Abstract
Background
Chlamydia trachomatis (Ct) and Chlamydia pneumoniae (Cp) are medically significant infectious agents associated with various chronic human pathologies. Nevertheless, specific roles in disease progression or initiation are incompletely defined. Both pathogens infect established cell lines in vitro and polymerase chain reaction (PCR) has detected Chlamydia DNA in various clinical specimens as well as in normal donor peripheral blood monocytes (PBMC). However, Chlamydia infection of other blood cell types, quantification of Chlamydia infected cells in peripheral blood and transmission of this infection in vitro have not been examined.
Methods
Cp specific titers were assessed for sera from 459 normal human donor blood (NBD) samples. Isolated white blood cells (WBC) were assayed by in vitro culture to evaluate infection transmission of blood cell borne chlamydiae. Smears of fresh blood samples (FB) were dual immunostained for microscopic identification of Chlamydia-infected cell types and aliquots also assessed using Flow Cytometry (FC).
Results
ELISA demonstrated that 219 (47.7%) of the NBD samples exhibit elevated anti-Cp antibody titers. Imunofluorescence microscopy of smears demonstrated 113 (24.6%) of samples contained intracellular Chlamydia and monoclonals to specific CD markers showed that in vivo infection of neutrophil and eosinophil/basophil cells as well as monocytes occurs. In vitro culture established WBCs of 114 (24.8%) of the NBD samples harbored infectious chlamydiae, clinically a potentially source of transmission, FC demonstrated both Chlamydia infected and uninfected cells can be readily identified and quantified.
Conclusion
NBD can harbor infected neutrophils, eosinophil/basophils and monocytes. The chlamydiae are infectious in vitro, and both total, and cell type specific Chlamydia carriage is quantifiable by FC.
Background
Chlamydiae, obligate intracellular bacterial pathogens, cause an array of medically and economically important infectious diseases. Chlamydia trachomatis (Ct), the most common cause of sexually transmitted bacterial disease, is also the world's leading cause of infectious blindness [1]. Chlamydophila pneumoniae (Cp) is a ubiquitous respiratory pathogen responsible for sinusitis, bronchitis, and 10–15% of community acquired pneumonia cases worldwide[2]. By age 20, ~50% of the population exhibits evidence of past infection by C. pneumoniae and re-infection is common throughout life [3]. Cp has attracted increasing interest because it is associated with an array of chronic human diseases that are not restricted to mucosal surfaces. C. pneumoniae has been implicated in the pathobiology of atherosclerosis [4-7] multiple sclerosis [8,9], Alzheimer's disease (AD) [10], reactive arthritis [11], and asthma [12].
Although C. pneumoniae has been implicated as a factor in this diverse array of chronic human diseases, it remains unknown whether it is the causative agent or is simply important in exacerbating these pathologies. By PCR, evidence of these organisms has been found in the peripheral blood mononuclear cells (PBMCs) of healthy blood donors, and in patients with coronary artery disease [13-16]. However, evidence of organism infectivity and information outlining the intra-host spread of either organism from an initial infection site in lung or genital tract to widely disseminated sites where pathology occurs remains to be fully defined. Using a rabbit model, and cell culture, studies have shown that alveolar macrophages serve as host cells for Cp [17], transporting it through the mucosal barrier to the lymphatic system, then beyond into the systemic circulation [18]. Chlamydia infected macrophages have been found in atherosclerotic plaques [19], and recent studies have revealed that in vitro Cp can infect human neutrophil granulocytes, and then initiate delays in their spontaneous apoptosis [20]. Evidence has been presented that in vitro Cp can infect B and T lymphocyte cell lines as well as human peripheral blood mononuclear cells and induce a cytokine response [21,22]. As demonstrated by PCR, Ct also disseminates within the host following experimental genital tract infection in a murine model, but the mechanisms involved have not yet been fully clarified [23-29].
We initiated an exploration of viable, infectious Chlamydia carriage in human blood cells. The current study was directed at assessing the prevalence of Chlamydia in samples from a Normal Blood Donor (NBD) population as well as identifying the different types of white blood cells that harbor this pathogen in vivo and quantifying their numbers by flow cytometry. Results from our study examining a random cohort of 459 normal donor samples indicated that (i) the average blood borne carriage rate of Chlamydia for this cohort is 24.6%, (ii) Chlamydia can be present in NBD peripheral blood granulocytic neutrophils and eosinophil/basophil cells, as well as monocytes, (iii) chlamydiae in 24.8% of NBD peripheral WBC are infectious as demonstrated by in vitro culture and (iv) FC can quantify both total infected cell load (ICL) and infected cell type specific load (ICSL) in the peripheral WBC population.
Methods
Sample collection
Approval for using residual material from routinely collected peripheral blood specimens from healthy normal blood donors was obtained from the Institutional Review Board at Baystate Medical Center. De-identified NBD residuals collected in ethylenediaminetetraacetic acid (EDTA) tubes were obtained from the Blood Bank at Baystate Medical Center in Springfield, MA from November 2001 to October 2002. Samples were collected monthly and stored at 4°C until tested. For other assessments, de-identified fresh blood sample residuals (FB), collected in EDTA tubes, were obtained on a weekly basis from patients seen at the University Health Services (UHS) and used immediately. Unless otherwise specified, conjugated secondary antibody reagents were obtained from Jackson ImmunoResearch Laboratories, West Grove PA; the standard incubation was 1 h at room temperature (RT) unless otherwise specified.
C. pneumoniae specific peptide ELISA
A 14 amino acid C. pneumoniae specific peptide sequence deriving from the gene encoding the major outer membrane porin protein of C. pneumoniae AR 39 [30] was synthesized (Sigma Genosys, The Woodlands, TX). It served as the antigen in ELISA tests for C. pneumoniae specific antibody in NBD plasma samples. ELISA wells (NUNC™, Nunc A/S, Roskilde, Denmark) were coated for 1 h with 100 ul containing 1 μg/ml peptide in phosphate buffered saline pH 7.2 (PBS). Plasma samples were serially diluted in ELISA wells using 0.1% BSA in PBS and standard protocols for blocking (3% bovine serum albumin: BSA, Sigma, St. Louis, MO), incubations, and washes (X3 with 0.1%BSA/PBS). Detection of bound human antibody used alkaline phosphatase (AP)-conjugated goat anti-human IgG and after 45 min incubation and multiple rinses, 100 μl/well of P-Nitrophenyl Phosphate substrate (pNPP, Sigma, St. Louis, MO) was added. Absorbances at 405 nm were measured with a VERSAmax Microplate reader (Molecular Devices Sunny Vale, CA). Titers were determined by comparison of absorbance readings of known positive and negative controls with those of the test samples.
