Abstract
Signaling pathways respond to stimuli in a variety of ways, depending on the magnitude of the input and the physiological status of the cell. For instance, yeast can respond to pheromone stimulation in either a binary or graded fashion. Here we present single cell transcription data indicating that a transient binary response in which all cells eventually become activated is typical. Stochastic modeling of the biochemical steps that regulate activation of the mitogen-activated protein kinase Fus3 reveals that this portion of the pathway can account for the graded-to-binary conversion. To test the validity of the model, genetic approaches are used to alter expression levels of Msg5 and Ste7, two of the proteins that negatively and positively regulate Fus3, respectively. Single cell measurements of the genetically altered cells are shown to be consistent with predictions of the model. Finally, computational modeling is used to investigate the effects of protein turnover on the response of the pathway. We demonstrate that the inclusion of protein turnover can lead to sustained oscillations of protein concentrations in the absence of feedback regulation. Thus, protein turnover can profoundly influence the output of a signaling pathway.
INTRODUCTION
The pheromone response of yeast is one of the best-characterized signaling pathways (1). Much is known about the proteins that transmit the pheromone signal, as well as about the mechanisms by which events at the cell surface are linked to subsequent biochemical changes in the cytoplasm and nucleus. A diagram of the pathway is given in Fig. 1 A. Activation of the pathway is initiated by binding of mating pheromones to specific cell surface receptors, and ends with the fusion of a- and α-haploid cell types to form an a/α-diploid (mating). In the a-cell type, signaling is initiated by binding of the pheromone α-factor to its receptor Ste2. Receptor activation in turn leads to the exchange of GDP for GTP on the G-protein α-subunit Gpa1, and subsequent dissociation from the G-protein βγ-subunit dimer composed of Ste4 and Ste18. The signal is then transmitted and amplified through effector proteins that bind to Gβγ. A major target of the Gβγ-subunits is a cascade of four protein kinases that begins with Ste20 and ends with the mitogen-activated protein kinase (MAPK) Fus3. Ste20 phosphorylates and activates Ste11, which phosphorylates and activates Ste7, which in turn phosphorylates and activates the MAPK Fus3 (on Tyr-182 and Thr-180). Fus3 has a number of substrates including the transcription factor Ste12, which is responsible for induction of genes required for mating. Inactivation of signaling requires that Fus3 is dephosphorylated on both Tyr and Thr. Both sites are recognized by the dual-specificity phosphatase Msg5. Tyr is also dephosphorylated by at least two other phosphatases, Ptp2 and Ptp3. In the absence of Fus3, however, a closely related MAP kinase Kss1 can carry out most of the functions of Fus3 (1).
FIGURE 1.
(A) The pheromone signaling pathway. Shown are the pheromone receptor (Ste2), G-protein α-, β-, and γ-subunits (Gpa1, Ste4, Ste18), the Regulator of G-protein signaling (RGS, Sst2) which accelerates Gpa1 GTPase activity, effector kinases including the MAPKKK kinase (Ste20), MAPKK kinase (Ste11), MAPK kinase (Ste7), MAP kinases (Kss1, Fus3), and nuclear transcription factor (Ste12). Three of the kinases bind to a kinase scaffold protein (Ste5). Several other effectors and regulatory components are not shown, for clarity. Pheromone-dependent transcriptional induction of SST2 represents a negative feedback loop. (B) A schematic diagram of Fus3 regulation. Fus3 activation and deactivation are assumed to occur through a distributive kinetic mechanism in which two collisions with Ste7 (MAPK kinase) and Msg5 (MAPK phosphatase) are required. The MAPK Kss1 also activates the pheromone response pathway, and the phosphatases Ptp2 and Ptp3 also inactivate kinase activity. However, to simplify the models the kinase and phosphatase activities of these proteins are not distinguished from those of Fus3 and Msg5.
Most of the components of the pheromone response pathway have been identified genetically, through the isolation of mating-defective or sterile gene mutations. Further genetic, biochemical, and molecular biological analysis of the pathway revealed the order of each signaling event, and has established many basic principles of G-protein and MAPK signaling relevant to all eukaryotes (2). Having now determined the essential components and events in G-protein-coupled receptor signaling, an emerging goal is to develop mathematical models that describe their behavior over time.
Cell signaling pathways can vary their response to a stimulus in a variety of different ways. At the receptor level, pathway activation is determined by the number of liganded receptors and therefore is proportional to the concentration of the input stimulus. This graded binding event can propagate through the pathway and produce a graded transcriptional response that is proportional to the input signal. However, many pathways convert a graded input instead into a binary transcriptional response (3–5). A binary response refers to an all-or-none situation in which the probability of an individual cell responding to a stimulus is proportional to the strength of the signal. Binary responses are often attributed to multiple steady states that arise from feedback regulation (4,6). However, graded and binary outputs are not the only time-dependent responses of which signaling pathways are capable. Sustained oscillations and more complicated dynamics are also possible.
