Abstract
We demonstrate the existence of a large endoplasmic reticulum (ER)-localized multiprotein complex that is comprised of the molecular chaperones BiP; GRP94; CaBP1; protein disulfide isomerase (PDI); ERdj3, a recently identified ER Hsp40 cochaperone; cyclophilin B; ERp72; GRP170; UDP-glucosyltransferase; and SDF2-L1. This complex is associated with unassembled, incompletely folded immunoglobulin heavy chains. Except for ERdj3, and to a lesser extent PDI, this complex also forms in the absence of nascent protein synthesis and is found in a variety of cell types. Cross-linking studies reveal that the majority of these chaperones are included in the complex. Our data suggest that this subset of ER chaperones forms an ER network that can bind to unfolded protein substrates instead of existing as free pools that assembled onto substrate proteins. It is noticeable that most of the components of the calnexin/calreticulin system, which include some of the most abundant chaperones inside the ER, are either not detected in this complex or only very poorly represented. This study demonstrates an organization of ER chaperones and folding enzymes that has not been previously appreciated and suggests a spatial separation of the two chaperone systems that may account for the temporal interactions observed in other studies.
INTRODUCTION
To travel along the secretory pathway and eventually reach their appropriate cellular destinations, newly synthesized secreted and membrane-bound proteins must fold and assemble correctly. Failure to do so results in their retention in the endoplasmic reticulum (ER) and eventual degradation. The proper conformational maturation of nascent secretory pathway proteins is both aided and monitored by a number of ER chaperones and folding enzymes in a complex process termed ER quality control (Hammond and Helenius, 1994). The components and mechanisms of action of two major chaperone systems have been best studied. The first system is dependent on the presence of both monoglucosylated N-linked glycans and unfolded regions on nascent glycoproteins. The resident ER protein UDP-glucosyltransferase (GT) binds to the unfolded regions and adds a single glucose to the deglucosylated glycan (Trombetta and Parodi, 1992), which in turns provides the binding site for the ER chaperones calnexin and calreticulin (Sousa et al., 1992; Hammond et al., 1994). Cleavage of this glucose by the resident ER protein glucosidase II (Kornfeld and Kornfeld, 1985) abrogates the calnexin/calreticulin binding site (Trombetta and Parodi, 1992; Hebert et al., 1995). If during the ensuing time the nascent chain folds, UDP-GT will not rebind and the protein will be released from the ER. However, if folding is not complete or correct folding is unable to occur, the cycle will repeat itself (Sousa et al., 1992; Hebert et al., 1995).
The second major ER chaperone system is only dependent on the presence of unfolded regions on proteins containing hydrophobic residues, which are recognized by the ER chaperone BiP (Flynn et al., 1991; Blond-Elguindi et al., 1993). In fact, some calnexin/calreticulin substrates can bind to BiP instead, if N-linked glycosylation is blocked (Balow et al., 1995; Zhang et al., 1997). BiP is the ER Hsp70 family member (Haas and Wabl, 1983; Munro and Pelham, 1986), and like all Hsp70 proteins, it binds both ADP and ATP, which serve to regulate its binding and release from nascent chains (Kassenbrock and Kelly, 1989; Wei and Hendershot, 1995). The hydrolysis of ATP to ADP causes Hsp70 proteins to bind tightly to substrates, and the exchange of ATP for ADP induces a conformational change in Hsp70, which in turn causes the release of bound substrates (Kassenbrock and Kelly, 1989; Palleros et al., 1993; Buchberger et al., 1995; Wei and Hendershot, 1995). The ATPase cycle of Hsp70 proteins is both positively and negatively regulated by a number of chaperones and cofactors, including DnaJ, GrpE, Hip, Hop, and Bag-1 (Liberek et al., 1991; Frydman and Hohfeld, 1997; Hohfeld and Jentsch, 1997; Cheetham and Caplan, 1998); however, to date mammalian ER homologues of most of these proteins have not been identified. Like the calnexin/calreticulin system, Hsp70 proteins are thought to undergo cycles of binding and release from unfolded proteins (Gamer et al., 1996; Bukau and Horwich, 1998), with folding occurring during the release cycle (Hendershot et al., 1996). A number of other resident ER chaperones and folding enzymes, such as GRP94 (Melnick et al., 1992; Kuznetsov et al., 1994; Chavany et al., 1996), GRP170 (Lin et al., 1993; Kuznetsov et al., 1997), ERp72 (Mazzarella et al., 1990; Lin et al., 1993; Reddy et al., 1996), protein disulfide isomerase (PDI) (Roth and Pierce, 1987; Bulleid and Freedman, 1988; Reddy et al., 1996), and peptidyl-prolyl isomerases (Bose et al., 1994; Bush et al., 1994) have been identified and shown to bind to some nascent ER proteins. However, the role of most of these proteins in ER quality control and their relationship to the two major chaperone systems have not been clearly elucidated.
Hetero-oligomeric Ig proteins have provided an excellent system for studying the interaction of nascent ER proteins with chaperones during folding and subunit assembly. Ig molecules interact with several molecular chaperones as they mature in the ER (Haas and Wabl, 1983; Bole et al., 1986; Roth and Pierce, 1987; Hochstenbach et al., 1992; Melnick et al., 1992; Lin et al., 1993; Lassoued et al., 1996). Among these, BiP has been shown to play a vital role in the folding and assembly of immunoglobulin heavy and light chains. Although BiP interacts very transiently with the variable domain of light chains (VL) (Hellman et al., 1999) and with some constant region domains of heavy chains (Kaloff and Haas, 1995), it remains bound to the first constant domain of the heavy chain (CH1) in the absence of light chain synthesis (Hendershot et al., 1987). This is because the CH1 domain does not fold until light chains assemble and release BiP (Lee et al., 1999). In this way, BiP retains unassembled heavy chains inside the ER and prevents their secretion or transport to the cell surface (Hendershot et al., 1987). Interestingly, when BiP is released from unassembled heavy chains in vitro with ATP, the CH1 domain can fold rapidly and form its intramolecular disulfide bond (Lee et al., 1999). However, in mouse myeloma cells that lack light chains, the heavy chains have a long half-life, remain very stably bound to BiP, and do not fold their CH1 domain (Vanhove et al., 2001), even though ATP is present in the ER (Clairmont et al., 1992). These observations led us to speculate that there may be a regulatory protein(s) in the heavy chain–BiP complex that prevents BiP from cycling on and off heavy chains in vivo, which is lost upon detergent lysis of cells (Lee et al., 1999).
In this report, we demonstrate by chemical cross-linking that a number of additional ER molecular chaperones and folding enzymes are part of the heavy chain–BiP complex. GRP94 is one of the most abundant proteins present in this complex. A number of other ER chaperones and folding enzymes (i.e., CaBP1 or protein disulfide isomerase P5, PDI, an ER Hsp40 cochaperone [ERdj3], GRP170, ERp72, cyclophilin B, UDP-GT and the SDF2-L1 protein) are also found in this complex. Calnexin and calreticulin, which are major ER proteins and which interact with nascent glycoproteins in the ER (Tatu and Helenius, 1997; Zhang et al., 1997), were either absent from this complex or only present in very small quantities. This large multiprotein complex, excluding ERdj3 and to a lesser extent PDI, also forms in the absence of heavy chain synthesis and may constitute the ER network that has been proposed by others (Kuznetsov et al., 1994, 1997; Reddy et al., 1996; Tatu and Helenius, 1997).
MATERIALS AND METHODS
Cell Lines and Antibodies
The human hepatoma cell line HepG2 and mouse lymphoma cell lines Ag8(8) (γ+, LC−) (Bole et al., 1986), G403 (γΔCH1+, LC−) (Hendershot et al., 1987), Ag8.653 (Ig−) (Kearney et al., 1979), and J558L (Oi et al., 1983) were grown in complete RPMI-1640 medium containing 10% fetal bovine serum, 2 mM l-glutamine, and 100 U/ml penicillin-streptomycin. NIH3T3 mouse fibroblasts were cultured in DMEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 100 U/ml penicillin-streptomycin. Polyclonal anti-rodent BiP (Hendershot et al., 1995), anti-GRP94 (Lawson et al., 1998), and anti-calnexin were raised against recombinant proteins in this laboratory. Anti-GRP170, anti-ERp29, anti-UDP-GT, and CaBP1 antibodies were kindly provided by Drs. R. Zimmermann (Universitat des Saarlandes, Homburg, Germany), S. Mkrtchiana (Karolinska Institute, Stockholm, Sweden), D. Thomas (McGill University, Montreal, Canada), and D. Ferrari (Max Plank Institute, Gottingen, Germany), respectively. Antibodies specific for protein disulfide isomerase, calreticulin, and ERp72 were purchased from Stressgen (Victoria, British Columbia, Canada).