Immunostain of normal blood donor smears
Smears prepared with ~20 μl/ slide of whole blood collected in EDTA were air dried then fixed 10 min in 70% cold methanol (MEOH). Following PBS rinses, slides were incubated in a moist chamber with a rabbit or guinea pig anti-Chlamydia EB serum (Biomeda Corp. Foster City, CA) then multiply rinsed in PBS. Bound anti-Chlamydia antibody was detected with fluoresein isothiocynate (FITC)-conjugated goat anti-rabbit or donkey anti-guinea pig IgG. Primary and secondary antibody incubations used the standard protocol noted above. After PBS rinsing X3, coverslips were mounted using Fluoromount G (BioWhittaker, Walkersville, MD), sealed, then examined and photographed using a Nikon Eclipse E600 epifluorescence microscope and a SPOT digital camera (Digital Instruments Inc.) or a Zeiss LSM 510 Meta Confocal System
Culture of normal blood donor samples
The buffy coat (BC) containing WBC, was removed from each 5 ml NBD sample, rinsed X4 with sterile PBS, treated to release EBs and cultured on J774A.1 monolayers as described previously [30]. At 96 h post infection (pi) monolayers on coverslips were rinsed with PBS, fixed with 70% MEOH, rinsed again and immunostained with a rabbit anti-Chlamydia EB, and then mounted as noted above.
Detection of Chlamydia infected Monocyte, Eosinophil/Basophil and Neutrophil
Smears of BC cell pellets from each 5 ml fresh blood samples (FB), average donor age 28 years, were fixed with 70% MEOH. Except as specified, immunostaining used rabbit polyclonal anti-Chlamydia EB antibody followed by a TRITC conjugated goat anti-rabbit antibody. Specific cell type detection used mouse monoclonals: anti-Human CD14 (0.2 μg/106cells) (Sigma, Saint Louis, MO), or anti-Human CD16b (1.0 μg/106cells) (BD Biosciences Pharmigen, San Diego, CA.). Binding was detected with FITC-conjugated goat anti-mouse IgG. For samples incubated with PE-conjugated anti-Human CDw125 (1.0 μg/106cells), bound anti-Chlamydia antibody detection used FITC conjugated anti-rabbit antibody. Slides were incubated, rinsed, mounted and sealed as described above and digital images of optical sections through these samples were acquired with a Zeiss LSM 510 Meta Confocal System.
Flow Cytometry assessment of a WBC dilution series
Flow cytometry (FC) quantification of Chlamydia-infected peripheral blood WBC used PBS rinsed BC cells from smear positive NBD samples. The WBC pool was fixed and permeabilized (1% paraformaldehyde and 1% Triton X-100, 10 min. at RT; Aldrich Chemical Company, Inc., Milwaukee, WI). PBS rinsed RBCs from the same samples were also pooled. BC and RBC were separately incubated with pre-diluted guinea pig anti-Chlamydia primary antibody followed by incubation with PE-conjugated F(ab')2 donkey anti-guinea pig IgG, PBS rinsed X3, and mono-dispersed using a nylon mesh filter (Lab-Line Instruments Inc, Melrose Park, IL). A WBC dilution series was prepared in separate tubes as follows: 100%, 90%, 80%, 70%, 60%, 50%, 40%, 30%, 20%, 10%, 5%, 2%, 1%, 0.5%, and 0% WBC/tube. Sufficient RBCs were added to obtain a total of 50,000 cells/tube and the series analyzed by FACScan (Becton, Dickinson and Company, Franklin Lakes, NJ).
Flow Cytometry of FB WBCs dual stained for Monocytes, Eosinophil/Basophil and Neutrophils
BC isolated from FB samples was pre-screened for Chlamydia infected cells by anti-Chlamydia immunostaining as described above. BC from four smear positive samples and separately, BC from four smear negative samples were pooled. Each pool was washed, fixed and permeabilized as described above. After counting, aliquots of the cell pools were incubated with the polyclonal guinea pig anti-Chlamydia primary antibody and one of the three monoclonal antibodies, at the concentrations noted above. To detect Chlamydia and CD14 or CD16b, BC were incubated with PE-conjugated F(ab')2 donkey anti-guinea pig IgG and FITC-conjugated rabbit anti-mouse IgG. Chlamydia in BC immunostained with PE-conjugated anti-CDw125, were detected with FITC conjugated F(ab')2 donkey anti-guinea pig IgG. After standard incubations and rinsing, samples were mono-dispersed, counted, diluted to obtain 1 × 106 cells/ml of sample and analyzed using the FACScan instrument.
Statistics
NBD sample data were analyzed using the SPSS® 11.5 graduate pack statistics program [SPSS, Inc., Chicago, IL]. Comparisons of culture and smear positivity used Cross-tabs with the Fisher exact test to uncover nonrandom associations between these two categorical variables and Chi-Squared test and Kappa measure of agreement to determine significance. To verify the reproducibility of culture and smear assessments all samples were initially tested in duplicate. Subsequently, approximately 20% of the samples were re-tested by each method. The SD and CV were calculated based on the differences in the positive and negative percentages determined by the original vs. the repeat tests. All tests were two sided, and the level of significance was p ≤ 0.05.
Results
Preliminary observations of cells in blood smears of normal donor sample residuals and our studies of pediatric blood sample residuals [30] showed each could contain cells that were infected with Chlamydia. To screen normal donors for Chlamydia carriage in circulating PBMCs, and to assess infectivity, we utilized ELISA, immunofluorescence microscopy, in vitro culture and flow cytometry
Presence of anti-C. pneumoniae antibody in NBD Samples
A random cohort of blood donor sample residuals was obtained from the Blood Bank unit at Baystate Medical Center over a 12 month period. Donor demographics are presented in Table 1. As shown, the average age of this cohort was 44 years and consisted of 55% male and 45% female donors. Plasma was removed from the samples, and an aliquot assayed by ELISA for anti-C. pneumoniae antibody. As Table 2 shows, 219 (47.7%) of 459 samples tested exhibited a positive titer against the 14 amino acid C. pneumoniae-specific peptide. This level of sero-positive status is fully consistent with published reports for this age group and indicates this cohort is not anomalous in this respect.
Table 1.
Demographics: Random Blood Donor Cohort
| Average Donor Age | 44 years | ||
| Male | 252 (55%) | ||
| Female | 207 (45%) | ||
| Blood Type Distribution in Donor Cohort | |||
| A | 40% | B | 10% |
| AB | 4% | O | 46% |
Table 2.
Chlamydia Related Statistics: Normal Blood Donor Cohort
| Assessment | Positive | Negative | Total |
| Cp Serologic Status | 219 (47.7%) | 240(52.3%) | 459 |
| Smear Status | 113 (24.6%) | 346(75.4%) | 459 |
| Culture Status | 114 (24.8%) | 345(75.2%) | 459 |
| Culture and Smear Positive | 76 (67.3% of total Smear Positive) | N/A | |
| Culture and Smear Negative | 308 (89% of total Smear Negative) | N/A | |
| Culture Pos/Smear Negative | 38 (33.3% of total Culture positive) | N/A | |
| Culture Neg/Smear Positive | 37 (32.7%-of total Smear positive) | N/A | |
Microscopic demonstration of peripheral blood cells infected by Chlamydia
Samples, including blood, are routinely assessed for Chlamydia by PCR and RT-PCR [5,13-15] but inherently PCR cannot demonstrate whether this DNA represents fragments or viable, infectious organisms [13-16] and immunostaining to detect infected PBMCs is not routinely used. This study therefore included immunofluorescent staining as described above to identify Chlamydia-infected NBD cells. As Figure 1A-C shows, Chlamydia positive material can be readily detected within lymphocyte-like and monocyte-like cells and examination of all 459 samples demonstrated 113 (24.6%) were positive for Chlamydia within peripheral WBCs (Table 2). The blood residuals, normally ≥30/month, were obtained over a 12 month period (Table 3) and comparison by month indicated that samples collected in December exhibited distinctly higher carriage of intracellular Chlamydia: 42% smear positive and 33% culture positive. In contrast, for August and November carriage was notably lower. Culture and smear positive results for these two months were 15%, 15% and 17%, 12% respectively. These divergent percentages may reflect a variable carriage rate in this cohort during the year since the sample numbers tested for each of these months were similar: 34, 35 and 39, respectively.