In yeast, pheromone signaling can produce either a graded or binary transcriptional response depending on the dose of pheromone, the time of treatment, and the intracellular activation event being measured. A binary response may be appropriate in some physiological situations but not in others. For instance in yeast, pheromones initiate a process leading to mating, an inherently irreversible process where an all-or-none decision is appropriate. Binary outputs are also appropriate during cell division, cell differentiation, and cellular apoptosis. Thus, establishing the mechanisms by which the graded-to-binary conversion is accomplished is a fundamental problem in cell biology. Here we seek to identify components of the pheromone response pathway that mediate the graded-to-binary conversion and to uncover the mechanism by which this conversion is accomplished. We employ an approach that combines experimental analysis with computational modeling. Data from fluorescence-based transcriptional induction assays in single cells are used as the experimental basis for a stochastic model of the biochemical steps that regulate MAPK activation. The mathematical model is motivated by the theoretical work of Markevich et al. (7) on multisite phosphorylation of protein kinases. Computational analysis is used to validate the model and generate testable hypotheses, which are in turn confirmed experimentally.
Our stochastic modeling reveals that small changes in protein abundances can have large effects on MAPK activation. Additionally, data on protein turnover suggests that protein degradation plays an important role in regulating the pheromone pathway. Pheromone-stimulated degradation has been documented previously for the receptor (Ste2) (8), a regulator of G-protein signaling (Sst2) (9), components of the effector kinase cascade (Ste11, Ste7) (9–11), and the transcription factor (Ste12) (12). However, the functional consequences of accelerating protein turnover have not been well characterized. These observations motivate extending the computational model to include protein turnover. We demonstrate that the qualitative behavior of the pathway depends on the mechanism through which protein degradation occurs. In particular, we show that protein degradation can generate sustained oscillations in protein concentrations. Finally, stochastic modeling is used to demonstrate that biochemical fluctuations increase the parameter range over which the oscillations occur, and that the oscillations can generate an apparent binary response in flow cytometry experiments.
MATERIALS AND METHODS
Model description
In this study, we focus on the MAPK portion of the pheromone response pathway. The biochemical steps involved in the regulation of the MAP kinase Fus3 are shown in Fig. 1 B. Transmission of the intracellular signal involves phosphorylation of Fus3 by Ste7. Pathway activation can also occur via the MAPK Kss1. However, in this study we do not distinguish between Fus3 and Kss1. Ste7 is a dual-specificity kinase that modifies Fus3 at Thr-180 and Tyr-182 sites and stimulates its catalytic activity. Msg5 is a dual-specificity phosphatase that inactivates Fus3. Ptp2 and Ptp3 are tyrosine phosphatases that also inactivate Fus3. For simplicity, we do not distinguish between these three proteins. As shown in Fig. 1 B, we assume a distributive kinetic mechanism for the dual phosphorylation and dephosphorylation reactions (7,13). A distributive mechanism refers to one in which the kinase and phosphatase release the monophosphorylated substrate intermediate and a second interaction is required to generate the final product. We use [KK], [K], and [P] to denote the concentrations of Ste7 (MAPK kinase), Fus3 (MAPK), and Msg5 (phosphatase), respectively. [Kp] and [Kpp] denote the concentrations of the singly phosphorylated and doubly phosphorylated forms of Fus3, respectively. Protein/protein complexes are denoted with a dot (·). For example, [K·KK] denotes the concentration of the Fus3/Ste7 complex. The biochemical reactions used in the stochastic and rate equation models are given in Appendix A. Where available, we use experimentally measured values for the model parameters. For the parameters that have not been measured, we use biologically realistic values. A summary of the model parameters and the values used in the simulations is given in Table 1.
TABLE 1.
Model parameters and the values used in the simulations
| Parameter | Description | Value (Cases I, II, III) | Reference |
|---|---|---|---|
| k1 | Association rate constant K/KK binding | 0.00275, 0.0011, 0.0198 | (7) |
| k−1 | Dissociation rate constant for K·KK | 2.5, 1, 1 | (7) |
| k2 | kcat for first phosphorylation event | 0.025, 0.01,0.01 | (7) |
| k3 | Association rate constant for Kp /KK binding | 0.00445, 0.00178, 0.03204 | (7) |
| k−3 | Dissociation rate constant for Kp·KK | 2.5, 1, 1 | (7) |
| k4 | kcat for second phosphorylation event | 37.5, 15, 15 | (7) |
| h1 | Association rate constant Kpp/P binding | 0.00625, 0.0025, 0.045 | (7) |
| h−1 | Dissociation rate constant for Kpp·P | 2.5, 1, 1 | (7) |
| h2 | Rate constant for phosphate release of the phosphotyrosine | 0.23, 0.092, 0.092 | (7) |
| h3 | Dissociation rate constant for Kp·P | 2.5, 1, 1 | (7) |
| h−3 | Association rate constant for Kp/P | 0.0014, 5.6e-4, 0.0099 | (7) |
| h4 | Association rate constant for Kp/P binding | 0.0014, 5.6e-4, 0.0099 | (7) |
| h−4 | Dissociation rate constant for (Kp·P) | 2.5, 1,1 | (7) |
| h5 | Rate constant for phosphate release of the phosphothreonine | 1.25, 0.5,0.5 | (7) |
| h6 | Dissociation rate constant for K·P | 0.215, 0.086, 0.086 | (7) |
| h−6 | Association rate constant for K/P binding | 1.525e-4, 6.1e-5, 0.099 | (7) |
| γk | Synthesis rate of Fus3 | N/A, 0.9, 0.095 | Determined from abundance and degradation rate |
| γkk | Synthesis rate of Ste7 | N/A, 0.33504, 0.016 | Determined from abundance and degradation rate |
| γp | Synthesis rate of Msg5 | N/A, 0.828, 0.023 | Determined from abundance and degradation rate |
| δk | Degradation rate of Fus3 | N/A,1e-4, 1e-4 | Experiment |
| δkk | Degradation rate of Ste7 | N/A, 3.2e-4, 3.2e-4 | (9) |
| δp | Degradation rate of Msg5 | N/A, 4.6e-4, 4.6e-4 | Experiment |
| NKT | Molecular abundance of Fus3 | 9000, N/A, N/A | (18) |
| NKKT | Molecular abundance of Ste7 | 900, N/A, N/A | (18) |
| NPT | Molecular abundance of Msg5 | 1800, N/A, N/A | (7,18,22) |
All rate constants have units of s−1. K denotes Fus3 (MAPK), KK denotes Ste7 (MAPKK), and P denotes Msg5 (phosphatase).