Metabolic Labeling and Cross-Linking of Proteins
Cells (20–40 × 106) were metabolically labeled with 35S-TransLabel (ICN Pharmaceuticals, Costa Mesa, CA) (50 μCi/ml) in 8 ml of methionine-free RPMI-1640 medium supplemented with 10% complete RPMI-1640 medium. After 16 h of labeling, an additional 0.1 mCi of 35S-TransLabel was added to the cell culture for an extra 30-min incubation. The labeled cells were washed with cold HEPES buffer (25 mM HEPES-KOH, pH 8.3, and 125 mM KCl) three times, resuspended at 10 × 106 cells/ml in HEPES buffer, and aliquoted into tubes. A 5-mg/ml solution of the membrane-permeable, thiol-cleavable cross-linker dithiobis(succinimidylpropionate) (DSP) was freshly prepared in dimethyl sulfoxide and added to the cells to achieve a final concentration of 150 μg/ml. The cells were incubated on ice for 1 h with occasional shaking and then incubated with 1 M glycine (100 mM final concentration) and 1 M N-ethylmaleimide (40 mM final concentration) for an additional 15 min on ice to quench the cross-linking reaction. Control incubations were treated in an identical manner except no cross-linker was added. The cells were collected by centrifugation at 2500 rpm in a microcentrifuge for 3 min at 4°C and lysed in 1 ml of NP-40 lysing buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% deoxycholic acid, and 0.5% NP-40). The resulting lysates were clarified by centrifugation at 14,000 rpm for 10 min at 4°C. Ig heavy chains were precipitated with protein A-Sepharose for 2 h, because the γ heavy chains bind directly to protein A-Sepharose and do not required a primary antibody for immunoprecipitation. Resident ER proteins were immunoprecipitated by incubating cell lysates with the appropriate antisera for 90 min followed by a 30-min incubation with protein A-Sepharose. Immune precipitates were washed and prepared for SDS-PAGE analysis as described previously (Hendershot et al., 1995).
In an attempt to isolate the multiprotein complex without using a cross-linker, seven different methods were used to disrupt the ER vesicles contained in the postnuclear fraction. These included 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS) lysing buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, and 1% CHAPS), digitonin lysing buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, and 1% digitonin), dodecylmaltoside lysing buffer (50 mM Tris-HCl, pH 7.4, 165 mM NaCl, 2 mM EDTA, and 1% dodecylmaltoside), NP-40 lysing buffer, a cycle of freeze-thawing, followed by homogenization of the postnuclear fraction in HFTP buffer (25 mM Tris-HCl, pH 8.2, 1 mM EDTA, 50 mM NaCl, 10% [vol/vol] glycerol, 10 mM Na2MoO4, 1 mM phenylmethylsulfonyl fluoride, 20 μg/ml leupeptin, and 20 μg/ml aprotinin), Triton X-100 lysing buffer (20 mM HEPES, pH 7.4, 50 mM NaCl, 20 mM imidazole, and 1% Triton X-100, diluted to 0.02% before immunoprecipitation), and resuspension of the postnuclear fraction in TESV buffer (50 mM Tris-HCl, pH 7.5, 2 mM EDTA, 50 mM NaCl, and 1 mM NaVO3) followed by sonication three times on ice for 10 s each as described previously (Chavany et al., 1996). In each case, insoluble matter was pelleted by centrifugation at 14,000 rpm for 10 min at 4°C, and Ig heavy chains [Ag8(8)] were precipitated from the resulting supernatant with protein A-Sepharose beads that had been washed in the appropriate buffer and BiP (Ag8.653) was immunoprecipitated with a specific antibody. The precipitated protein complexes were washed in their respective lysing buffers containing 400 mM NaCl.
Two-Dimensional (2D) SDS-PAGE Analysis
To examine direct protein–protein interactions, 6 × 106 cells were treated with 150 μg/ml DSP. Protein complexes were immunoprecipitated as described above. The samples were first electrophoresed under nonreducing conditions to separate different cross-linked complexes that might be present. The gel strip corresponding to a single sample was cut from the first gel and equilibrated in 5 ml of reducing SDS sample buffer for 40 min at room temperature on a rocker to reduce DSP and liberate the various proteins in the complex. The gel strip was then placed on the top of a second gel and run at a 90° angle to the first. After electrophoresis, gels were stained with Coomassie Blue, destained, treated with Amplify Reagent (Amersham Biosciences, Piscataway, NJ), and dried for autoradiography.
Western Blot Analysis
To identify proteins in the heavy chain complex, 20 × 106 unlabeled Ag8(8) cells (γ+, LC−) were either cross-linked with 150 μg/ml DSP or kept on ice, untreated. After lysis, Ig heavy chains were precipitated from the samples with protein A-Sepharose, and complexes were fractionated on 10% SDS-PAGE gels under reducing conditions. As a positive control for the various antisera, whole cell lysates were prepared from 2 × 106 cells and loaded directly onto the gels. Ag8.653 cells [an Ig− subclone of the Ag8(8) cell line] were treated similarly and served as a negative control for proteins that bind nonspecifically to protein A-Sepharose instead of to the heavy chains. After electrophoresis, proteins were transferred to a nitrocellulose membrane and probed with the indicated primary antibodies in gelatin wash buffer (0.1% gelatin, 15 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.1% Triton X-100, and 0.002% NaN3). After washing, the blots were incubated for 90 min with the appropriate secondary antibodies (goat anti-mouse Ig for anti-calnexin, anti-PDI, and anti-ERp29 antibodies and goat anti-rabbit Ig for anti-calreticulin, anti-GRP170, anti-GRP94, anti-ERp72, anti-CaBP1, anti-UDP-GT, and anti-BiP). Blots were then incubated with horseradish peroxidase-protein A for 30 min and developed with the enhanced chemiluminescence reagent (Amersham Biosciences).
Postnuclear Fraction Preparation and Vesicle Lysis
To increase the efficiency of cross-linking, the postnuclear fraction, which contained ER vesicles, was prepared before cross-linking and isolated before lysis in some experiments as indicated. The method for vesicle production and purification represents a crude fractionation allowing removal of most of the contaminants. The labeled cells (20 × 106) were washed twice with 8 ml of cold HEPES buffer and resuspended in 2 ml of HEPES buffer and disrupted with 50 strokes in a Teflon homogenizer. The resulting sample was centrifuged at 2500 rpm in Microfuge for 10 min at 4°C to separate cell debris from the vesicles, which remained in the supernatant. The supernatant was divided into two tubes; one was treated with 150 μg/ml DSP for 1 h on ice, and the other received only dimethyl sulfoxide. After quenching with 100 mM glycine and 40 mM N-ethylmaleimide for 15 min on ice, the postnuclear fraction was pelleted at 14,000 rpm for 10 min at 4°C. The vesicle pellet was lysed with NP-40 lysing buffer and prepared for immunoprecipitation.
Preparation of Proteins for Sequencing
To purify proteins bound to Ig heavy chains for identification, the postnuclear fraction was prepared from 300 × 106 Ag8(8) cells. The vesicle suspension was treated with 150 μg/ml DSP, quenched, pelleted, and lysed as described above. The Ig heavy chain complex was isolated with protein A-Sepharose and subjected to reducing SDS-PAGE analysis. Coomassie-stained bands were excised, reduced with dithiothreitol, and cysteine residues were alkylated with iodoacetmide. The proteins were then digested with sequencing-grade trypsin (Promega, Madison, WI), and peptides were extracted with 0.1% trifluoroacetic acid plus 5% acetonitrile for analysis by combined liquid chromatography/tandem mass spectrometry. Peptides were isolated and sequenced on the basis of their ion fragmentation patterns and then compared with protein sequence databases. Briefly, separation was performed on a capillary high-performance liquid chromatography system from Waters (Milford, MA) by using a 0.32 × 150-mm column of Waters Delta-Pak C8 packed by MicroTech Scientific (Sunnyvale, CA). Acetic acid (1%) was used as mobile phase and elution was accomplished at a flow rate of 3 μl/min with a gradient of 0–45% acetonitrile >40 min. Mass spectrometry was performed using an LCQ-Deca ion-trap mass spectrometer from ThermoFinnigan (San Jose, CA) with an electrospray ion source. Peptides were assigned to known proteins by searching the uninterpreted spectra acquired by collision-induced dissociation of peptides against the National Center for Biotechnology Information nonredundant protein sequence database using the SEQUEST program provided by ThermoFinnigan.
Glycerol Gradients
Ten million Ag8(8) and Ag8.653 cells were metabolically labeled and postnuclear supernatants were prepared as described previously. After quenching, Triton X-100 (1% vol/vol) was added to disrupt the vesicles, and samples were made 3% glycerol (vol/vol) before layering on to 20–40% glycerol gradients (20 mM HEPES, 150 mM NaCl, and 0.2% Triton X-100). Gradients were centrifuged in an SW41 rotor at 45,000 rpm for 16 h. Fractions (15×333 μl) were collected from the bottom of the tube and immunoprecipitated as described above. Molecular weight standards were purchased from Pharmacia (Peapack, NJ) and sedimented with each run. Fractions were collected, analyzed by SDS-PAGE, and visualized by Coomassie staining.
