAP2α contains a chromosome binding domain and its association with chromosomes is regulated by protein phosphorylation (Dechat et al., 1998; Vlcek et al., 1999). It also exists as oligomers on the surface of chromosomes in cell extracts (Dechat et al., 1998). Similarly, lamins A and C have distinct self-assembly properties and are also capable of forming oligomers on the surface of chromosomes in vitro (Glass and Gerace, 1990). Finally, lamin A/C localization influences the distribution of LAPα in the nucleus (Dechat et al., 2000), suggesting that both proteins interact at the same sites. Therefore, one possible mechanism by which LAP2α and lamin A/C may function is by forming local oligomeric complexes on chromosome surfaces. Lamin-LAP2α oligomers would have the capacity to bind Rb at a site (in pocket C) adjacent to its E2F binding domains in pockets A and B. Lamin/LAP2α oligomers may thus provide a mechanism for enhanced silencing. The presence of lamin/LAP2α oligomers on the surface of chromosomes might provide a platform that tethers one or more Rb molecules beside or across a promoter. These oligomers would be expected to be highly stable and therefore may be capable of forming an effective silencing complex.
Nuclear Tethering and Tumor Formation
Nuclear tethering of Rb seems to be important for its function because forms of Rb identified in certain tumors carry deletions spanning pockets B and C. These forms of Rb are not tethered in the nucleus (Mittnacht et al., 1991) and do not associate with LAP2α and lamins A/C (Figure 5). If anchorage of Rb to chromatin is necessary for its silencing activities, loss of this activity might be oncogenic. Because lamins A/C and LAP2α are involved in nuclear anchorage of Rb, loss of expression of one or more of these proteins might also be oncogenic. Consistent with this hypothesis is the loss of expression of lamin A in a wide range of human cancers (Venables et al., 2000) We hope to test this hypothesis directly using LAP2α −/− mice, which are currently being produced or with the existing lamin A/C −/− mice. The nuclear anchorage domain in Rb is also the site of interaction with the c-Abl tyrosine kinase (Welch and Wang, 1993, 1995) and MDM2 (Xiao et al., 1995). Therefore, LAP2α and lamins A/C might compete with c-Abl and MDM2 for binding to Rb. If this is true, an alternative function for nuclear anchorage may be to regulate the interactions between Rb and c-Abl and/or MDM2.
ACKNOWLEDGMENTS
We thank Prof. Howard Worman (Columbia University, New York, NY), Prof. David Lane and Dr. Joost Zomerdick (University of Dundee, Dundee, United Kingdom) for the supply of lamin and Rb constructs and for the gift of antibodies. We also thank Dr. William Whitfield for help and advice. This work was generously supported by grants from the Cancer Research Campaign and the Wellcome Trust (to C.J.H.), with a grant from The International Association for Cancer Research (to C.J.H. and R.F.), and by a grant from the Austrian Science Research Fund (FWF P13374) (to R.F.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–07–0450. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–07–0450.
REFERENCES
- Berger R, Theodor L, Shoham J, Gokkel E, BrokSimoni F, Avraham KB, Copeland NG, Jenkins NA, Rechavi G, Simon AJ. The characterization and localization of the mouse thymopoietin lamina-associated polypeptide 2 gene and its alternatively spliced products. Genome Res. 1996;6:361–370. doi: 10.1101/gr.6.5.361. [DOI] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Cao L, Faha B, Dembski M, Tsai LH, Harlow E, Dyson N. Independent binding of the retinoblastoma protein and p107 to the transcription factor E2F. Nature. 1992;355:176–179. doi: 10.1038/355176a0. [DOI] [PubMed] [Google Scholar]
- Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- Cohen M, Lee KK, Wilson KL, Gruenbaum Y. Transcriptional repression, apoptosis, human diseases, and the functional evolution of the nuclear lamins. Trends Biochem Sci. 2001;26:41–48. doi: 10.1016/s0968-0004(00)01727-8. [DOI] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Gotzmann J, Stockinger A, Harris CA, Talle MA, Siekierka JJ, Foisner R. Detergent-salt resistance of LAP2 alpha in interphase nuclei and phosphorylation-dependent association with chromosomes early in nuclear assembly implies functions in nuclear structure dynamics. EMBO J. 1998;17:4887–4902. doi: 10.1093/emboj/17.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Korbei B, Vaughan OA, Vlcek S, Hutchison CJ, Foisner R. Lamina-associated polypeptide 2 alpha binds intranuclear A-type lamins. J Cell Sci. 2000;113:3473–3484. doi: 10.1242/jcs.113.19.3473. [DOI] [PubMed] [Google Scholar]