Computational methods
Stochastic and deterministic modeling
Biochemical reactions are inherently random processes. The Gillespie algorithm can be used to generate single realizations of biochemical networks (14). We have implemented an efficient version of the Gillespie algorithm in our software package BioNetS (15). Most of the stochastic modeling was done using BioNetS. The BioNetS scripts used to generate the results are available upon request.
When protein abundances are sufficiently large, the law of mass action can be used to construct rate equations for the concentrations of the various chemical species. Here we present a very simple example to illustrate the connection between the stochastic models and the rate equations. Consider the following two biochemical reactions:
 |
(1) |
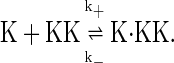 |
(2) |
When Eq. 1 proceeds in the forward direction, a new molecule of Fus3 (K) is synthesized. The reverse reaction represents the degradation of a single Fus3 molecule. Eq. 2 represents the binding and dissociation of a Fus3 molecule (K) with a Ste7 molecule (KK). In the stochastic models the system is described in terms of molecule numbers. Therefore the rate constants, δk, γk, k+, and k−, all have units of inverse time. The second-order rate constant k+ is inversely proportional to the effective volume V. That is, 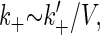 where the constant
where the constant  is independent of the volume and has units of [concentration]−1[time]−1. The effective volume might be that of a yeast cell or smaller if the proteins are spatially restricted in their localization. Let NK(t), NKK(t), and NK·KK(t) denote the number of molecules of the proteins Fus3, Ste7, and the complex Fus3·Ste7, respectively, at time t. Note that in the example given by Eqs. 1 and 2, the total number of Ste7 molecules, NKK+NK·KK, remains constant in time and that Fus3 cannot be degraded when it is in a complex with Ste7. Concentrations are formed by dividing the number of molecules by the volume. To convert to molar concentration, the molecule number also must be divided by Avogadro's number NA. For example, [K] = NK/(V NA) is the Fus3 concentration. Using the law of mass action, we can write rate equations for the concentrations:
is independent of the volume and has units of [concentration]−1[time]−1. The effective volume might be that of a yeast cell or smaller if the proteins are spatially restricted in their localization. Let NK(t), NKK(t), and NK·KK(t) denote the number of molecules of the proteins Fus3, Ste7, and the complex Fus3·Ste7, respectively, at time t. Note that in the example given by Eqs. 1 and 2, the total number of Ste7 molecules, NKK+NK·KK, remains constant in time and that Fus3 cannot be degraded when it is in a complex with Ste7. Concentrations are formed by dividing the number of molecules by the volume. To convert to molar concentration, the molecule number also must be divided by Avogadro's number NA. For example, [K] = NK/(V NA) is the Fus3 concentration. Using the law of mass action, we can write rate equations for the concentrations:
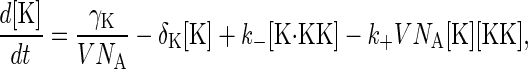 |
(3) |
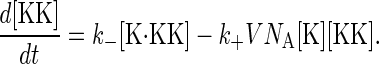 |
(4) |
The conservation of total Ste7 number can now be written as d/dt([KK] + [K·KK]) = 0. Therefore, once the total Ste7 concentration, [KK]T, has been specified, [K·KK] can be found from the relation [K·KK] = [KK]T – [KK]. Equations 3 and 4 represent a macroscopic description of the process, because they ignore biochemical fluctuations. The macroscopic limit is reached as the molecule numbers and volume become large with their ratios (concentrations) remaining finite. Fluctuations in concentration typically scale like 1/V1/2, so that Eqs. 3 and 4 are valid in limit of large volume. When investigating the range of validity of these equations through comparisons with stochastic simulations, the synthesis rates must be scaled with the volume, whereas the second-order rate constants must scale inversely with volume. This scaling ensures that Eqs. 3 and 4 remain unchanged as the volume is increased. The numerical simulations of the rate equations were carried out in MatLab (The MathWorks, Natick, MA) and the bifurcation analysis was done in MatLab and XPPAUT (16).
Protein degradation and synthesis
We investigate several different models of protein degradation and synthesis.