RESULTS
Identification of Additional Proteins in Heavy Chain–BiP Complex
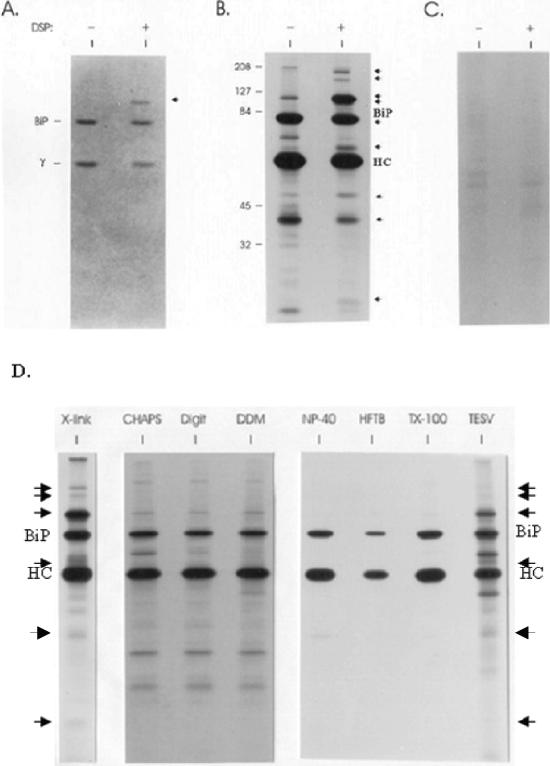
To determine whether additional ER proteins that were not stable to detergent-lysing conditions might be bound to unassembled Ig heavy chains, we isolated the postnuclear fraction from metabolically labeled cells and treated them with DSP, a membrane-permeable, thiol-cleavable cross-linker. Two cell lines were used, the first one, Ag8(8), only expresses the Ig heavy chain. Without light chains the heavy chains cannot be secreted and accumulate in the ER bound to BiP. The second one, Ag8.653, does not express either light or heavy chains and served as a negative control. Ig heavy chains were isolated from Ag8(8) cells, and the pattern of proteins binding to heavy chains after cross-linking was compared with that obtained with heavy chains isolated from nontreated vesicles. Coomassie-stained gels that had been run under reducing conditions revealed an additional protein in the heavy chain–BiP complex after cross-linking that migrated with apparent molecular mass of ∼94 kDa and was present in amounts that seemed similar to BiP and heavy chains (Figure 1A). The autoradiograph obtained from the same gel revealed that, in addition to BiP, at least nine other proteins were part of the unassembled Ig heavy chain complex. Their molecular masses were ∼170, 150, 94, 90, 72, 55, 46, 43, and 23 kDa (Figure 1B). The 72-kDa band was only detectable on a shorter exposure of this autoradiograph and was masked by the BiP signal on the longer exposure (our unpublished data). Heavy chains isolated in the absence of DSP coprecipitated either very small or nondetectable quantities of these same proteins. The 94-kDa band showed the most dramatic increase among the seven proteins after cross-linking (Figure 1B) and was present in substantial quantities as observed by Coomassie staining, whereas the other proteins seemed to be present in much lower amounts because they were not readily detected by this method. To confirm that these additional proteins bound specifically to heavy chains rather than nonspecifically to protein A-Sepharose, the postnuclear fraction from the Ag8.653 Ig− subclone was treated with DSP and protein precipitation was carried out using protein A-Sepharose as described above. There was no detectable binding of these proteins to protein A-Sepharose when labeled lysates from the DSP-treated Ag8.653 postnuclear fraction was used (Figure 1C). The amounts of several proteins coprecipitating with heavy chains actually decreased after the cells were treated with the cross-linker (Figure 1B). One of these, the 60-kDa protein, was identified as mitochondrial hsp60 by microsequencing. We hypothesize that it is binding opportunistically to the heavy chains after NP-40 lysis when the other proteins of the complex are no longer present. Several different nonionic detergents were used in an attempt to isolate the complex, but other than cross-linking only the disruption of ER vesicles by sonication allowed some of the complex to be preserved (Figure 1D). However, trace amounts of some additional heavy chain-associated proteins could be detected with some of the nonionic detergents (CHAPS, digitonin, and dodecylmaltoside). The use of a cross-linker provided the greatest and most reproducible recovery of the additional proteins associated with the heavy chain, so all the following experiments were performed with it.
Figure 1.
Unassembled Ig heavy chains associate with a number of ER proteins in addition to BiP. The postnuclear fraction was obtained from metabolically labeled Ag8(8) cells and divided into two tubes: one for a negative control and the other treated with 150 μg/ml DSP. After quenching, vesicles were solubilized and heavy chains were precipitated with protein A-Sepharose for SDS-PAGE analysis under reducing conditions. The gel was first stained with Coomassie Blue (A) and then exposed to film for an autoradiograph (B). BiP and a γ heavy chain are indicated on the left of the gel. Additional proteins detected after cross-linking are marked with arrows. Ag8.653 cells were labeled and treated similarly (− and +DSP) and then immunoprecipitated with protein A-Sepharose (C). The cell lysates were obtained by using different detergents to release the ER proteins (1% CHAPS, 1% digitonin, 1% dodecylmaltoside [DDM], 0.5% deoxycholic acid, 0.5% NP-40, and 0.2% Triton X-100) or no detergent (a cycle of freeze-thawing followed by homogenization of the postnuclear fraction in HFTB buffer (HFTB) or 10-s sonication in TESV buffer (TESV) as described in MATERIALS AND METHODS (D). Additional proteins detected after cross-linking and after sonication are indicated with arrows.
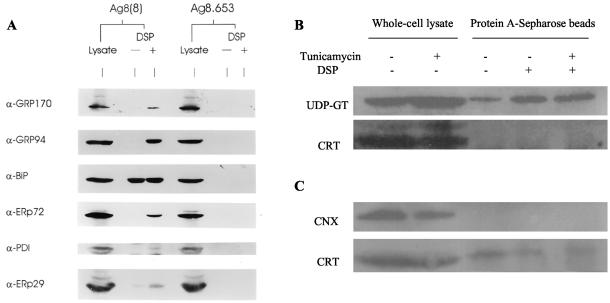
Because the molecular masses of some of the proteins in the complex were similar to those of known ER chaperones and folding enzymes, Western blot analysis was performed using antibodies specific for these resident proteins. As a positive control for the ability of the various antibodies to detect the murine proteins, whole cell lysates of Ag8(8) and Ag8.653 cells were also tested. Western blot analysis demonstrated that the 150-kDa protein bound to heavy chains after cross-linking was GRP170, and the 94-kDa band was identified as GRP94 (Figure 2). Although there were trace amounts of GRP170 and GRP94 coprecipitating with heavy chains before cross-linking, the amount of both proteins markedly increased after cross-linking, which is consistent with the SDS-PAGE analysis of metabolically labeled bands corresponding to these molecular masses (Figure 1B). BiP binds heavy chains very stably in the absence of cross-linkers, so there was no detectable difference in the amount of BiP coprecipitated before and after DSP treatment. The 72- and 58-kDa band was identified as ERp72 and protein disulfide isomerase, respectively (Figure 2A). In an attempt to identify the 23-kDa protein, an antibody to ERp29 was used. ERp29 is a recently identified ER protein that interacts with BiP and contains a thioredoxin-like domain (Mkrtchian et al., 1998). Although the 23-kDa protein observed on our autoradiograph did not comigrate with ERp29 (our unpublished data), we did find that small amounts of ERp29 cross-linked specifically to heavy chains by Western blotting (Figure 2A). Either the incorporation of isotope into ERp29 or its relative pool size may have contributed to our inability to detect it bound to heavy chains by metabolic labeling. Antibodies to both calnexin and calreticulin were also used for Western blotting analysis. First, both calnexin and calreticulin did not seem to be components of the multiprotein complex (Figure 2B). However, if larger numbers of cells were used to isolate the complex, trace amounts of calreticulin coprecipitated with heavy chains both in the presence and absence of the cross-linking agent (Figure 2C). This small amount of calreticulin was still present after tunicamycin treatment, suggesting either that it was not interacting with the N-linked glycan on the heavy chain or, due to the long half-life of heavy chains, that it interacts with the remaining glycosylated heavy chain. Although there was little calreticulin and no calnexin associated with the complex, UDP-GT (p170), which catalyzes the monoglucosylation that is essential for their binding, was readily detected in the complex (Figure 2B).
Figure 2.
Heavy chain–BiP complexes contain substantial quantities of GRP94 and smaller amounts of other chaperones and folding enzymes. Total cell lysates from 1 × 106 Ag8(8) and Ag8.653 cells were used as a positive control for the antibodies (lanes 1 and 4). The postnuclear fraction from 10 × 106 cells was either treated with DSP (lanes 3 and 6) or left untreated (lanes 2 and 5) and lysates were prepared as described in MATERIALS AND METHODS. Lysates from Ag8(8) (lanes 2 and 3) and from Ag8.653 (lanes 5 and 6) were incubated with protein A-Sepharose and precipitated proteins were electrophoresed and transferred for blotting. Segments of the nitrocellulose were reacted with the antibodies indicated and developed as described (A). Then 10 × 106 Ag8(8) cells were treated overnight with 2 (+) or 0 (−) μg/ml tunicamycin, incubated with DSP, and heavy chains were isolated from lysates with protein A-Sepharose and processed as in A. Total cell lysate from 10 × 106 cells was used as a positive control for the antibodies (B). Cells were treated as in B except that 40 × 106 Ag8(8) cells per lane were used, and only 1/10 of the lysate was used as a positive control for the antibodies (C).