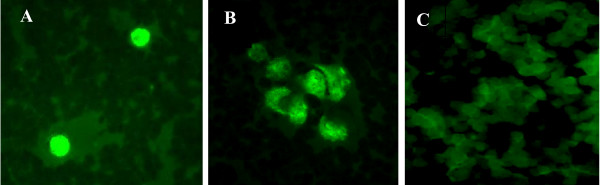
Figure 1.
Immunostained smears of NBD whole blood samples. Smears were prepared by standard methods, immunostained using an anti-Chlamydia EB antibody and an FITC-conjugated secondary antibody. Epifluorescence images in Panels A and B show representative examples of Chlamydia positive smears exhibiting characteristic inclusion bodies. Panel C shows a representative smear negative sample and does not contain inclusion bodies or other Chlamydia positive material. Samples were photographed using a Nikon Eclipse E600 epifluorescence microscope and a SPOT digital camera. Original magnification 400×
Table 3.
Sample Distribution: Normal Donor Cohort Positive by Month
| Month 2001–2002 | Sero-Positive | Smear Positive | Culture Positive |
| November | 50 | 17 | 12 |
| December | 97 | 42 | 43 |
| February | 63 | 22 | 28 |
| March | 24 | 20 | 22 |
| April | 55 | 30 | 30 |
| May | 37 | 31 | 24 |
| June | 40 | 27 | 24 |
| July | 70 | 24 | 30 |
| August | 10 | 15 | 15 |
| September | 33 | 24 | 27 |
| October | 70 | 20 | 25 |
Demonstration in vitro of viable, infectious Chlamydia in WBC of peripheral blood samples
Immunostain for Chlamydia antigens demonstrated their presence within PBMC. Therefore we next assessed whether infectious organisms per se are present. Chlamydiae are known to infect macrophages in vivo [17-19] and in vitro the J774A.1 macrophage monolayers are readily infected by Chlamydia and support growth for ~120 h pi. Using an effective culture method we developed previously and have described [30-33], WBC lysates were prepared and tested in duplicate on J774A.1 monolayers. At 96 h pi the monolayers were rinsed, fixed, and immunostained to detect Chlamydia. The results demonstrated that of the 459 samples, 114 (24.8%) were positive for growth of Chlamydia (Table 2). Figure 2 shows representative immunostained monolayers incubated with WBC derived inoculum from Chlamydia smear positive (Figure 2A) or negative samples (Figure 2B). At 96 h pi the cultures of smear positive samples contain large, late-stage inclusions as well as small, newly forming inclusions and released extracellular EBs. Table 2 shows that of the 459 samples tested 76 were positive by both smear and culture tests. Of the total positive samples, 37 were positive by smear but negative for cultivable organisms, potentially an indicator of persistence. Conversely 38 were negative by smear and positive by culture; this last finding may reflect the small sample size (20 μl) used in smear preparations. Together however the smear and in vitro culture findings demonstrated that not only do NBD samples carry PBMC infected by Chlamydia and visually identifiable by fluorescent microscopy, but frequently these WBC borne chlamydiae are infectious.
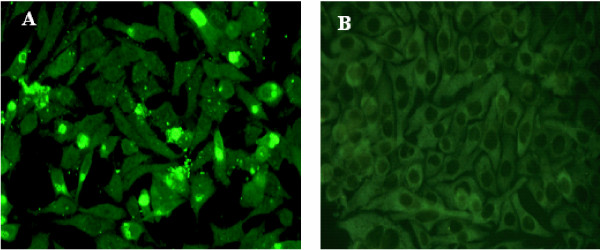
Figure 2.
Chlamydiae within Peripheral Blood Transfer Infection when Cultured in vitro on J774A.1 host cell monolayers. BC from NBD peripheral blood samples were isolated, lysed and layered onto host cell monolayers as described. Epifluorescence images in Panel A show a representative example of a 96 h pi culture of a BC lysate from a Chlamydia smear positive sample and demonstrate these monolayers have remained healthy. Panel B shows a representative example of 96 h pi culture of a BC lysate from a Chlamydia negative smear sample. Note: panel A contains numerous cells with large inclusions, as well as cells with small inclusions and extracellular chlamydiae. These are characteristic of multiple replicative rounds in culture. Photographed using a Nikon Eclipse E600 epifluorescence microscope and a SPOT digital camera Original magnification 400×
Identification of Chlamydia infected Monocytes, Eosinophils/Basophils and Neutrophils in smears
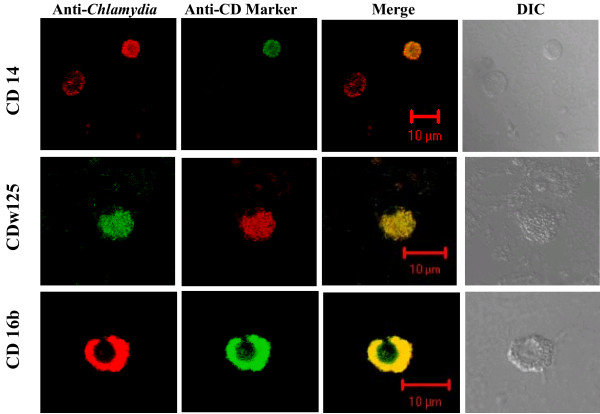
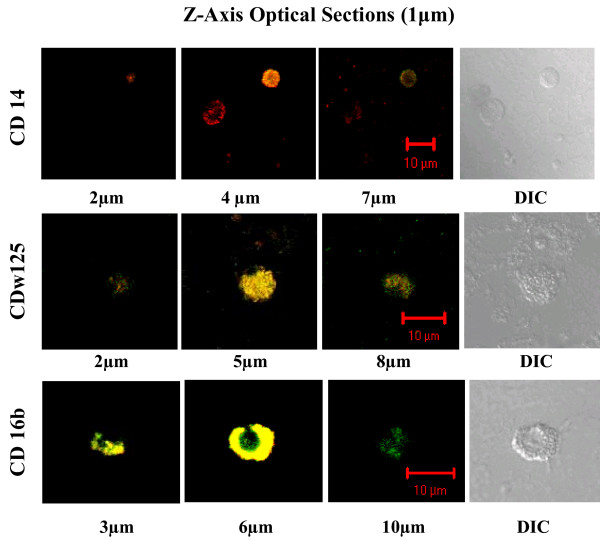
Despite the demonstration of Chlamydia DNA in PBMC samples, and demonstrations that in vitro numerous cell lines can be infected by different Chlamydia species and serovars, the specific types of WBC infected in vivo have not been fully examined. We selected three specific CD markers to identify WBC types and used them in conjunction with anti-Chlamydia antibody to immunostain WBCs in peripheral blood sample residuals. Since WBC inherently exhibit different life spans and can exhibit differential fragility, these assessments utilized fresh blood sample residuals. Smears of these FB samples were prepared, fixed and dual immunostained to detect Chlamydia and either CD14 (monocytes), CDw125 (eosinophils/basophils) or CD16b (neutrophil). Using Meta Confocal microscopy, optical sections through cells were examined and digital images acquired. Figure 3 shows representative examples of Chlamydia-infected monocyte (CD14), eosinophil/basophil (CDw125) and neutrophil (CD16b) cells. These can exhibit punctate fluorescent staining (row CD14 and CD125w) that reflects the expected uneven distribution of antigens. This is especially evident in non adherent cells and readily detectible in an optical section through a chlamydial inclusion. The anti-Chlamydia column shows the presence and distribution of Chlamydia positive material within the individual optical section, while the CD marker column identifies the cell type specific marker. The yellow or orange/yellow merge demonstrates the co-localization of the immunoprobes in that same optical section. The final column displays the DIC images and shows the position and general morphology of the individual cells. A comparison of the DIC and Merge images of the same row shows additional cells can be present that are not immunostained by either antibody (Figure 3-CD14 and CDw125). This observation serves to indicate the specificity of the immunoprobes. Figure 4 presents merged images obtained at three different levels from the same Z-axis optical sections through the cells shown in Figure 3. As the optical sections in the CD16b series show, the CD16b protein co-associates with the chlamydial antigens (6 μ) but also is present at the cell surface (10 μ).