Case I—conservation of enzyme concentrations. The simplest model assumes that proteins are neither degraded nor synthesized. In this case the total concentration of all three enzymes is constant in time. That is,
 |
(5) |
 |
(6) |
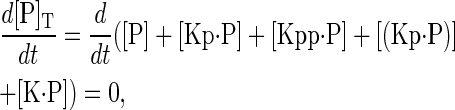 |
(7) |
where the subscript T stands for total. The chemical species (Kp·P) in Eq. 5 results from the assumption that the phosphotyrosine is dephosphorylated first (7,17). That is, the chemical species (Kp·P) indicates the phosphatase attacking the phosphothreonine, whereas Kp·P is the product after phosphotyrosine has been dephosphorylated. The assumption of conserved enzyme concentrations is generally justified by the observation that protein synthesis and degradation occur on timescales that are considerably longer than phosphorylation/dephosphorylation reactions. However, it was recently shown for the pheromone response pathway that the degradation rate of Ste7 is large and increases upon exposure to pheromone (9). Below we present experimental results that show the degradation rate of Msg5 is also large, and slightly increases with pheromone stimulation. The fact that steady-state levels of these proteins do not change appreciably after exposure to pheromone implies that protein synthesis must also increase (data not shown). There are many possibilities for how to incorporate protein degradation into the model. We will investigate two cases that illustrate that the dynamics of the system depends critically on this choice.
Case II—no protection from degradation. In addition to Eq. 1 for Fus3, the reactions for the synthesis and degradation of free Ste7 and Msg5 are
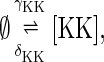 |
(8) |
 |
(9) |
In this model, we also allow proteins to be degraded regardless of their chemical state. That is, all reactions of the form
 |
(10) |
 |
(11) |
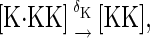 |
(12) |
are included. The total concentration of each protein species is no longer conserved, and Eqs. 5–7 become
 |
(13) |
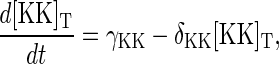 |
(14) |
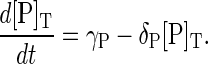 |
(15) |
At steady state, the synthesis and degradation rates balance, and the steady-state values of the total protein concentrations are 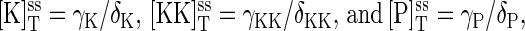 where the superscript ss denotes steady state.
where the superscript ss denotes steady state.
Case III—degradation of free protein only. In this scenario, only the free inactive form of the protein is degraded. That is, protein degradation only occurs via the reactions given in Eqs. 1, 8, and 9, so that proteins are protected against degradation when they are in a protein-protein complex or phosphorylated. In this case, Eqs. 13–15 become
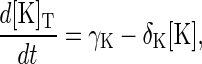 |
(16) |
 |
(17) |
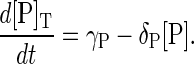 |
(18) |
At steady state, the free enzyme concentrations are given by [K]ss = γK/δK, [KK]ss = γKK/δKK, and [P]ss = γP/δP. Below we show that this model can produce sustained oscillations in concentration levels. These oscillations persist if the phosphorylated species of Fus3 are not protected from degradation.
For simplicity, we will focus on the three cases described here. However, many other possibilities exist. For example, it is possible that degradation rates are different for free proteins versus those in complexes or those that are phosphorylated.
Experimental methods
Strains and plasmids
The S. cerevisiae strains used in this study were BY4741 (MATa leu2Δ met15Δ his3Δ ura3Δ), BY4741-derived strains containing the tandem-affinity purification (TAP) tag fused to FUS3 or MSG5 (18), or BY4741-derived deletion mutants lacking SST2 or MSG5 (Research Genetics, Huntsville, AL). The single copy SST2 expression plasmid pRS316-SST2 (2X SST2) was described previously (19). The single-copy MSG5 plasmid was constructed by PCR amplification of the MSG5 gene, using flanking PCR primers that anneal ∼600-bp upstream (GAGGATCCGACGATGATGACGATGATGATG) or ∼600-bp downstream (GAGGATCCTGCAGCAACACCTTTGG) of the open reading frame. The PCR product was then subcloned into pRS316 (American Type Culture Collection, Manassas, VA) by BamHI digestion and ligation to yield pRS316-MSG5. The STE7 overexpression plasmid was constructed by PCR amplification (forward primer: ATGTTTCAACGAAAGACTTTA; reverse primer: AATGGGTTGATCTTTCCGATTG) of the STE7 gene and then ligated into pYES2.1/V5-His-TOPO (Invitrogen, Carlsbad, CA) to yield pGAL-STE7. The FUS1-green fluorescent protein (GFP) reporter (containing destabilized PEST-domain containing variant of GFP) was subcloned from pDS30 (20) into pRS303 (American Type Culture Collection) using EcoRI and NotI, and then linearized by XcmI to drive genomic integration at the FUS1 locus.
Protein degradation time course and immunoblot detection
Cells were treated with 3 μM α-factor for indicated times. To monitor the degradation of proteins over time, midlog cell cultures were treated with cycloheximide (10 μg/ml in 0.1% ethanol, final concentrations) for up to 90 min before harvesting. The cell growth and treatment was stopped by the addition of 10 mM NaN3 and transfer to an ice bath. Cells were washed and resuspended directly in boiling SDS-PAGE sample buffer for 10 min, disrupted by glass-bead homogenization, and clarified by microcentrifugation. After SDS-PAGE and transfer to nitrocellulose, the membrane was probed with antibodies to Sst2 or protein A (Sigma-Aldrich, St. Louis, MO). Immunoreactive species were visualized by enhanced chemiluminescence detection (Pierce Biotechnology, Rockford, IL) of horseradish peroxidase-conjugated anti-rabbit IgG (Bio-Rad, Hercules, CA).