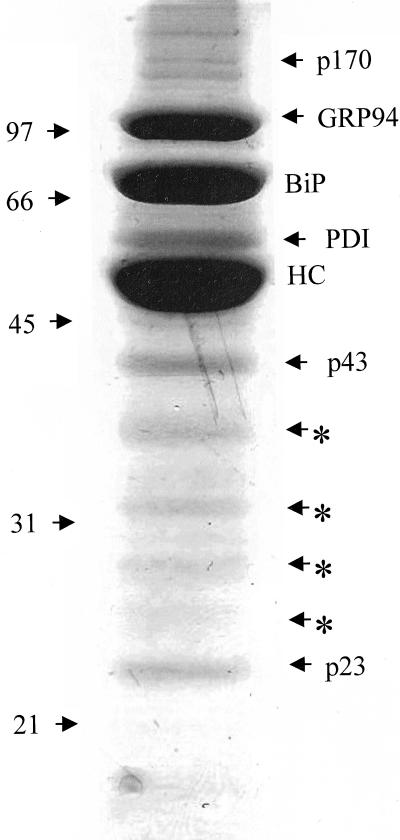
In an attempt to identify all the additional proteins in this complex, the postnuclear fraction was isolated from 300 × 106 unlabeled Ag8(8) cells, treated with DSP, and prepared for precipitation with protein A-Sepharose. The multiprotein complex associated with the heavy chain was resolved under reducing conditions by SDS-PAGE. Individual proteins were visualized by Coomassie Blue stain, and most bands indicated by arrows were identified by mass spectrometry (Figure 3). The results (Table 1) confirmed the presence of the proteins identified by Western blot including PDI, which showed a weak signal by Western blot (Figure 2). The presence of UDP-GT was confirmed (∼170-kDa band) and p43 was identified as ERdj3, a mammalian ER DnaJ cochaperone (Bies et al., 1999; Yu et al., 2000). The ∼23-kDa band contained cyclophilin B, an ER peptidyl-prolyl-isomerase, and the SDF2-L1 protein (stromal cell-derived factor 2-like1), a member of the protein O-mannosyltransferase family (Fukuda et al., 2001) (Figure 4). Although the 94-kDa band appeared as a doublet on some gels, the mass spectrometry data of this band revealed the presence of only GRP94. It is possible that the faster migrating band represents a pool of unglycosylated GRP94. The mass spectrometry data for the 55-kDa band revealed that it primarily contained PDI (50 peptides assigned to this protein) and confirmed that only trace amounts of calreticulin and ERp57 were present (three and eight peptides, respectively, assigned from this band). The bands indicated by asterisks were sequenced and found to entirely correspond to degradation products of the heavy chains (Figure 3). In summary, unassembled heavy chains can be found in the ER associated with a number of different ER chaperones and folding enzymes. Unlike BiP, the association of these other proteins with heavy chains was not particularly stable to NP-40 lysis.
Figure 3.
Coomassie staining of the proteins of the complex for isolation and identification. Then 300 × 106 Ag8(8) cells were treated with 150 μg/ml DSP. The postnuclear fraction was prepared, and Ig heavy chains and associated proteins were isolated with protein A-Sepharose, which were separated by reducing SDS-PAGE on a 10% acrylamide gel. The gel was then Coomassie stained and protein bands were isolated and submitted to mass spectrometry analysis. The arrows shows all the proteins analyzed and the asterisk (*) shows degradation products from the heavy chain.
Table 1.
Identification of the additional proteins by mass spectrometry
| Proteins identified | No. of peptides assigned* | % of protein covered |
|---|---|---|
| UDP-glucosyltransferase | 7 | 9 |
| GRP94 | 37 | 26 |
| PDI | 50 | 40 |
| CaBPI | 15 | 24 |
| ERdj3 | 10 | 25 |
| SDF2-L1/cyclophilin B | 7/2 | 19/13 |
All sequences uniquely identify the proteins shown.
Figure 4.
Sequences of four other proteins of the complex. The following sequences were obtained from the p170, p43, and p23 bands associated with heavy chains as described in MATERIALS AND METHODS. The sequences of UDP-GT (A), the ER Hsp40 cochaperone-ERdj3 (B), cyclophilin B (C), and SDF2-L1 (D). The peptide sequences obtained for each protein are shown in bold.
ER Chaperones Exist as Multiprotein Complexes
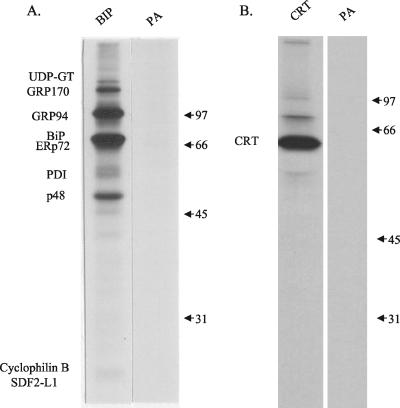
We wished to determine whether the individual proteins of the chaperone complex formed on the unassembled heavy chains or whether the heavy chains associated with a preformed chaperone network. Ag8.653 cells (Ig−) were treated with DSP, and proteins were immunoprecipitated with a polyclonal anti-BiP serum and analyzed by reducing SDS-PAGE. Most of the proteins found in the complex, with the exception of ERdj3, were coprecipitated with BiP (Figure 5A). It is also noticeable that the relative amount of PDI in the complex was decreased in the anti-BiP immunoprecipitated material. A new protein that migrated at ∼48 kDa was detected. Because p48 migrates similar to the heavy chain band in Ag8(8) cells, it is possible that it was also present in the complex associated with heavy chains but was masked by them. As a control, proteins were also immunoprecipitated with a polyclonal anti-calreticulin antibody. Because only a trace amount of calreticulin was found in the complex associated with the heavy chain, we expected not to see the same complex. The data showed that calreticulin does not interact with this complex, and only a trace amount of a 75-kDa protein, which might be BiP, coprecipitated with calreticulin (Figure 5B). Similar results were obtained when NIH3T3 fibroblasts, HepG2 hepatoma cells, or ER vesicles from rat liver were examined, demonstrating that these chaperone complexes are a normal feature of the ER organization (our unpublished data). Western blots were done on the various BiP-associated proteins from Ag8.653 cells to confirm their identity. However, no antibodies were available for cyclophilin B or SDF2-L1, so we can only say that a band at ∼23 kDa is present or absent in the different immunoprecipitations. These data suggest, first, that the chaperones exist together in complexes in the ER, and second, that their assembly with each other is not dependent on the presence of unassembled, unfolded heavy chains but rather that unassembled heavy chains may bind to this preformed ER chaperone network. It is notable that nascent protein substrates of BiP seem to be absent from the immune isolates from Ag8.653 cells. We believe that unlike the unassembled Ig heavy chains, which are a major product of the myeloma cell lines, have a long half-life, and remain incompletely folded, the amount of any other given nascent protein is too small compared with the ER chaperones to detect as single bands and would instead appear as trace smears.
Figure 5.
ER molecular chaperones exist as a complex in the absence of heavy chains. Ag8.653 cells were labeled for 16 h. The postnuclear fraction was isolated and treated with DSP as described in MATERIALS AND METHODS. Lysate was precipitated as indicated with anti-BiP antibody (A) or anti-calreticulin antibody (B). Protein A-Sepharose only was used as a control for nonspecific precipitation.