Figure 3.
Infected and uninfected WBCs dual immunostained with CD Specific monoclonal and anti-Chlamydia antibody. Representative medial confocal microscopic images from the Z-axis series of optical sections through Chlamydia-infected human PBMC cells. Immunostaining used anti-Chlamydia EB antibody and either anti-CD14(monocyte), anti-CD 16b(neutrophil), or anti-CDw125 (eosinophil/basophil) mAb as indicated. Respectively, the Merge and DIC columns show the combined FITC and TRITC images, and the distribution and morphology of the cells in the field. Note: In vivo each of the three cell types can be infected by Chlamydia and the CD14 row includes a Chlamydia infected cell (TRITC positive) that is not immunostained by the ant-CD14 monoclonal (FITC negative).
Figure 4.
Merged images at different depths from the confocal Z-axis series of dual immunostained WBCs. These merged TRITC-FITC confocal images are from different depths in the same series of Z-axis optical sections acquired for each of the cell types shown in Figure 3. The position of individual sections relative to the cell surface is indicated in μm below each image. Optical section images were acquired with the Zeiss Metaconfocal System.
Flow Cytometry of serially diluted buffy coat pool
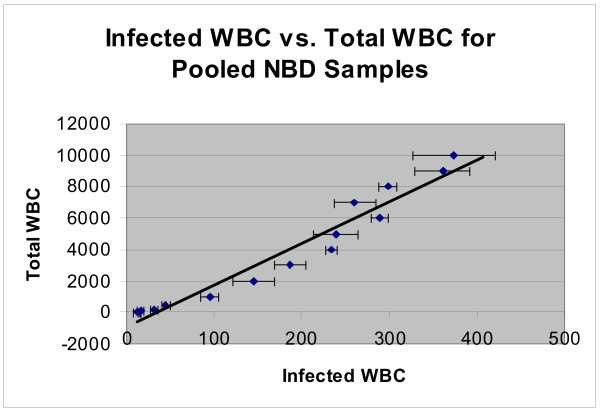
Immunofluorescence allows detection of chlamydial antigens by microscopy but smears are inherently susceptible to significant sampling error. They also can not support quantification of total Chlamydia infected cell numbers in peripheral blood samples. We therefore utilized FC to quantify potentially 'sticky' Chlamydia infected BC samples. After pooling WBC pre-screened by immunostain for intracellular Chlamydia, a dilution series was prepared and FACScan assessments for each dilution analyzed (Figure 5). The data shows the series is linear over a broad range of infected cell numbers and that numeric differences between dilutions in the series are readily evident. As Figure 5 also shows, even in the most dilute sample that contains only 0.5% WBC, 3 ± 1.5 Chlamydia positive cells above background were detectible in the 10,000 cell sample. In addition to indicating the utility of FC for detecting Chlamydia in peripheral WBC samples, the series also serves as a control for subsequent FC directed towards quantifying specific WBC types infected by Chlamydia in vivo.
Figure 5.
FACScan Analysis of WBC Dilution Series from Donor Peripheral Blood Immunostained for Chlamydia. A buffy coat pool prepared from fresh human samples was assessed as positive for Chlamydia by immunostaining. The pool was immunostained for Chlamydia, serially diluted with RBCs pooled from the same samples then analyzed with a FACScan flow cytometer. The graphed results of the series demonstrates detection of Chlamydia infected cells in this non clinical down to 0.5% WBC/sample, a level of 3 ± 1.5 infected cells in the total of 10,000 cells counted.
Presence of Chlamydia in Monocytes, Eosinophils/Basophils and Neutrophils detected by Flow Cytometry
Given the demonstrated linearity and sensitivity, we utilized dual channel FC as a rapid, sensitive tool to quantify both the infected cell load (ICL) and the specific WBC types infected in vivo. Using fresh blood samples, two WBC pools were prepared and immunostained. One pool contained WBC from samples positive by blood smear anti-Chlamydia immunostain examination. The second pool, termed weakly positive, was prepared from samples Chlamydia negative by immunostain but positive by initial FC assessment. Aliquots of BC also were immunostained with a single antibody and used to establish the appropriate compensation for signal overlap between the two fluorescence channels and to establish appropriate, conservative demarcations of fluorescence positive.