Fluorescence-activated cell sorting
The measurement of GFP in individual yeast cells was described previously (19,21). Briefly, cells containing the integrated FUS1-GFP reporter were treated with indicated concentrations of α-factor for indicated times. The cell growth and α-factor treatment were stopped by the addition of 10 mM NaN3 and transfer to an ice bath. The resulting fluorescence in each cell was monitored by cell sorting.
RESULTS
The pheromone response can be binary or graded
Transcriptional induction can occur in a graded or binary fashion (5). A binary response can exist transiently with all the cells eventually becoming activated or can be permanent with a persistent subpopulation of cells in the inactivate state. To investigate the temporal response of the pheromone pathway, we conducted single cell fluorescence measurements of transcription. Fig. 2 presents flow cytometry data using the destabilized green fluorescent protein (GFP) as a reporter for the transcription induction activity of individual cells after exposure to pheromone. The distributions shown as solid lines in the first column of Fig. 2 A are results at different time-points for wild-type cells exposed to 1 μM pheromone. The cells display a transient binary response with all cells eventually responding after 120 min. The distributions shown as solid lines in the top row of Fig. 2 B are flow cytometry data for wild-type cells at various pheromone doses taken 60 min after exposure to pheromone. At this time-point, the binary response is most pronounced at 1 μM of pheromone.
FIGURE 2.
Pheromone-dependent transcriptional induction was measured for four different strains of cells: wild-type cells (WT), an sst2 gene deletion mutant (sst2Δ), cells transformed with a single copy plasmid (pRS316) containing genomic clone of SST2 (2XSST2) and sst2Δ containing a single copy plasmid (pRS316) containing genomic clone of MSG5 (2XMSG5). An integrated pheromone-responsive FUS1 promoter-GFP reporter was used to monitor expression. Cells were then treated with various concentrations of α-factor (0.3, 1, 3, and 10 μM) and the resulting fluorescence in each cell was monitored by cell sorting at 45, 60, 75, 90, and 120 min. All experiments were repeated at least twice with similar results. Fluorescence measurements are reported on a log scale (x axis). (A) The rows in this figure correspond to fluorescence measurements made at different time-points after treatment with 1 μM of pheromone. In each column the distributions shown as dotted lines are data for the sst2Δ mutant that responds in a graded fashion. In contrast the distributions shown as solid lines are for the WT (first column), 2XSST (second column) and 2XMSG5 in sst2Δ (third column) strains and show a transient binary response. The shaded dashed line in the first two columns is a guide for the eye to help illustrate the delayed response of the 2XSST2 strain relative to the WT case. (B) The columns in this figure correspond to fluorescence measurements made at different pheromone concentrations 60 min after exposure. Again the distributions shown as dotted lines are data for the sst2Δ mutant and the distributions shown as solid lines are for the WT (first row), 2XSST2 (second row), and 2XMSG5 in sst2Δ (third row) strains. The delayed response of the 2XSST2 strain can again be observed for α-factor concentrations of 0.3, 1, and 3 μM.
Sst2 is a regulator of G-protein signaling that accelerates G-protein-catalyzed GTP hydrolysis, and in this way inhibits pathway activation. Sst2 transcription and synthesis is induced by pheromone producing a negative feedback loop. We recently demonstrated that Sst2 degradation is also increased upon exposure to pheromone (19). Computational modeling revealed that this positive feedback mechanism can counteract the negative effects of Sst2 and generate a binary response. A prediction of the computational model is that the deletion of Sst2 should remove the transient binary response resulting in a graded temporal response. To test this possibility we conducted single cell fluorescence measurements on cells lacking Sst2 (sst2Δ). These cells exhibited a graded response for all doses and time-points tested. The distributions shown as dotted lines in Fig. 2, A and B, are a subset of these results. Again the columns of Fig. 2 A are the results for different time-points using a pheromone concentration of 1 μM and the rows of Fig. 2 B are the results for different doses at 60 min after exposure. Because the sst2Δ mutant always produced a grade response, we have included these results on all the plots as a reference. To further investigate how Sst2 attenuates the pheromone response, we performed fluorescence measurement on cells containing an extra copy of the SST2 gene (2XSst2). The results are shown in the second column of Fig. 2 A and second row of Fig. 2 B. These data indicate that the 2XSst2 strain also exhibits a transient binary response and at pheromone doses of 0.3, 1, and 3 μM, this response lags that of wild-type cells (compare column 1 with 2 in Fig. 2 A and row 1 with 2 in Fig. 2 B). Fig. 2 B also indicates that the binary response is more pronounced at higher pheromone concentrations than in wild-type cells.
The results presented in Fig. 2 demonstrate that the graded-to-binary conversion is regulated by the G-protein; however, these data do not establish that the conversion is mediated by the G-protein itself or by another downstream signaling component. Previous results suggest that it is the MAPK that mediates the switch. Evidence for this comes from the work of Poritz et al. (21). Using sst2Δ mutants, they demonstrated over a range of pheromone concentrations that inhibition of the pathway downstream of the G-protein converts a graded response to a binary one. Their measurements were made 4–6 h after pheromone treatment, indicating that the binary response is permanent. This suggests that inhibiting the pathway downstream of the G-protein generates bistability. Recent theoretical work of Markevich et al. (7) also suggests that regulation of MAPK is sufficient to generate bistability, and thus might account for the graded-to-binary conversion observed by Poritz et al. (21). To test if regulation of MAPK is also sufficient to explain the temporal response of the pathway described here, we constructed a stochastic model of Fus3 activation. Stochastic models treat biochemical reactions as random processes and therefore provide information about the effects of concentration fluctuations on the response of the pathway.