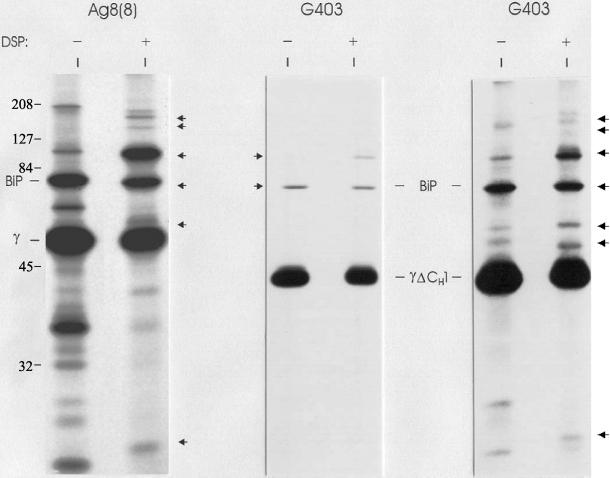
Because the ER chaperones seemed to be present in the ER as a preformed complex, we wished to determine whether their association with Ig heavy chains was dependent on the interaction of BiP with heavy chains. For these experiments, we used the G403 cell line, which synthesizes a γ heavy chain that has deleted its CH1 domain, and therefore no longer possesses a permanent BiP binding site (Hendershot et al., 1987). Heavy chains are normally retained inside the ER and degraded when expressed without light chains, but the same heavy chains, lacking the CH1 domain (e.g., G403 cell line), are transported and secreted very rapidly. Metabolically labeled cell lysates of G403 cells were either subjected to DSP cross-linking or left untreated, and heavy chains were isolated with protein A-Sepharose. As expected, decreased amounts of BiP were associated with the CH1-deleted heavy chains compared with full-length γ heavy chains (Figure 6). Of interest, the other proteins of the complex are also no longer bound to the heavy chain. This provides an additional control for the specificity of binding observed after cross-linking and suggests that either the binding of these proteins is dependent on the presence of BiP or that they all bind to the unfolded CH1 domain. We did observe small amounts of BiP binding, which is a result of its transient association with the other Ig domains (Kaloff and Haas, 1995). After a prolonged exposure of the same film (G403, far right), we observed bands corresponding to the sizes of all the additional proteins of the complex, suggesting that transient association of BiP with heavy chain domains also occurs as part of the same complex (Figure 6). The identification of the various proteins in the complex were confirmed by Western blotting with the same antibodies as in Figure 2 (our unpublished data). In addition, the 48-kDa band found in the anti-BiP precipitated material was also observed, suggesting that it might be also present in the complex with full length heavy chains but masked due to its similarity in size. This protein was identified as CaBP1, also called protein disulfide isomerase P5, both by mass spectrometry (Figure 7 and Table 1) and by Western blot analysis (our unpublished data).
Figure 6.
Additional ER chaperones bind to the same domain of heavy chain as BiP. Ag8(8) (lanes 1 and 2) and G403 (lanes 3–6) cells (heavy chain [HC] ΔCH1+, LC−) were labeled overnight, split in two aliquots, and treated with 0 or 100 μg/ml DSP. Gamma heavy chains were precipitated with protein A-Sepharose and analyzed by reducing SDS-PAGE. The right panel is the same thing that the middle panel but are from an autoradiograph that was exposed 15 times longer.
Figure 7.
Sequence of CaBP1. Sequence of CaBP1 was obtained from 200 × 106 G403 cells. Cells were cross-linked, lysed in NP-40 lysing buffer, and precipitated with protein A-Sepharose. The precipitate was then resolved by reducing SDS-PAGE and the gel Coomassie stained. The sequence was obtained as described in MATERIALS AND METHODS.
Organization of Multiprotein Complex
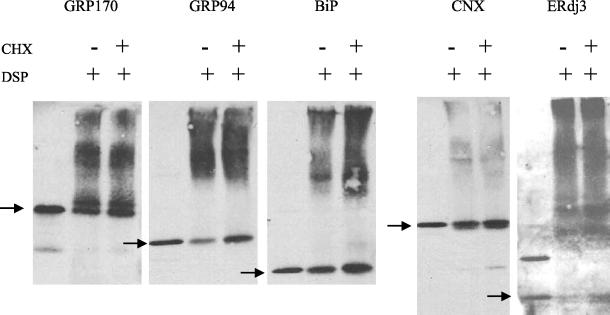
To assess whether these complexes can form without any unfolded protein substrates and to determine what portion of the ER pool of the various chaperones is part of the complex, Ag8.653 cells were treated with cycloheximide to inhibit the translation of new proteins and to allow newly synthesized proteins to exit the ER. This dose of cycloheximide was shown to inhibit translation by labeling a separate pool of treated cells with [35S]methionine and cysteine. Less than 2% of total protein synthesis remained at this dose (our unpublished data). In addition, the amount of time it took to empty the ER of a secreted protein was examined by labeling the J558L plasmacytoma cells, chasing in the presence of cycloheximide, and determining the amount of λI light chains remaining at various times. After 90 min of treatment, only trace amounts of the secreted light chains were still detected inside cells (our unpublished data). Although different proteins leave the ER at various rates, we reasoned that 2 h of cycloheximide treatment should be sufficient to empty the ER of a significant pool of newly synthesized proteins. Cycloheximide-treated and untreated Ag8(8) cells were mechanically disrupted and the postnuclear fraction was treated with the cross-linker. The total lysate was then resolved under nonreducing conditions on a 5–15% gradient SDS gel before transferring and blotting with the indicated antisera (Figure 8). The postnuclear fraction from 1/10 as many untreated cells was removed before cross-linking to serve as a control for the mobility of the free pool of each chaperone. Under normal conditions, the majority of GRP170, GRP94, and BiP were present in high-molecular-weight complexes (Figure 8). Calnexin was also present in larger complexes, although our immunoprecipitation data show that these complexes are distinct from those containing the other three chaperones. The lack of a strong distinct signal in the top portion of the gel may suggest that the calnexin complexes are more heterogeneous than the one containing BiP. Only a small fraction of these chaperones was released from their respective complexes after 2 h of cycloheximide treatment, with <10% of the total pool of each migrating as a free protein. This demonstrates that the majority of BiP, GRP94, ERdj3, and GRP170 are present as large complexes even in the absence of ongoing protein synthesis. It is possible that the other chaperones are also mostly present in the complex but we did not have the reagents to examine this directly. It could be noticed that the antibody raised against ERdj3 recognized an unidentified protein migrating around 63 kDa, which one can contribute to the signal observed for the complexes.
Figure 8.
Multiprotein complex remains upon treatment with cycloheximide to inhibit the translation of new proteins. Ag8.653 cells (8 × 106) were left untreated or treated with 50 μg/ml cycloheximide for 2 h. The postnuclear fraction was prepared from cells, cross-linked with DSP, and lysed in NP-40 lysing buffer. The two cross-linked ER lysates were divided into five aliquots, each of which were directly applied to 5–15% gradient gels before transferring to nitrocellulose membranes for blotting. Untreated Ag8.653 cells (8 × 105) were lysed without prior cross-linking to identify the mobility of the free pool of the various proteins. The nitrocellulose membranes were reacted with the indicated polyclonal antisera. The mobility of the free pool of each protein is indicated with an arrow.
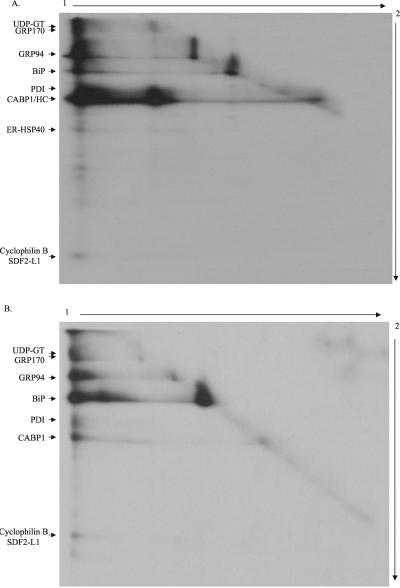
In an attempt to better characterize the chaperone complexes, labeled Ag8(8) cells were directly treated with the cross-linking agent and heavy chain complexes were isolated and resolved by 2D gels; the first dimension run under nonreducing condition and the second dimension under reducing condition. Similarly, BiP-containing complexes were isolated from labeled Ag8.653 cells and analyzed by the same method. In both cases, most of the proteins isolated previously were found in a high-molecular-weight complex(es) that migrated near the top of the first-dimension gel (Figure 9, A and B). The large complex associated with the heavy chain contained among other proteins UDP-GT, GRP170, GRP94, BiP, PDI, ERdj3, cyclophilin B, and SDF2-L1 (Figure 9A). In this gel, a band comigrating with the heavy chain in the second dimension but migrating at ∼120 kDa in the first dimension corresponds to a heavy chain dimer. The presence of free heavy chain dimers, BiP, and GRP94 on the diagonal suggests that cross-linking was not complete when the whole cells were treated, because all of the heavy chains are bound to BiP under nonreducing conditions and should at the very least migrate as heavy chain–BiP complexes after DSP treatment (Figure 1; Hendershot, 1990). The complex associated with BiP in Ag8.653 cells contained readily detectable quantities of UDP-GT, GRP170, GRP94, CaBP1, and the cyclophilin B/SDF2-L1 band. In the case of Ag8.653 cells, it is not determined whether both of the last two proteins were present in the complex. At this moment, it is not clear whether all the ER chaperones are part of a single complex or whether several different high-molecular-weight complexes exist that contain different chaperone complements. However, there is no evidence on these gels for the formation of smaller complexes of individual chaperones with heavy chains or each other.
Figure 9.
Visualization of the high-molecular-weight complexes by 2D gels. Ag8(8) or Ag8.653 cells (6 × 106) were metabolically labeled overnight and then treated with 150 μg/ml DSP. Protein complexes were immunoprecipitated with protein A-Sepharose alone for Ag8(8) or anti-BiP and protein A-Sepharose for Ag8.653. The samples were first electrophoresed under nonreducing conditions to separate different cross-linked complexes that might be present (1). The gel strip corresponding to a single sample was cut from the first gel and equilibrated in 5 ml of reducing SDS sample buffer for 40 min at room temperature on a rocker to reduce DSP and liberate the various proteins in the complex. The gel strip was then placed on the top of a second gel and run at a 90° angle to the first (2). After staining the gel was dried and a film exposed. A and B is obtained from Ag(8) and Ag8.653 cells, respectively.