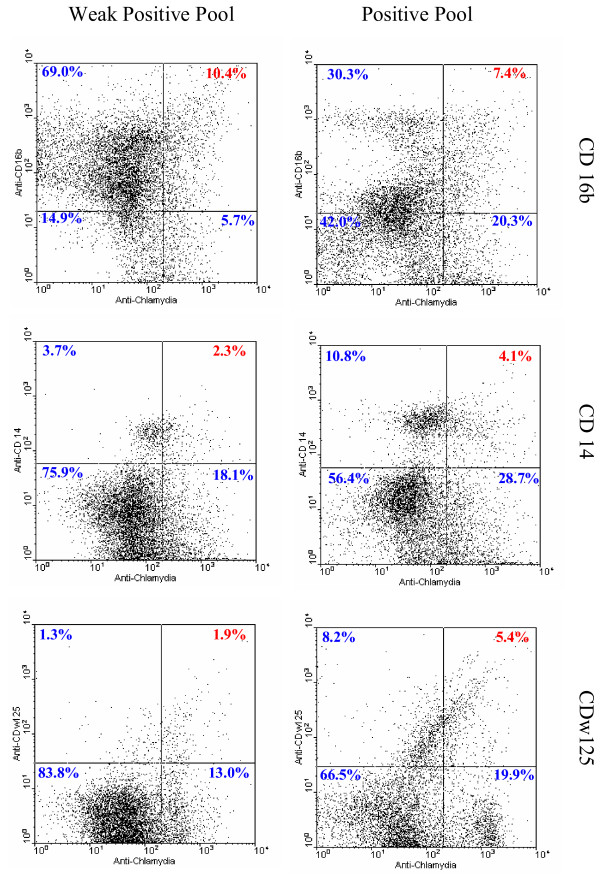
Peripheral WBC populations contain neutrophils (CD16b), monocytes (CD14) and eosinophil/basophils (CDw125) typically present at levels of 60–70%, 3–8%, and 1–4% respectively. Aliquots of each WBC pool were dual immunostained with anti-Chlamydia antibody and monoclonal antibody specific for CD16b, CD14, or CDw125 as described above. The dual channel FC data (Figure 6) indicate that in the population of 10,000 cells counted for the weakly positive pool, 10.4% were positive for both Chlamydia and the neutrophil marker, CD16b, while in the positive pool 7.4% exhibited dual positive fluorescence. Notably, the dot plot for the positive pool also contains a distinct distribution ('cloud') of cells lying somewhat above the CD16b line of demarcation that clearly is absent from the dot plot of the weakly positive sample. In this 'cloud' region, numerous cells in the positive sample exhibit a lower level of CD16b expression, as indicated by their location relative to the CD16b positive demarcation line. This low CD16b expression level would be consistent with the cells being immature neutrophils. Dual immunostaining of Chlamydia-positive monocytes (CD14) demonstrated that in the weakly positive pool, 2.3% of the 10,000 cells was dual positive, while in the positive pool, 4.1% of cells, nearly double, was dual positive. Finally, in aliquots immunostained to detect Chlamydia infected eosinophil/basophils (CDw125), dual positives were identified in 1.9% of cells (weakly positive sample) and 5.4% (positive sample), more than double the number of cells.
Figure 6.
Detection, identification and quantification of Chlamydia infected cells in buffy coat pools from FB samples using flow cytometry. Fresh blood smears were assessed for Chlamydia-positive cells by immunostain. Aliquots of the pooled BC were dual immunostained to detect intracellular Chlamydia and identify CD16b, CD14, or CDSw125 cells. For each of the dual-immunostained BC pairs, aliquots of 10,000 cells were assessed. Left panels: results for the weakly positive pool. Right panels: results for the positive pool. The dot plots indicate: in vivo each cell type can be infected with Chlamydia, the different cell-types exhibit different patterns with different percentages of Chlamydia-infected cells and inherently the same preparation provides enumeration of uninfected as well as infected cells, thereby providing numerical totals for infection of each cell type/10,000 cells.
This quantitative dual channel FC data indicates that in vivo all three of these cell types can become infected, and is consistent with our identification in dual immunostained smears. The data also shows FC can quantify each cell type and that both the dot plot patterns and cell type specific numbers clearly differ for the weakly positive vs. positive samples. Therefore even at the minimal level of Chlamydia-infected cell carriage in these samples, cells of each type become infected in vivo and both the total infected cell load (ICL) and the infected cell type specific load (ICSL) can be quantified.
Discussion
The studies presented here examined blood residuals obtained throughout the year and provide an assessment of blood cell borne Chlamydia carriage in a random cohort of normal blood donors. The aims were to determine the average rate of carriage, identify the key cell types infected in vivo, test whether Chlamydia infected cells can transmit infection to host cell monolayers, and assess quantification of the infected cell load in samples by using flow cytometry.
Our serological results demonstrated that in this donor population, 47.7% was sero-positive for C. pneumoniae. Given the average age of 44 years, this finding is consistent with the CDC report of serologic evidence of a previous infection by C. pneumoniae by the age of 20 (50%) [3] and demonstrates this donor cohort is not anomalous with respect to sero-positivity. Our assessments also showed a blood borne carriage rate for Chlamydia-infected cells of 24.6% positive. However, some notable differences were observed in the rate of Chlamydia-infected WBC, depending on the time of year (Table 3). Although only a multiyear study can verify seasonal variation, our finding suggests that the season of collection could potentially be a factor in the Chlamydia DNA detection rates of 18.5% to 46.15 % reported by others for PBMC. These PCR studies have not concomitantly tested for infectious organisms, nor is the time of year known [15,34-37]. Nevertheless, given published reports, the carriage rate we observed was not surprising, although the initial detection of blood borne Chlamydia carriage in immunofluorescent stained blood smears was unexpected. In addition, we also tested WBC from these samples for infectious organisms using an in vitro macrophage monolayer system, rather than the non-phagocytic HEp-2 or HeLa cells often used by others. As previously reported [30], this macrophage system is readily phagocytic and thus will take up the rare chlamydial EB in a more efficient manner. Moreover, using the J774A.1 cell monolayers and IMEMZO, we routinely cultured for 96 h pi, a time frame that supported re-infection within the monolayers. As Figure 2 shows, these cells continue to look healthy and the amplification achieved at 96 h pi allowed rapid identification of samples with infectious chlamydiae. Importantly, it also showed that the Chlamydia in these NBD cells can be infectious (Table 2, 24.8%).
The presence and especially the infectivity of blood cell borne chlamydiae is inherently relevant and potentially significant in medical contexts as diverse as tissue and organ transplantation [38,39], transfusion medicine and blood banking. As demonstrated previously by PCR [5,6,25,27] and here by immuno-detection and in vitro culture of intracellular organisms per se, a level of carriage clearly occurs. The demonstration that these organisms can be infectious suggests that removal of WBC from banked blood, for example, would be important and simple pre-storage leukoreduction is very effective [32]. In addition, where possible an assessment of the infected cell load in peripheral blood from an organ donor could be highly relevant to transplant surgery.
Our data also shows that not all samples positive for Chlamydia by examination of immunofluorescent smears were positive for infectious Chlamydia by in vitro culture (Table 2). This result may reflect the presence of persistently infected cells. In vivo, this pathogen can enter a persistent phase in response to various micro-environmental conditions. In this state it would be detected in immunofluorescent stained smears but produce few, if any infectious EBs. Although reversion from this state has not been directly demonstrated in vivo, studies have shown that in vitro Chlamydia can revert and give rise to infectious EBs [40-46]. Conversely, some immunostain negative samples were positive by culture. However, this result is not surprising and likely reflects the 20 μl sample size used for smear preparation vs. the much larger BC aliquot (50% of the total BC isolated from a 5 ml whole blood sample) used for the replicate in vitro cultures.
Recent experiments have shown in vitro infection of primary human neutrophils by Chlamydia as well as a concomitant delay of spontaneous apoptosis for infected neutrophils but not uninfected ones [20]. In contrast, the current study sought to identify blood borne WBC that had become infected by Chlamydia in vivo. Both immunofluorescence microscopy and flow cytometry demonstrate additional infective capababilities for this stealth pathogen. These studies used commercially available CD specific monoclonal antibody and both techniques demonstrated that non-clinical samples contain in vivo infected neutrophils and eosinophil/basophil cells as well as monocytes. PCR screening for Chlamydia of a portion of these samples verified Chlamydia DNA is present and in two instances where species specific primers were used, C. trachomatis and C. pneumoniae were present in the same sample (data not shown).