Stochastic modeling, bistability, and the transient binary response
A diagram of the MAPK portion of the pheromone response pathway is shown in Fig. 1 B. Ste7 is a dual-specificity kinase that phosphorylates Fus3 at both threonine and tyrosine residues and stimulates its catalytic activity. Msg5 is a dual-specificity phosphatase that inactivates Fus3. As shown in Fig. 1 B, we assume a distributive kinetic mechanism for the dual phosphorylation and dephosphorylation reactions. That is, two collisions between Fus3 and Ste7 are required for the dual phosphorylation of Fus3. Evidence for a distributive phosphorylation mechanism comes from work on MAPKK-1 phosphorylation of p42 MAPK in Xenopus (13), where it was shown that during the phosphorylation process the amount of monophosphorylated MAPK exceeded the amount of dually phosphorylated MAPK. This result is only possible if two collisions between the MAPKK and MAPK are required for full phosphorylation. We also assume that two collisions between Fus3 and Msg5 are required to convert the doubly phosphorylated Fus3 back to the unphosphorylated state (7). The biochemical reactions used in the computational model are given in Appendix A. Where available, we use experimentally measured values for the model parameters. For the parameters that have not been measured, we use biologically realistic values. A summary of the model parameters and the values used in the simulations is given in Table 1.
A surprising property of the reaction scheme illustrated in Fig. 1 B is that the system can exhibit bistability in the absence of feedback regulation (7). Bistability refers to the situation in which the system possesses two stable steady states and is generally attributed to feedback regulation. Fig. 3 shows a plot of the steady-state values of the doubly-phosphorylated (active) Fus3 concentration [Kpp]ss as a function of the total (phosphorylated and unphosphorylated) Ste7 concentration [KK]T. This type of graph, often referred to as a bifurcation diagram, illustrates where qualitative changes to the steady-state values of [Kpp]ss occur. If an effective cell volume of 30 μm3 is assumed, then the concentrations of Fus3, Ste7, and Msg5 used to generate Fig. 3 correspond to total molecular abundances of 9000, 700∼1200, and 1800, respectively, which are similar to experimentally determined values (18,22). The solid portions of the curve indicate stable steady states and the dashed portion indicates unstable steady states. As is typical of bistable systems, in the bistable region there exist three steady states for each value of the total Ste7 concentration [KK]T, two stable and one unstable. Because the unstable steady state is unstable against all perturbations, it is not experimentally observable.
FIGURE 3.
A bifurcation diagram illustrating bistability in the reactions shown in Fig. 1 B. The steady-state values of the activated Fus3 concentration [Kpp]ss are plotted as a function of the total Ste7 concentration [KK]T. The solid portion of the curve indicates stable steady states and the dashed portion unstable steady states. The bistable region occurs between 44.6 nM and 58.4 nM. Assuming a cell volume of 30 μm3, this corresponds to 802 and 1051 Ste7 molecules, respectively. The values of the model parameters used to produce this plot are given in Table 1.
The effects of random fluctuations in protein concentrations can be investigated by considering a stochastic model of the system. Concentration fluctuations enable the system to undergo random transitions between the two stable states in the bistable region. Therefore, a histogram of protein concentration taken from a population of cells exhibiting bistability would be bimodal with a subpopulation of cells in an activated state and another subpopulation in the inactive state. Equivalently, histograms generated from a sufficiently long time-series from a single cell would also be bimodal. However, stochastic modeling revealed that when realistic protein abundances are used, the average time for spontaneous transitions between stable steady states to occur is much longer than all biologically relevant timescales. This is illustrated in Fig. 4. This figure shows simulated time-series of the activated Fus3 molecule number for cases near the bifurcation point at 58.4 nM (1051 Ste7 molecules/cell). Proceeding from top to bottom, each panel of this figure corresponds to the addition of one molecule of Ste7. In each panel, two different sets of initial conditions were used; one set was chosen to be near the steady state corresponding to low levels of activated Fus3 and the other set corresponds to high levels of activated Fus3. In each graph only a single time-series started near the high state is shown, because multiple runs did not generate any transitions to the low state. This indicates that the activated state is very stable against fluctuations in protein concentration. Each graph in Fig. 4 shows 10 time-series started near the low state. The top panel corresponds to a case in which two stable steady states exist, and the transitions from low to high occur infrequently. The bottom panel corresponds to a case with a unique steady state. In this case, the system transitions from low to high levels of activated Fus3 relatively quickly. That is, the average transition time is noticeably shorter even though the top and bottom panels only differ by two Ste7 molecules. Although this sensitivity to molecular abundance is surprising and of theoretical interest, it seems unlikely to have biological significance and does not underlie any of the results presented here. Note, however, that there is considerable variability in the transition time even when a single steady state exists (Fig. 4, middle and bottom). This variability continues for Ste7 concentrations well beyond the region of bistability and is sufficient to generate a binary response. The model shows a similar sensitivity on the deactivation time to Msg5 levels (data not shown).