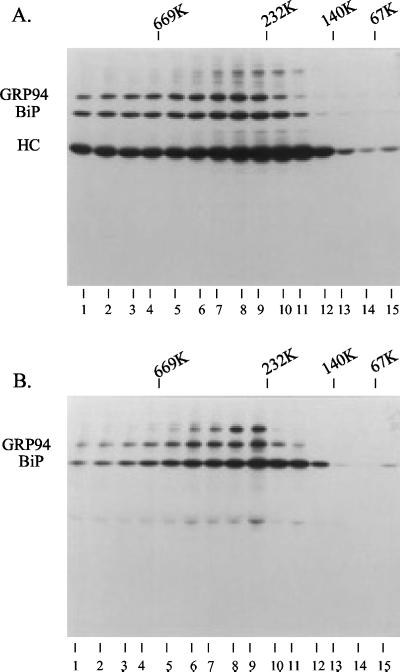
To determine the relative size of the ER chaperone complex, we resolved it by gradient density centrifugation. After fractionation, heavy chain complexes from Ag8(8) cells were precipitated with protein A-Sepharose, and BiP-containing complexes were immunoprecipitated from Ag8.653 cells and separated by reducing SDS-PAGE (Figure 10). The complexes seem to be somewhat heterogeneous ranging from ∼140 to >700 kDa. However, the major BiP-containing complex resolved at ∼232 kDa for the Ag8.653 (Figure 10B, lanes 8 and 9), whereas the heavy chain-containing complex(es) from the Ag8(8) cells fractionated at a slightly larger size (Figure 10A, lanes 7–11), which is in keeping with the chaperone complex binding to the heavy chains. Most proteins fractionate together and the size of the complex is not huge, which may be more compatible with discrete complexes as opposed to a very large and continuous matrix. However, the heterogeneity observed could be due to noncomplete cross-linking of a large network rather than more discrete complexes, making it difficult for us to draw definite conclusions at this point.
Figure 10.
Velocity gradient centrifugation to determine the size of the cross-linked complex. Ag8(8) and Ag8.653 cells (10 × 106) were metabolically labeled and the postnuclear fraction was prepared as described previously. Cross-linking was performed by including 150 μg/ml DSP in the HEPES buffer containing 0.25 M sucrose. Triton X-100 (1%) was added to the cross-linked sample and then the lysates and high-molecular-weight markers were centrifuged through 20–40% glycerol gradients. Each gradient was then separated into 15 fractions and the complex was precipitated with protein A-Sepharose beads for the Ag8(8) complexes (A) or immunoprecipitated with a polyclonal anti-BiP antibody for the Ag8.653 complexes (B).
DISCUSSION
Data obtained from a number of studies have demonstrated that multiple ER chaperones can associate with a given nascent protein. Sitia and coworkers demonstrated that unoxidized Ig light chains form disulfide bonds transiently with both PDI and ERp72 and suggested that these proteins may form a kind of affinity matrix in the ER that impedes the transport of unoxidized nascent proteins (Reddy et al., 1996). Similarly, both thyroglobulin (Kuznetsov et al., 1997) and HCGβ (human chorionic gonadotropin beta) (Feng et al., 1995) can be cross-linked to BiP, GRP94, and ERp72 during their maturation, and the influenza hemagglutinin protein binds to a number of ER proteins, including BiP, GRP94, calreticulin, and calnexin when cross-linking agents are added to the cells (Tatu and Helenius, 1997). However, it was not clear from these studies whether the chaperones were binding as a complex or whether the individual chaperones were binding to distinct unfolded regions on these proteins. Our data provide direct evidence that molecular chaperones exist as large complexes in the ER and provide new insights into the nature of this network. First, our data reveal that a large fraction of BiP, GRP94, and GRP170 exist as components of multichaperone complexes, even in the absence of unfolded substrates, which strongly suggests they are preformed instead of forming on unfolded proteins. Second, most of the known ER chaperones and folding enzymes are present in the ER chaperone complex. Third, the assembly of the complexes and their binding to heavy chains are very sensitive to detergent, but can be isolated by using a cross-linker or nondetergent-based methods for disrupting the ER (our unpublished data). Calnexin, calreticulin, ERp57, which functions as a cochaperone for calreticulin and calnexin (Oliver et al., 1999), and the glycosidases are conspicuously absent or very poorly associated with the complex. However, the inclusion of UDP-GT provides a link between these chaperones and the BiP–GRP94-based chaperone system. It is not clear whether the chaperones are components of a single or multiple complexes, but only one large complex can be isolated by both the 2D gel electrophoresis and the density centrifugation analysis (our unpublished data). Based on the composition of the ER chaperone complexes, we hypothesize that not only do they serve, by virtue of their size, to prevent incompletely folded or assembled proteins from continuing through the secretory pathway but also that they also act to concentrate folding enzymes and chaperones onto the unfolded protein. Although nascent secretory pathway proteins are translocated into a concentrated mixture of structural elements, molecular chaperones, folding enzymes, and other nascent unfolded proteins, in most cases they fold rapidly and efficiently making it almost implicit that such an organization of chaperones and folding enzymes should exist.
GRP94 is one of the most abundant ER resident proteins and is thought to be the cytosolic homologue of Hsp90 based on strong sequence homology. However, unlike Hsp90, which has been well studied and shown to be essential for the maturation of numerous proteins, including steroid receptors (Bresnick et al., 1989; Smith et al., 1990), kinases (Schulte et al., 1995), and p53 (Blagosklonny et al., 1996), the function of GRP94 remains somewhat of an enigma. This may be due, in part, to the detergent sensitivity of GRP94–chaperone protein complexes. Hsp90 complexes are also very sensitive to detergents (Smith et al., 1990), but cytosolic Hsp90 can be isolated from reticulocytes by hypotonic lysis, whereas most methods for disrupting ER membranes rely on detergents. Together with Hsp70, Hsp90 binds to unfolded cytosolic proteins and acts as a scaffold to recruit a number of additional chaperones, folding enzymes, and regulators of Hsp70 function, which form a series of dynamic complexes that cycle on and off unfolded proteins (Smith, 1993; Buchner, 1999). It is of interest to note that Hsp90 and Hsp70 also form these dynamic complexes in the absence of unfolded proteins (Buchner, 1999).
At this time, we have no evidence that GRP94 acts as the ER scaffold for assembling chaperones. Further experiments are needed to better characterize the role of GRP94 in this complex. However, there are a number of similarities between hsp90 and GRP94. First, like Hsp90, GRP94 is present as a major component of the chaperone complex containing BiP (an Hsp70). Second, its association with the various chaperones and heavy chains is extremely sensitive to detergent. Third, its assembly into chaperone complexes is not dependent on the presence of unfolded proteins. And fourth, the complexes isolated with GRP94 also contain regulators of BiP function like the ERdj3 (Bies et al., 1999; Yu et al., 2000). The ER DnaJ cochaperone is a homologue of the cytosolic Hsp40 protein that regulates the ATPase activity of Hsp70 and is present in the Hsp90/Hsp70 complex associated with the hormone receptor (Smith, 1993; Buchner, 1999). It is noticeable that much larger amounts of ERdj3 are present in the complex when BiP is associated with the Ig heavy chain, which is in agreement with its proposed function (i.e., the stimulation of BiP's ATPase activity; Yu et al., 2000). This is also consistent with the function of its cytosolic homologue Hsp40, which binds both Hsp70 and the unfolded substrate to control the ATPase cycle of Hsp70 and provides a good control for the specificity of the cross-linking procedure, because this protein is absent from the complex without substrates. Finally, our identification of cyclophilin B, an ER immunophilin protein, as part of the ER chaperone complex is in keeping with the presence of cytosolic immunophilins in the Hsp90–Hsp70 complex. In addition, several proteins that are not found in the cytosolic Hsp70–Hsp90 complex are present in the ER complex. The UDP-GT enzyme, which catalyzes the monoglucosylation reaction, GRP170, an ER Hsp70 family member whose function is not yet well characterized, and several members of the protein disulfide isomerase family (ERp72, PDI, and CaBP1) are also present, suggesting that if the ER complex is analogous to the cytosolic one, modifications have been made to fit the needs of protein folding in the ER. It is also the case of SDF2-L1, an ER stress-inducible protein, showing significant similarities to the central hydrophilic part of proteins O-mannosyltransferase (Fukuda et al., 2001). This large complex, with the exception of ERdj3, is not only detected in the presence of the heavy chain but also in absence of any substrates and suggests that the ER is organized as a network able to bind nascent proteins as soon as they translocate into the lumen. The existence of such a network(s) could also explain why some molecular chaperones are so efficiently retained inside the endoplasmic reticulum even when they do not possess a KDEL retention sequence (Sonnichsen et al., 1994; Monnat et al., 2000).