- Dowdy SF, Hinds PW, Louie K, Reed SI, Arnold A, Weinberg RA. Physical interaction of the retinoblastoma protein with human D-cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- Durfee T, Mancini MA, Jones D, Elledge SJ, Lee WH. The amino-terminal region of the retinoblastoma gene-product binds a novel nuclear matrix protein that colocalizes to centers for RNA processing. J Cell Biol. 1994;127:609–622. doi: 10.1083/jcb.127.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JA, Kill IR, Pugh G, Quinlan RA, Lane EB, Hutchison CJ. Cell cycle changes in A-type lamin associations detected in human dermal fibroblasts using monoclonal antibodies. Chromosome Res. 1997;5:383–394. doi: 10.1023/a:1018496309156. [DOI] [PubMed] [Google Scholar]
- Ellis DJ, Jenkins H, Whitfield WGF, Hutchison CJ. GST-lamin fusion proteins act as dominant negative mutants in Xenopus egg extract and reveal the function of the lamina in DNA replication. J Cell Sci. 1997;110:2507–2518. doi: 10.1242/jcs.110.20.2507. [DOI] [PubMed] [Google Scholar]
- Ewen ME, Sluss HK, Sherr CJ, Matsushime H, Kato JY, Livingston DM. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- Foisner R, Gerace L. Integral membrane-proteins of the nuclear-envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- Gant TM, Harris CA, Wilson KL. Roles of LAP2 proteins in nuclear assembly and DNA replication: truncated LAP2 beta proteins alter lamina assembly, envelope formation, nuclear size, and DNA replication efficiency in Xenopus laevis extracts. J Cell Biol. 1999;144:1083–1096. doi: 10.1083/jcb.144.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JR, Gerace L. Lamins A and C bind to and assemble on the surface of mitotic chromosomes. J Cell Biol. 1990;111:1047–1057. doi: 10.1083/jcb.111.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J, Osborn M. Does NuMa have a scaffold function in the interphase nucleus? Crit Rev Eukaryot Gene Exp. 1999;9:319–328. doi: 10.1615/critreveukargeneexpr.v9.i3-4.160. [DOI] [PubMed] [Google Scholar]
- Harris CA, Andryuk PJ, Cline S, Chan HK, Natarajan A, Siekierka JJ, Goldstein G. 3 distinct human thymopoietins are derived from alternatively spliced messenger-RNAs. Proc Natl Acad Sci USA. 1994;14:6283–6287. doi: 10.1073/pnas.91.14.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CA, Andryuk PJ, Cline SW, Mathew S, Siekierka JJ, Goldstein G. Structure and mapping of the human thymopoietin (TMPO) gene and relationship of human TMPO-beta to rat lamin-associated polypeptide-2. Genomics. 1995;28:198–205. doi: 10.1006/geno.1995.1131. [DOI] [PubMed] [Google Scholar]
- Hensey CE, Hong F, Durfee T, Qian YW, Lee EYHP, Lee WH. Identification of discrete structural domains in the retinoblastoma protein-amino-terminal domain is required for its oligomerization. J Biol Chem. 1994;269:1380–1387. [PubMed] [Google Scholar]
- Hozak P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to a nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- Hutchison CJ, Alvarez-Reyes M, Vaughan OA. Lamins in disease. why do ubiquitously expressed nuclear envelope proteins give rise to tissue-specific disease phenotypes? J Cell Sci. 2001;114:9–19. doi: 10.1242/jcs.114.1.9. [DOI] [PubMed] [Google Scholar]
- Izumi M, Vaughan OA, Hutchison CJ, Gilbert DM. Head and/or CaaX domain deletions of lamin proteins disrupt preformed lamin A, and C but not lamin B structure in mammalian cells. Mol Biol Cell. 2000;11:4323–4337. doi: 10.1091/mbc.11.12.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins H, Holman T, Lyon C, Lane B, Stick R, Hutchison C. Nuclei that lack a lamina accumulate karyophilic proteins and assemble a nuclear matrix. J Cell Sci. 1993;106:275–285. doi: 10.1242/jcs.106.1.275. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of the bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1999;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lees JA, Buchkovich KJ, Marshak DR, Anderson CW, Harlow E. The retinoblastoma protein is phosphorylated on multiple sites by human cdc2. EMBO J. 1991;10:4279–4290. doi: 10.1002/j.1460-2075.1991.tb05006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees JA, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13:7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz-Bohme B, Wismar J, Fuchs S, Reifegerste R, Buchner E, Betz H, Schmitt B. Insertional mutation of the Drosophila nuclear lamin Dmo gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J Cell Biol. 1997;137:1001–1016. doi: 10.1083/jcb.137.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rolef Ben-Shahar T, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11:3938–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- Mancini MA, Shan B, Nickerson JA, Penman S, Lee WH. The retinoblastoma gene-product is a cell cycle-dependent, nuclear matrix-associated protein. Proc Natl Acad Sci USA. 1994;91:418–422. doi: 10.1073/pnas.91.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J, Campbell KHS, Ford CC, Stick R, Hutchison CJ. The role of lamin-LIII in nuclear assembly and DNA replication, in cell-free extracts of Xenopus eggs. J Cell Sci. 1991;98:271–279. doi: 10.1242/jcs.98.3.271. [DOI] [PubMed] [Google Scholar]