FIGURE 4.
Time-series of the number of activated Fus3 molecules from stochastic simulations. Moving down the column corresponds to adding one molecule of Ste7 to the system. The plots illustrate pathway activation as the system moves out of the bistable regime through the bifurcation at 58.4 nM of Ste7 (1051 molecules/cell). Each graph shows 10 time-series started with identical initial conditions near the low state and one time-series started near the high state. As can be seen, the transition time from the low to high state is very sensitive to Ste7 molecule number. Also shown in the two lower panels are the results from the rate equation for [Kpp] (solid black lines). To convert to molecule number, the concentration was multiplied by the volume and Avogadro's number. In the top panel, the system is in the bistable regime so the solution to the rate equation (not shown) remains near the low state.
The solid lines shown in the bottom and middle panels of Fig. 4 are time-series from the rate equations for the protein concentrations (see Materials and Methods and Appendix A). Notice that these trajectories show a long time delay before the system moves from the low to high state. This time lag is referred to as a bottleneck (23). The bottleneck occurs for values of the total Ste7 concentration just beyond the bistable region shown in Fig. 3, because at low activated Fus3 concentrations Fus3 phosphorylation and dephosphorylation rates nearly balance. However, the dephosphorylation rate is slightly smaller and not able to maintain Fus3 in a deactivated state.
The time delay in Fus3 activation produced by the model might underlie the delay in Sst2 induction observed experimentally in cells overexpressing Sst2 (19). We next investigated if a transient binary response could be generated even for values of the total Ste7 concentration that do not generate bistability. In Fig. 5, we present results from stochastic simulations using a total Ste7 molecule number of 1062. For this value, the system is still close enough to the transition to bistability so that the rate equations produce a time lag (black solid line, Fig. 5 A). To simulate pathway activation the system was started near the low activated Fus3 steady state. Next 1250 sample paths were generated, 10 of which are shown in Fig. 5 A. The dashed line shown in this figure is the result of averaging the sample paths. This curve agrees fairly well with the rate equation result. However, the fluctuations cause the response to become less sharp. Next the sample paths were used to generate histograms of the activated Fus3 at several different times. The results are shown in Fig. 5 B. As can be seen, the system exhibits a transient binary response. That is, at intermediate times the distribution of activated Fus3 is bimodal, whereas at long times the distribution is centered at the high state, indicating that all the cells have become activated. This transient binary response can be observed for Ste7 concentrations well beyond the bistable region; however, the time lag becomes less pronounced.
FIGURE 5.
(A) Simulation time-series for the activated Fus3 molecule number NKpp. In this figure, the total Ste7 molecule number is 1062 (59 nM) The solid line is the result from the rate equations and the dashed line is the result from averaging 1250 realizations of the process. (B) Histograms from the stochastic simulations at times 50, 65, 69, and 80 min. Panels A and B illustrate that the Fus3 activation can account for the delay in the Sst2 dynamics and the transient binary response that are observed experimentally.
We note that a transient binary response is also observed if the pathway is operating within the bistable regime where the activated state is sufficiently stable so that transitions back to the deactivated state do not occur on biologically relevant timescales (<2 h). In this case, the time-series of Fus3 activation does not show the S-shape seen in Fig. 5 A. That is, the initial lag phase leading up to Fus3 activation is not present. Also, unless the system is very close to the transition to bistability at 44.6 nm, the lower stable steady state is sufficiently stable that pathway activation does not occur on timescales consistent with the experimental results.
The stochastic model can now be used to interpret the results of the single cell fluorescence experiments shown in Fig. 2. We postulate that under normal conditions in wild-type cells, the pathway is not bistable, but operating sufficiently close to a bistable region to generate a transient binary response. For the sst2Δ strain, the pathway is operating well beyond the bistable regime and therefore always responds in a graded fashion. For the 2XSst2 strain at moderate pheromone concentrations (1–3 μM), the pathway is just to the right of the bistable regime. This produces the transient binary response as well as the previously documented delay in Sst2 induction (19). For 2XSst2 strain, low pheromone concentrations (<0.3 μM) are insufficient to generate a response (data not shown). This indicates that the pathway is operating in the bistable region or to the left of it. If this model is correct, and if the binary-to-graded response is truly mediated at the level of Fus3, it should be possible to predict the outcome of experiments in which the proteins that regulate Fus3 are perturbed through deletion or twofold overexpression of the gene.
Experimental analysis of model predictions
Although the model described above lacks many biological details, it does make predictions that can be tested experimentally. Sst2 is a negative regulator of the pheromone response pathway. Deletion of Sst2 resulted in a temporally graded response for each concentration of pheromone we tested. Within the context of the computational model, deletion of Sst2 increases the amount of active Ste7 and moves the pathway away from the bistable regime shown in Fig. 3. Msg5 is a negative regulator of the pathway that dephosphorylates Fus3. Therefore, increasing the activity or expression of Msg5 should counteract the effect of deleting Sst2 and restore the binary response. To test this prediction, we engineered cells lacking Sst2 to express twice the normal amount of Msg5 (sst2Δ/2XMsg5). We then performed single cell fluorescence measurements of transcription at various pheromone concentrations. Whereas the parent sst2Δ strain shows the typical graded response, the sst2Δ/2XMsg5 strain exhibits a transient binary response for all pheromone concentrations tested. The results for the sst2Δ/2XMsg5 strain are shown in the third column of Fig. 2 A and the third row of Fig. 2 B. These results indicate that modest overexpression of Msg5 can restore the transient binary response in cells lacking Sst2, supporting the hypothesis that the graded-to-binary conversion occurs at the level of the MAPK Fus3.