In a study of immunoglobulin light chain (LC) association with BiP and GRP94, Melnick and Argon concluded that BiP binds to an early intermediate of LC folding, whereas GRP94 associates preferentially with a more mature form of the protein and suggested that these two chaperones might act in tandem to fold the LC (Melnick et al., 1994). Our data demonstrating that both proteins bind to unfolded Ig heavy chains as a single complex are not consistent with a “hand-off” mechanism between these two chaperones for folding. It should be noted that in the Argon studies, it was not possible to use cross-linkers to stabilize GRP94 association with LC, because the oxidation status of the LC was being examined on nonreducing gels. Thus, weaker interactions of GRP94 with the LC might have been lost. Alternatively, the discrepancies between these two studies may reflect the difference between a protein that can fold (λLC) and one that does not (γ heavy chain). However, our data on the secreted heavy chain from the G403 cell line, suggest this is probably not the case. Finally, it is possible that unfolded substrates might bind first to one member of the chaperone complex and then “roll over” to the next chaperone it requires. Our data due not allow us to determine which proteins other than BiP have direct contact with the unfolded heavy chain.
It has been proposed that during the translocation of a given glycoprotein into the ER, a choice is made between chaperone systems (Molinari and Helenius, 2000); one comprised of BiP/GRP94 and one consisting of calnexin/calreticulin. However, transfer from one system to the other can clearly occur. The binding of vesicular stomatitis virus glycoprotein G first to BiP and then to calnexin (Hammond and Helenius, 1994) demonstrates a temporal organization to chaperone interactions. Our data suggest this could be accomplished via a spatial organization of the two chaperone systems. We propose that the ER is organized into different networks containing distinct compositions of chaperone proteins. As the secreted proteins mature, they are transported inside the ER from one network (i.e., the BiP/GRP94/other proteins in our complex) to the other (i.e., calnexin/calreticulin/and perhaps glucosidases). Retention of some malfolded or incompletely folded proteins in the first network would prevent them from being transported to another subregion of the ER that contains calnexin/calreticulin. This might explain why the glycosylated heavy chains examined herein do not readily interact with calnexin/calreticulin even though UDP-GT is part of the chaperone complex associated with them. Release of proteins from this complex would allow them to next interact with calnexin/calreticulin, because their modification by UDP-GT would provide them with the appropriate recognition structures. It is also very possible that UDP-GT pools exist outside the BiP–GRP94 complex to allow continual interactions of some substrates with calnexin/calreticulin. Further support for this type of suborganellar organization to the ER comes from a recent study. By using fluorescence microscopy, the precursor of human asialoglycoprotein receptor, H2a, and the free heavy chains of major histocompatibility complex class I molecules were shown to accumulate in a compartment containing calnexin and calreticulin, but not BiP, PDI, or UDP-GT, when proteosomal degradation was inhibited (Kamhi-Nesher et al., 2001). Thus, not only does their study demonstrate physically distinct subregions of the ER but also the subdivision of the two chaperone systems observed by Kamhi-Nesher et al. (2001) is completely consistent with the results we have reported herein.
In summary, we present data that support the existence of a previously unrecognized physical organization of chaperones inside the ER. The majority of the chaperones and folding enzymes found in this organelle are assembled into an ER network or complex. Calnexin and calreticulin are conspicuously absent from this complex. These preformed chaperone complexes can associate both transiently with proteins that are folding and more stably with unfolded proteins that will ultimately be degraded.
ACKNOWLEDGMENTS
We thank Melissa Doyle for very helpful technical assistance. We thank Ashutosh Mishra and Clive Slaughter (Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Hospital) for protein identification. This work was supported by National Institutes of Health grant GM-54068, the Cancer Center CORE grant CA-21765, and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–05–0311. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–05–0311.
REFERENCES
- Balow JP, Weissman JD, Kearse KP. Unique expression of major histocompatibility complex class I proteins in the absence of glucose trimming and calnexin association. J Biol Chem. 1995;270:29025–29029. doi: 10.1074/jbc.270.48.29025. [DOI] [PubMed] [Google Scholar]
- Bies C, Guth S, Janoschek K, Nastainczyk W, Volkmer J, Zimmermann R. A Scj1p homolog and folding catalysts present in dog pancreas microsomes. Biol Chem. 1999;380:1175–1182. doi: 10.1515/BC.1999.149. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV, Toretsky J, Bohen S, Neckers L. Mutant conformation of p53 translated in vitro or in vivo requires functional HSP90. Proc Natl Acad Sci USA. 1996;93:8379–8383. doi: 10.1073/pnas.93.16.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- Bole DG, Hendershot LM, Kearney JF. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J Cell Biol. 1986;102:1558–1566. doi: 10.1083/jcb.102.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Mucke M, Freedman RB. The characterization of a cyclophilin-type peptidyl prolyl cis-trans-isomerase from the endoplasmic-reticulum lumen. Biochem J. 1994;300:871–875. doi: 10.1042/bj3000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick EH, Dalman FC, Sanchez ER, Pratt WB. Evidence that the 90-kDa heat shock protein is necessary for the steroid binding conformation of the L cell glucocorticoid receptor. J Biol Chem. 1989;264:4992–4997. [PubMed] [Google Scholar]
- Buchberger A, Theyssen H, Schroder H, McCarty JS, Virgallita G, Milkereit P, Reinstein J, Bukau B. Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J Biol Chem. 1995;270:16903–16910. doi: 10.1074/jbc.270.28.16903. [DOI] [PubMed] [Google Scholar]
- Buchner J. Hsp90 & Co. - a holding for folding. Trends Biochem Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Bulleid NJ, Freedman RB. Defective co-translational formation of disulphide bonds in protein disulphide-isomerase-deficient microsomes. Nature. 1988;335:649–651. doi: 10.1038/335649a0. [DOI] [PubMed] [Google Scholar]
- Bush KT, Hendrickson BA, Nigam SK. Induction of the FK506-binding protein, FKBP13, under conditions which misfold proteins in the endoplasmic reticulum. Biochem J. 1994;303:705–708. doi: 10.1042/bj3030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavany C, Mimnaugh E, Miller P, Bitton R, Nguyen P, Trepel J, Whitesell L, Schnur R, Moyer J, Neckers L. p185erbB2 binds to GRP94 in vivo. Dissociation of the p185erbB2/GRP94 heterocomplex by benzoquinone ansamycins precedes depletion of p185erbB2. J Biol Chem. 1996;271:4974–4977. doi: 10.1074/jbc.271.9.4974. [DOI] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clairmont CA, De Maio A, Hirschberg CB. Translocation of ATP into the lumen of rough endoplasmic reticulum-derived vesicles and its binding to luminal proteins including BiP (GRP 78) and GRP 94. J Biol Chem. 1992;267:3983–3990. [PubMed] [Google Scholar]
- Feng W, Matzuk MM, Mountjoy K, Bedows E, Ruddon RW, Boime I. The asparagine-linked oligosaccharides of the human chorionic gonadotropin beta subunit facilitate correct disulfide bond pairing. J Biol Chem. 1995;270:11851–11859. doi: 10.1074/jbc.270.20.11851. [DOI] [PubMed] [Google Scholar]
- Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Frydman J, Hohfeld J. Chaperones get in touch: the Hip-Hop connection. Trends Biochem Sci. 1997;22:87–92. doi: 10.1016/s0968-0004(97)01005-0. [DOI] [PubMed] [Google Scholar]
- Fukuda S, et al. Murine and human SDF2L1 is an endoplasmic reticulum stress-inducible gene and encodes a new member of the Pmt/rt protein family. Biochem Biophys Res Commun. 2001;280:407–414. doi: 10.1006/bbrc.2000.4111. [DOI] [PubMed] [Google Scholar]
- Gamer J, Multhaup G, Tomoyasu T, McCarty JS, Rudiger S, Schonfeld HJ, Schirra C, Bujard H, Bukau B. A cycle of binding and release of the DnaK, DnaJ and GrpE chaperones regulates activity of the Escherichia coli heat shock transcription factor sigma32. EMBO J. 1996;15:607–617. [PMC free article] [PubMed] [Google Scholar]
- Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- Hammond C, Braakman I, Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C, Helenius A. Quality control in the secretory pathway: retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment, and Golgi apparatus. J Cell Biol. 1994;126:41–52. doi: 10.1083/jcb.126.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- Hellman R, Vanhove M, Lejeune A, Stevens FJ, Hendershot LM. The in vivo association of BiP with newly synthesized proteins is dependent on the rate and stability of folding and not simply on the presence of sequences that can bind to BiP. J Cell Biol. 1999;144:21–30. doi: 10.1083/jcb.144.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot L, Bole D, Kohler G, Kearney JF. Assembly and secretion of heavy chains that do not associate posttranslationally with immunoglobulin heavy chain-binding protein. J Cell Biol. 1987;104:761–767. doi: 10.1083/jcb.104.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot L, Wei J, Gaut J, Melnick J, Aviel S, Argon Y. Inhibition of immunoglobulin folding and secretion by dominant negative BiP ATPase mutants. Proc Natl Acad Sci USA. 1996;93:5269–5274. doi: 10.1073/pnas.93.11.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot LM. Immunoglobulin heavy chain and binding protein complexes are dissociated in vivo by light chain addition. J Cell Biol. 1990;111:829–837. doi: 10.1083/jcb.111.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot LM, Wei JY, Gaut JR, Lawson B, Freiden PJ, Murti KG. In vivo expression of mammalian BiP ATPase mutants causes disruption of the endoplasmic reticulum. Mol Biol Cell. 1995;6:283–296. doi: 10.1091/mbc.6.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstenbach F, David V, Watkins S, Brenner MB. Endoplasmic reticulum resident protein of 90 kilodaltons associates with the T- and B-cell antigen receptors and major histocompatibility complex antigens during their assembly. Proc Natl Acad Sci USA. 1992;89:4734–4738. doi: 10.1073/pnas.89.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohfeld J, Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaloff CR, Haas IG. Coordination of immunoglobulin chain folding and immunoglobulin chain assembly is essential for the formation of functional IgG. Immunity. 1995;2:629–637. doi: 10.1016/1074-7613(95)90007-1. [DOI] [PubMed] [Google Scholar]
- Kamhi-Nesher S, Shenkman M, Tolchinsky S, Fromm SV, Ehrlich R, Lederkremer GZ. A novel quality control compartment derived from the endoplasmic reticulum. Mol Biol Cell. 2001;12:1711–1723. doi: 10.1091/mbc.12.6.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassenbrock CK, Kelly RB. Interaction of heavy chain binding protein (BiP/GRP78) with adenine nucleotides. EMBO J. 1989;8:1461–1467. doi: 10.1002/j.1460-2075.1989.tb03529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney JF, Radbruch A, Liesegang B, Rajewsky K. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J Immunol. 1979;123:1548–1550. [PubMed] [Google Scholar]
- Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Kuznetsov G, Chen LB, Nigam SK. Several endoplasmic reticulum stress proteins, including ERp72, interact with thyroglobulin during its maturation. J Biol Chem. 1994;269:22990–22995. [PubMed] [Google Scholar]
- Kuznetsov G, Chen LB, Nigam SK. Multiple molecular chaperones complex with misfolded large oligomeric glycoproteins in the endoplasmic reticulum. J Biol Chem. 1997;272:3057–3063. doi: 10.1074/jbc.272.5.3057. [DOI] [PubMed] [Google Scholar]
- Lassoued K, Illges H, Benlagha K, Cooper MD. Fate of surrogate light chains in B lineage cells. J Exp Med. 1996;183:421–429. doi: 10.1084/jem.183.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson B, Brewer JW, Hendershot LM. Geldanamycin, an hsp90/GRP94-binding drug, induces increased transcription of endoplasmic reticulum (ER) chaperones via the ER stress pathway. J Cell Physiol. 1998;174:170–178. doi: 10.1002/(SICI)1097-4652(199802)174:2<170::AID-JCP4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Lee YK, Brewer JW, Hellman R, Hendershot LM. BiP and immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol Biol Cell. 1999;10:2209–2219. doi: 10.1091/mbc.10.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HY, Masso-Welch P, Di YP, Cai JW, Shen JW, Subjeck JR. The 170-kDa glucose-regulated stress protein is an endoplasmic reticulum protein that binds immunoglobulin. Mol Biol Cell. 1993;4:1109–1119. doi: 10.1091/mbc.4.11.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzarella RA, Srinivasan M, Haugejorden SM, Green M. ERp72, an abundant luminal endoplasmic reticulum protein, contains three copies of the active site sequences of protein disulfide isomerase. J Biol Chem. 1990;265:1094–1101. [PubMed] [Google Scholar]
- Melnick J, Aviel S, Argon Y. The endoplasmic reticulum stress protein GRP94, in addition to BiP, associates with unassembled immunoglobulin chains. J Biol Chem. 1992;267:21303–21306. [PubMed] [Google Scholar]
- Melnick J, Dul JL, Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994;370:373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- Mkrtchian S, Baryshev M, Matvijenko O, Sharipo A, Sandalova T, Schneider G, Ingelman-Sundberg M, Mkrtchiana S. Oligomerization properties of ERp29, an endoplasmic reticulum stress protein. FEBS Lett. 1998;431:322–326. doi: 10.1016/s0014-5793(98)00786-8. [DOI] [PubMed] [Google Scholar]
- Molinari M, Helenius A. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 2000;288:331–333. doi: 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- Monnat J, Neuhaus EM, Pop MS, Ferrari DM, Kramer B, Soldati T. Identification of a novel saturable endoplasmic reticulum localization mechanism mediated by the C-terminus of a Dictyostelium protein disulfide isomerase. Mol Biol Cell. 2000;11:3469–3484. doi: 10.1091/mbc.11.10.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Pelham HR. An Hsp70-like protein in the ER: identity with the 78 kD glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- Oi VT, Morrison SL, Herzenberg LA, Berg P. Immunoglobulin gene expression in transformed lymphoid cells. Proc Natl Acad Sci USA. 1983;80:825–829. doi: 10.1073/pnas.80.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JD, Roderick HL, Llewellyn DH, High S. ERp57 functions as a subunit of specific complexes formed with the ER lectins calreticulin and calnexin. Mol Biol Cell. 1999;10:2573–2582. doi: 10.1091/mbc.10.8.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleros DR, Reid KL, Shi L, Welch WJ, Fink AL. ATP-induced protein-Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature. 1993;365:664–666. doi: 10.1038/365664a0. [DOI] [PubMed] [Google Scholar]
- Reddy P, Sparvoli A, Fagioli C, Fassina G, Sitia R. Formation of reversible disulfide bonds with the protein matrix of the endoplasmic reticulum correlates with the retention of unassembled Ig light chains. EMBO J. 1996;15:2077–2085. [PMC free article] [PubMed] [Google Scholar]
- Roth RA, Pierce SB. In vivo cross-linking of protein disulfide isomerase to immunoglobulins. Biochemistry. 1987;26:4179–4182. doi: 10.1021/bi00388a001. [DOI] [PubMed] [Google Scholar]
- Schulte TW, Blagosklonny MV, Ingui C, Neckers L. Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J Biol Chem. 1995;270:24585–24588. doi: 10.1074/jbc.270.41.24585. [DOI] [PubMed] [Google Scholar]
- Smith DF. Dynamics of heat shock protein 90-progesterone receptor binding and the disactivation loop model for steroid receptor complexes. Mol Endocrinol. 1993;7:1418–1429. doi: 10.1210/mend.7.11.7906860. [DOI] [PubMed] [Google Scholar]
- Smith DF, Schowalter DB, Kost SL, Toft DO. Reconstitution of progesterone receptor with heat shock proteins. Mol Endocrinol. 1990;4:1704–1711. doi: 10.1210/mend-4-11-1704. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, Fullekrug J, Nguyen Van P, Diekmann W, Robinson DG, Mieskes G. Retention and retrieval: both mechanisms cooperate to maintain calreticulin in the endoplasmic reticulum. J Cell Sci. 1994;107:2705–2717. doi: 10.1242/jcs.107.10.2705. [DOI] [PubMed] [Google Scholar]
- Sousa MC, Ferrero-Garcia MA, Parodi AJ. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 1992;31:97–105. doi: 10.1021/bi00116a015. [DOI] [PubMed] [Google Scholar]
- Tatu U, Helenius A. Interactions between newly synthesized glycoproteins, calnexin and a network of resident chaperones in the endoplasmic reticulum. J Cell Biol. 1997;136:555–565. doi: 10.1083/jcb.136.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta SE, Parodi AJ. Purification to apparent homogeneity and partial characterization of rat liver UDP-glucose:glycoprotein glucosyltransferase. J Biol Chem. 1992;267:9236–9240. [PubMed] [Google Scholar]
- Vanhove M, Usherwood YK, Hendershot LM. Unassembled Ig heavy chains do not cycle from BiP in vivo but require light chains to trigger their release. Immunity. 2001;15:105–114. doi: 10.1016/s1074-7613(01)00163-7. [DOI] [PubMed] [Google Scholar]
- Wei J, Hendershot LM. Characterization of the nucleotide binding properties and ATPase activity of recombinant hamster BiP purified from bacteria. J Biol Chem. 1995;270:26670–26676. doi: 10.1074/jbc.270.44.26670. [DOI] [PubMed] [Google Scholar]
- Yu M, Haslam RH, Haslam DB. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J Biol Chem. 2000;275:24984–24992. doi: 10.1074/jbc.M000739200. [DOI] [PubMed] [Google Scholar]
- Zhang JX, Braakman I, Matlack KE, Helenius A. Quality control in the secretory pathway: the role of calreticulin, calnexin and BiP in the retention of glycoproteins with C-terminal truncations. Mol Biol Cell. 1997;8:1943–1954. doi: 10.1091/mbc.8.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]