In terms of infected cell quantification, earlier studies had applied FC in various contexts relating to Chlamydia e.g. quantifying EB binding to standard host cell lines in vitro, establishing Chlamydia sensitivity to antibiotics or examining the role of estrogen receptors in Chlamydia infections [47-51]. However quantification of peripheral WBC infected in vivo by Chlamydia has not been reported, quite likely because infected cell carriage in non-clinical blood samples has been under appreciated. The FC quantification of Chlamydia infected WBC (Figure 5) demonstrated both sensitivity and linearity and provided a basis for the cell type specific quantifications comparing weakly positive and positive samples shown in Figure 6. For neutrophils (CD16b+ cells) the FC data supports the suggestion that unusual numbers of putatively immature neutrophils may be present as a correlate of blood cell borne Chlamydia carriage. FC dot plots also demonstrated that the distribution patterns and the numbers of monocytes and eosinophil/basophil cells are distinctly different for these two pools. For the monocytes (CD14+ cells), there were totals of 149,000 vs. 60,000 cells in the positive vs. weakly positive pool and the detected level of Chlamydia positive monocytes was 4.1% vs. 2.3%, a 1.8 fold difference. Interestingly, the total numbers of CDw125+, eosinophil/basophil cells/10,000 cells was much higher for the positive sample (1,360) vs. the weakly positive one (320), a 4.25 fold difference, as was the percentage of each pool that was infected (5.4% vs.1.9%), a 2.8 fold difference. After establishing parameters for routine use, a logical next step would be to assess WBC from patient samples and focus on clinical conditions in which a Chlamydia involvement is known or suspected.
Our evidence from the blood smear studies, as well as the earlier PCR data of others that indicated Chlamydia DNA carriage in normal populations, underscore the potential relevance of these and additional FC quantification studies. The demonstration that these chlamydiae can be infectious has implications for specific diagnoses, as well as health care in general. Clearly even low carriage levels of Chlamydia-infected cells can be quantified by FC (Figure 5) supporting determination of the total infected cell load in the peripheral circulation. Additionally, the FC dual immunostain data (Figure 6) shows that for specific cell types, both the total and infected cell populations are readily and simultaneously quantifiable. Such information would support establishing individual cell type specific load and potentially identify any skewing of infected cell types that might be associated with parameters such as levels of Chlamydia-infected WBC carriage or specific clinical presentations.
The significance of these specific findings and dot plot patterns remain to be established and will require additional broad based assessments. However, FC already is a routine diagnostic tool employed to enumerate CD4+ T cells and CD34+ progenitor cells and to screen for HLA-B27 antigen and leukemia/lymphoma immuno-phenotyping [52]. Thus once optimized for this application, FC could be routinely available to provide a non-invasive means of detecting and monitoring WBC infection by Chlamydia and could be of considerable diagnostic and prognostic value as well as a screening tool to for routine health assessments.
Conclusion
Within the normal blood donor population tested, there is a Chlamydia carriage rate of 24.6%, contained in the WBC of the peripheral circulation. As demonstrated by fluorescent immunostaining combined with confocal microscopy and flow cytometric techniques, this pathogen is present within neutrophils and eosinophil/basophil cells, as well as in the monocyte/macrophage cells previously implicated. In addition, in vitro culture demonstrated Chlamydia in lysates of these non-clinical peripheral WBC can be infectious and readily transmits the Chlamydia to host cell monolayers. Therefore carriage of Chlamydia in the peripheral circulations could be significant in the context of transfusion or transplant recipients as well as routine health assessments.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
ESS supervised FC and NLC, directed the design and execution of the experiments and the draft manuscript and figures, and prepared the final version of the manuscript. WCW carried out the screening and statistical analysis of normal blood donor samples and the in vitro tests with isolated BC preparations to detect infectivity as a part of his doctoral work with ESS. WCW also provided training to NLC in the culture and in vitro infection of the cell lines, generation of pathogen stocks, and contributed to writing the final manuscript.
FC and CW carried out the flow cytometry, prepared and immunostained the smears of human peripheral blood cells to identify WBC types harboring intracellular Chlamydia and contributed to the manuscript preparation. FC carried out the confocal microscopy, image acquisition and figure preparation.
NLC carried out companion studies under controlled in vitro conditions and provided evidence supporting the observations of receptors initially detected in studies of cells from donor blood samples (manuscript in preparation).
CA coordinated NBD sample acquisition, provided expertise in Blood Banking and immunological related issues, as well as blood sample characterization, and provided valuable advice in manuscript preparation.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
We thank the staff at the Blood Bank Center at Baystate Medical Center for their assistance in this study. We also thank Dale A. Callaham and Alenka Lovy-Wheeler of the Massachusetts Central Microscopy Facility for training and assistance with the Zeiss Metaconfocal System. The Microscopy Facility is supported by a grant from the National Science Foundation (NSF BBS 8714235). This work was supported in part by an award from the CVIP Development Fund (ESS), University of Massachusetts, Amherst. We also thank Dr. Lisa Minter, Veterinary and Animal Sciences Department, University of Massachusetts, for her guidance and expertise during the FACS studies. Preliminary screening of PBMC utilized Pel-Freeze reagents, a gift from William R. and Lynda R. McDonald, Biotech Source Inc. 1 Inland Farm Drive Windham ME 04062 http://www.thebiotechsource.com.
Contributor Information
Frances Cirino, Email: fcirino@microbio.umass.edu.
Wilmore C Webley, Email: wilmore@microbio.umass.edu.
Corrie West, Email: esstuart@microbio.umass.edu.
Nancy L Croteau, Email: Nancy.Croteau@umassmed.edu.
Chester Andrzejewski, Jr, Email: chester.andrzejewski@bhs.org.
Elizabeth S Stuart, Email: esstuart@microbio.umass.edu.
References
- Schachter J. Infection and disease epidemiology. Washington, DC: ASM Press; 1999. [Google Scholar]
- Kuo CC, Jackson LA, Campbell LA, Grayston JT. Chlamydia pneumoniae (TWAR) Clin Microbiol Rev. 1995;8:451–461. doi: 10.1128/cmr.8.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Chlamydia pneumoniae Technical information. 2003.