- Mittnacht S, Weinberg RA. G1/S phosphorylation of the retinoblastoma protein is associated with an altered affinity for the nuclear compartment. Cell. 1991;65:381–393. doi: 10.1016/0092-8674(91)90456-9. [DOI] [PubMed] [Google Scholar]
- Moir RD, Goldman RD. Overexpression of the nuclear lamins alters lamina assembly, composition and DNA replication. Mol Biol Cell. 1995;6:1171–1171. [Google Scholar]
- Moir RD, Spann TP, Herrmann H, Goldman RD. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J Cell Biol. 2000;149:1179–1191. doi: 10.1083/jcb.149.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GE, Manilal S. Heart to heart: from nuclear proteins to Emery-Dreifuss muscular dystrophy. Hum Mol Genet. 1999;8:1847–1851. doi: 10.1093/hmg/8.10.1847. [DOI] [PubMed] [Google Scholar]
- Ozaki T, Saijo M, Murakami K, Enomoto H, Taya Y, Sakiyama S. Complex formation between lamin A and the retinoblastoma gene product- identification of the domain on lamin A required for its interaction. Oncogene. 1994;9:2649–2653. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatas T. Molecular cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1989. [Google Scholar]
- Smythe C, Jenkins HE, Hutchison CJ. Incorporation of the nuclear pore basket protein Nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 2000;19:3918–3931. doi: 10.1093/emboj/19.15.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan OA, Alvarez-Reyes M, Bridger JM, Broers JLV, Ramaekers FCS, Wehnert M, Morris GE, Whitfield WGF, Hutchison CJ. Both emerin, and lamin C depend on lamin. A for localization at the nuclear envelope. J Cell Sci. 2001;114:2577–2590. doi: 10.1242/jcs.114.14.2577. [DOI] [PubMed] [Google Scholar]
- Vaughan OA, Whitfield WGF, Hutchison CJ. Functions of the nuclear lamins. Protoplasma. 2000;211:1–7. [Google Scholar]
- Venables RS, McLean S, Luny D, Moteleb E, Morley S, Quinlan RA, Lane EB, Hutchison CJ. Expression of individual lamins in basal cell carcinomas of the skin. Br J Cancer. 2000;84:512–519. doi: 10.1054/bjoc.2000.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlcek S, Just H, Dechat T, Foisner R. Functional diversity of LAP2 alpha and LAP2 beta in postmitotic chromosome association is caused by an alpha-specific nuclear targeting domain. EMBO J. 1999;18:6370–6384. doi: 10.1093/emboj/18.22.6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JYJ, Knudsen ES, Welch PJ. The retinoblastoma tumor-suppressor protein. Adv Cancer Res. 1994;64:25–85. doi: 10.1016/s0065-230x(08)60834-9. [DOI] [PubMed] [Google Scholar]
- Welch PJ, Wang JYJ. A c-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-Abl tyrosine kinase in the cell cycle. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- Welch PJ, Wang JYJ. Abrogation of retinoblastoma protein function by c-Abl through tyrosine kinase dependent and tyrosine kinase-independent mechanisms. Mol Cell Biol. 1995;15:5542–5551. doi: 10.1128/mcb.15.10.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KL. The nuclear envelope, muscular dystrophy, and gene expression. Trends Cell Biol. 2000;10:125–129. doi: 10.1016/s0962-8924(99)01708-0. [DOI] [PubMed] [Google Scholar]
- Xiao ZX, Chen JD, Levine AJ, Modjtahedi N, Xing J, Sellers WR, Livingston DM. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- Yang L, Guan TL, Gerace L. Lamin-binding fragment of LAP2 inhibits increase in nuclear volume during the cell cycle and progression into S phase. J Cell Biol. 1997;139:1077–1087. doi: 10.1083/jcb.139.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacksenhaus E, Bremner R, Phillips RA, Gallie BL. A bipartite nuclear-localization signal in the retinoblastoma gene product and its importance for biological activity. Mol Cell Biol. 1993;13:4588–4599. doi: 10.1128/mcb.13.8.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma DD, Luo RX, Harbor JW, Dean DC. Exit from G1, and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF, and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]