- Wong YK, Dawkins KD, Ward ME. Circulating Chlamydia pneumoniae DNA as a predictor of coronary artery disease. J Am Coll Cardiol. 1999;34:1435–1439. doi: 10.1016/S0735-1097(99)00391-5. [DOI] [PubMed] [Google Scholar]
- Campbell LA, Kuo CC, Grayston JT. Chlamydia pneumoniae and cardiovascular disease. Emerg Infect Dis. 1998;4:571–579. doi: 10.3201/eid0404.980407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne GI, Kalayoglu MV. Chlamydia pneumoniae and atherosclerosis: links to the disease process. Am Heart J. 1999;138:S488–490. doi: 10.1016/S0002-8703(99)70282-6. [DOI] [PubMed] [Google Scholar]
- Gibbs RG, Carey N, Davies AH. Chlamydia pneumoniae and vascular disease. Br J Surg. 1998;85:1191–1197. doi: 10.1046/j.1365-2168.1998.00861.x. [DOI] [PubMed] [Google Scholar]
- Layh-Schmitt G, Bendl C, Hildt U, Dong-Si T, Juttler E, Schnitzler P, Grond-Ginsbach C, Grau AJ. Evidence for infection with Chlamydia pneumoniae in a subgroup of patients with multiple sclerosis. Ann Neurol. 2000;47:652–655. doi: 10.1002/1531-8249(200005)47:5<652::AID-ANA15>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Sriram S, Stratton CW, Yao S, Tharp A, Ding L, Bannan JD, Mitchell WM. Chlamydia pneumoniae infection of the central nervous system in multiple sclerosis. Ann Neurol. 1999;46:6–14. doi: 10.1002/1531-8249(199907)46:1<6::AID-ANA4>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Balin BJ, Gerard HC, Arking EJ, Appelt DM, Branigan PJ, Abrams JT, Whittum-Hudson JA, Hudson AP. Identification and localization of Chlamydia pneumoniae in the Alzheimer's brain. Med Microbiol Immunol (Berl) 1998;187:23–42. doi: 10.1007/s004300050071. [DOI] [PubMed] [Google Scholar]
- Cascina A, Marone Bianco A, Mangiarotti P, Montecucco CM, Meloni F. Cutaneous vasculitis and reactive arthritis following respiratory infection due to Chlamydia pneumoniae: report of a case. Clin Exp Rheumatol. 2002;20:845–847. [PubMed] [Google Scholar]
- Hahn DL, Peeling RW, Dillon E, McDonald R, Saikku P. Serologic markers for Chlamydia pneumoniae in asthma. Ann Allergy Asthma Immunol. 2000;84:227–233. doi: 10.1016/S1081-1206(10)62760-3. [DOI] [PubMed] [Google Scholar]
- Haranaga S, Yamaguchi H, Leparc GF, Friedman H, Yamamoto Y. Detection of Chlamydia pneumoniae antigen in PBMNCs of healthy blood donors. Transfusion. 2001;41:1114–1119. doi: 10.1046/j.1537-2995.2001.41091114.x. [DOI] [PubMed] [Google Scholar]
- Kaul R, Uphoff J, Wiedeman J, Yadlapalli S, Wenman WM. Detection of Chlamydia pneumoniae DNA in CD3+ lymphocytes from healthy blood donors and patients with coronary artery disease. Circulation. 2000;102:2341–2346. doi: 10.1161/01.cir.102.19.2341. [DOI] [PubMed] [Google Scholar]
- Boman J, Soderberg S, Forsberg J, Birgander LS, Allard A, Persson K, Jidell E, Kumlin U, Juto P, Waldenstrom A, Wadell G. High prevalence of Chlamydia pneumoniae DNA in peripheral blood mononuclear cells in patients with cardiovascular disease and in middle-aged blood donors. J Infect Dis. 1998;178:274–277. doi: 10.1086/517452. [DOI] [PubMed] [Google Scholar]
- Bodetti TJ, Timms P. Detection of Chlamydia pneumoniae DNA and antigen in the circulating mononuclear cell fractions of humans and koalas. Infect Immun. 2000;68:2744–2747. doi: 10.1128/IAI.68.5.2744-2747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass M, Gieffers J, Solbach W. Atherogenetically relevant cells support continuous growth of Chlamydia pneumoniae. Herz. 2000;25:68–72. doi: 10.1007/PL00001953. [DOI] [PubMed] [Google Scholar]
- Gieffers J, van Zandbergen G, Rupp J, Sayk F, Kruger S, Ehlers S, Solbach W, Maass M. Phagocytes transmit Chlamydia pneumoniae from the lungs to the vasculature. Eur Respir J. 2004;23:506–510. doi: 10.1183/09031936.04.00093304. [DOI] [PubMed] [Google Scholar]
- Kuroda S, Kobayashi T, Ishii N, Ikeda J, Shinohe Y, Houkin K, Iwasaki Y, Nagashima K. Role of Chlamydia pneumoniae-infected macrophages in atherosclerosis developments of the carotid artery. Neuropathology. 2003;23:1–8. doi: 10.1046/j.1440-1789.2003.00484.x. [DOI] [PubMed] [Google Scholar]
- van Zandbergen G, Gieffers J, Kothe H, Rupp J, Bollinger A, Aga E, Klinger M, Brade H, Dalhoff K, Maass M, et al. Chlamydia pneumoniae multiply in neutrophil granulocytes and delay their spontaneous apoptosis. J Immunol. 2004;172:1768–1776. doi: 10.4049/jimmunol.172.3.1768. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Haranaga S, Friedman H, Moor JA, Muffly KE, Yamamoto Y. A Chlamydia pneumoniae infection model using established human lymphocyte cell lines. FEMS Microbiol Lett. 2002;216:229–234. doi: 10.1111/j.1574-6968.2002.tb11440.x. [DOI] [PubMed] [Google Scholar]
- Kaukoranta-Tolvanen SS, Teppo AM, Laitinen K, Saikku P, Linnavuori K, Leinonen M. Growth of Chlamydia pneumoniae in cultured human peripheral blood mononuclear cells and induction of a cytokine response. Microb Pathog. 1996;21:215–221. doi: 10.1006/mpat.1996.0056. [DOI] [PubMed] [Google Scholar]
- Beutler AM, Whittum-Hudson JA, Nanagara R, Schumacher HR, Hudson AP. Intracellular location of inapparently infecting Chlamydia in synovial tissue from patients with Reiter's syndrome. Immunol Res. 1994;13:163–171. doi: 10.1007/BF02918277. [DOI] [PubMed] [Google Scholar]
- Beutler AM, Schumacher HR, Jr, Whittum-Hudson JA, Salameh WA, Hudson AP. Case report: in situ hybridization for detection of inapparent infection with Chlamydia trachomatis in synovial tissue of a patient with Reiter's syndrome. Am J Med Sci. 1995;310:206–213. doi: 10.1097/00000441-199511000-00006. [DOI] [PubMed] [Google Scholar]
- Gerard HC, Branigan PJ, Schumacher HR, Jr, Hudson AP. Synovial Chlamydia trachomatis in patients with reactive arthritis/Reiter's syndrome are viable but show aberrant gene expression. J Rheumatol. 1998;25:734–742. [PubMed] [Google Scholar]
- Whittum-Hudson JA, Gerard HC, Clayburne G, Schumacher HR, Hudson AP. A non-invasive murine model of chlamydia-induced reactive arthritis. Rev Rhum Engl Ed. 1999;66:50S–55S. discussion 56S. [PubMed] [Google Scholar]
- Schumacher HR, Jr, Arayssi T, Crane M, Lee J, Gerard H, Hudson AP, Klippel J. Chlamydia trachomatis nucleic acids can be found in the synovium of some asymptomatic subjects. Arthritis Rheum. 1999;42:1281–1284. doi: 10.1002/1529-0131(199906)42:6<1281::AID-ANR27>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Inman RD, Whittum-Hudson JA, Schumacher HR, Hudson AP. Chlamydia and associated arthritis. Curr Opin Rheumatol. 2000;12:254–262. doi: 10.1097/00002281-200007000-00004. [DOI] [PubMed] [Google Scholar]
- Villareal C, Whittum-Hudson JA, Hudson AP. Persistent Chlamydiae and chronic arthritis. Arthritis Res. 2002;4:5–9. doi: 10.1186/ar382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webley WC, Salva PS, Andrzejewski C, Cirino F, West CA, Tilahun Y, Stuart ES. The Bronchial Lavage of Pediatric Patients with Asthma contain Infectious Chlamydia. Am J Respir Crit Care Med. 2005 doi: 10.1164/rccm.200407-917OC. [DOI] [PubMed] [Google Scholar]
- Vora GJ, Stuart ES. A Role for the glycolipid exoantigen (GLXA) in chlamydial infectivity. Curr Microbiol. 2003;46:217–223. doi: 10.1007/s00284-002-3843-1. [DOI] [PubMed] [Google Scholar]
- Webley WC, Andrzejewski C, Stec T, Stuart ES. Successful removal of Chlamydophila pneumoniae from blood by Apheresis Leukoreduction. J Clinical Aphereis. 2003;18:80–81. (Published Abstract) [Google Scholar]
- Webley WC, Vora GJ, Stuart ES. Cell surface display of the chlamydial glycolipid exoantigen (GLXA) demonstrated by antibody-dependent complement-mediated cytotoxicity. Curr Microbiol. 2004;49:13–21. doi: 10.1007/s00284-003-4181-7. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Yamada M, Uruma T, Kanamori M, Goto H, Yamamoto Y, Kamiya S. Prevalence of viable Chlamydia pneumoniae in peripheral blood mononuclear cells of healthy blood donors. Transfusion. 2004;44:1072–1078. doi: 10.1111/j.1537-2995.2004.04005.x. [DOI] [PubMed] [Google Scholar]
- Rassu M, Lauro FM, Cazzavillan S, Bonoldi E, Belloni M, Bettini MC, Pilotto A, Mengoli C, Peron A, Zambello R, et al. Detection of Chlamydophila pneumoniae DNA in peripheral blood mononuclear cells of blood donors in the north-east of Italy. Med Microbiol Immunol (Berl) 2001;190:139–144. doi: 10.1007/s004300100092. [DOI] [PubMed] [Google Scholar]
- Haranaga S, Yamaguchi H, Friedman H, Izumi S, Yamamoto Y. Chlamydia pneumoniae infects and multiplies in lymphocytes in vitro. Infect Immun. 2001;69:7753–7759. doi: 10.1128/IAI.69.12.7753-7759.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smieja M, Mahony J, Petrich A, Boman J, Chernesky M. Association of circulating Chlamydia pneumoniae DNA with cardiovascular disease: a systematic review. BMC Infect Dis. 2002;2:21. doi: 10.1186/1471-2334-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville AR. Chlamydia pneumoniae is associated with graft dysfunction after lung transplantation. J Heart Lung Transplant. 2001;20:171. doi: 10.1016/S1053-2498(00)00339-9. [DOI] [PubMed] [Google Scholar]
- Glanville AR, Gencay M, Tamm M, Chhajed P, Plit M, Hopkins P, Aboyoun C, Roth M, Malouf M. Chlamydia pneumoniae infection after lung transplantation. J Heart Lung Transplant. 2005;24:131–136. doi: 10.1016/j.healun.2003.09.042. [DOI] [PubMed] [Google Scholar]
- Beatty WL, Morrison RP, Byrne GI. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol Rev. 1994;58:686–699. doi: 10.1128/mr.58.4.686-699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty WL, Morrison RP, Byrne GI. Reactivation of persistent Chlamydia trachomatis infection in cell culture. Infect Immun. 1995;63:199–205. doi: 10.1128/iai.63.1.199-205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne GI. Chlamydial treatment failures: a persistent problem? J Eur Acad Dermatol Venereol. 2001;15:381. doi: 10.1046/j.1468-3083.2001.00343.x. [DOI] [PubMed] [Google Scholar]
- Gerard HC, Kohler L, Branigan PJ, Zeidler H, Schumacher HR, Hudson AP. Viability and gene expression in Chlamydia trachomatis during persistent infection of cultured human monocytes. Med Microbiol Immunol (Berl) 1998;187:115–120. doi: 10.1007/s004300050082. [DOI] [PubMed] [Google Scholar]
- Gerard HC, Whittum-Hudson JA, Schumacher HR, Hudson AP. Differential expression of three Chlamydia trachomatis hsp60-encoding genes in active vs. persistent infections. Microb Pathog. 2004;36:35–39. doi: 10.1016/j.micpath.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Gieffers J, Durling L, Ouellette SP, Rupp J, Maass M, Byrne GI, Caldwell HD, Belland RJ. Genotypic differences in the Chlamydia pneumoniae tyrP locus related to vascular tropism and pathogenicity. J Infect Dis. 2003;188:1085–1093. doi: 10.1086/378692. [DOI] [PubMed] [Google Scholar]
- Koehler L, Nettelnbreker E, Hudson AP, Ott N, Gerard HC, Branigan PJ, Schumacher HR, Drommer W, Zeidler H. Ultrastructural and molecular analyses of the persistence of Chlamydia trachomatis (serovar K) in human monocytes. Microb Pathog. 1997;22:133–142. doi: 10.1006/mpat.1996.0103. [DOI] [PubMed] [Google Scholar]
- Dessus-Babus S, Belloc F, Bebear CM, Poutiers F, Lacombe F, Bebear C, de Barbeyrac B. Antibiotic susceptibility testing for Chlamydia trachomatis using flow cytometry. Cytometry. 1998;31:37–44. doi: 10.1002/(SICI)1097-0320(19980101)31:1<37::AID-CYTO5>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Levitt D, Zable B, Bard J. Binding, ingestion, and growth of Chlamydia trachomatis (L2 serovar) analyzed by flow cytometry. Cytometry. 1986;7:378–383. doi: 10.1002/cyto.990070413. [DOI] [PubMed] [Google Scholar]
- Montigiani S, Falugi F, Scarselli M, Finco O, Petracca R, Galli G, Mariani M, Manetti R, Agnusdei M, Cevenini R, et al. Genomic approach for analysis of surface proteins in Chlamydia pneumoniae. Infect Immun. 2002;70:368–379. doi: 10.1128/IAI.70.1.368-379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg AJ, Elsley WA, Pickett MA, Ward ME, Knight SC. Primary human T-cell responses to the major outer membrane protein of Chlamydia trachomatis. Immunology. 1993;79:1–9. [PMC free article] [PubMed] [Google Scholar]
- Guseva NV, Dessus-Babus SC, Whittimore JD, Moore CG, Wyrick PB. Characterization of estrogen-responsive epithelial cell lines and their infectivity by genital Chlamydia trachomatis. Microbes Infect. 2005 doi: 10.1016/j.micinf.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Keeney M, Barnett D, Gratama JW. Impact of standardization on clinical cell analysis by flow cytometry. J Biol Regul Homeost Agents. 2004;18:305–312. [PubMed] [Google Scholar]