Sst2 synthesis is induced upon exposure to pheromone (24). Induction of SST2 requires the transcription factor Ste12, which is itself phosphorylated and activated by Fus3 (25). Conversely, the activity of Fus3 is attenuated by Sst2 acting on the G-protein. We recently reported that cells engineered to overexpress Sst2 show a time delay in transcriptional induction (19). Computational modeling suggested that alterations in Fus3 activity could account for the observed delay in Sst2 induction. The model predicts that the delay results from a bottleneck that occurs near the transition to bistability. That is, for the 2XSst2 strain at 3 μM of pheromone, the pathway operates just to the right of bistable region shown in Fig. 3. If this mechanism is correct, then increasing the expression of a positive regulator of Fus3 should remove the time delay by moving the system further away from the bistable region. To test this possibility, we engineered cells to overexpress the MAPKK Ste7 in 2XSst2 strains (2XSst2/GAL-STE7). This was accomplished by inserting a plasmid containing the STE7 gene under the control of the galactose-inducible GAL1/10 promoter. Fig. 6, A and B, shows time-series for Sst2 induction in the 2XSst2/GAL-STE7 and 2XSst2 strains grown in galactose. In agreement with the model, the 2XSst2 strain shows a delayed response, whereas in the 2XSst2/GAL-STE7 strain the delay is absent. These results are in agreement with our model in which the biochemical steps that regulate MAPK activation mediate the graded-to-binary conversion.
FIGURE 6.
(A) Whole-cell extracts were prepared from wild-type cells transformed with a single copy plasmid (pRS316) containing genomic SST2 (2XSST2) and either an empty vector (pYES) or the same vector containing STE7 under control of the galactose-inducible GAL1/10 promoter (GAL-STE7). Cells treated with 3 μM α-factor for the indicated times, collected, resolved by 7.5% SDS-PAGE and immunoblotting, and probed using anti-Sst2 polyclonal antiserum as indicated (IB). The specificity of each antibody was confirmed using gene deletion or diploid cells lacking the indicated gene product (19). (B) To estimate the difference in protein expression the Sst2 band was analyzed by densitometric scanning.
Protein synthesis and degradation
The modeling results above indicate that small changes in Ste7 abundance can have profound effects on Fus3 phosphorylation. A similar sensitivity on protein abundance was found for the dephosphorylation of Fus3 by Msg5. It was recently demonstrated that exposure to pheromone increases the degradation rate of Ste7 (9). To further investigate the role of protein turnover in the regulation of the pheromone response pathway, we monitored the effects of pheromone on the degradation of Fus3 and Msg5. After 1 h of growth in the absence or presence of pheromone the cells were treated with cycloheximide to block new protein synthesis. Steady-state levels of Fus3 and Msg5 remaining were then monitored by immunoblotting. As shown in Fig. 7, Msg5 abundance declined quickly, and this decline was marginally faster when the cells were pretreated with pheromone. A different pattern of degradation was observed for Fus3. In this case degradation was fairly slow, and was slowed even further after pheromone treatment. These results are in contrast to Ste7, which is degraded more slowly than Msg5 and more rapidly than Fus3, and degradation is accelerated by pheromone treatment.
FIGURE 7.
Cells containing an integrated TAP-tagged form of (A) Fus3 or (B) Msg5 were treated with 3 μM α-factor for 60 min, and then treated with the protein synthesis inhibitor cycloheximide (CHX) for the indicated times. Cell extracts were analyzed by immunoblotting with anti-protein A antibodies. To estimate the difference in protein half-life, the intensity of each band was analyzed by densitometric scanning and expressed as a percentage of the amount of protein at the beginning of cycloheximide treatment.
Based on these experimental results and the sensitivity of Fus3 activation to Ste7 and Msg5 levels observed in the stochastic simulations, we expanded the computational model to include protein synthesis and degradation. Modeling protein degradation requires assumptions about when proteins are susceptible to proteolysis. We limit our investigations to the Case II and Case III scenarios discussed in the Materials and Methods. In Case I, protein synthesis and degradation are ignored. In Case II, each protein is degraded at a rate that is independent of its chemical state, and in Case III, each protein is protected from degradation when it forms a multimeric complex or is phosphorylated. Clearly, there are other possibilities for modeling protein degradation, and the mechanisms for protein stabilization and destabilization are not fully understood. However, these two scenarios represent extreme cases, and serve to illustrate the important effects protein synthesis and degradation have on the dynamics of the system.
We start with Case II, in which each protein is degraded at a rate that is independent of its chemical state. The data shown in Fig. 7 indicate that, in the presence of pheromone, Fus3 is stable for the duration of the experiment. Therefore, we only consider the degradation and synthesis of Ste7 and Msg5. At steady state, the total concentrations of Ste7  and
and 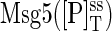 are given by the ratio of their synthesis rate to their degradation rate (see Materials and Methods). The degradation rate δP of Msg5 was estimated from the data presented in Fig. 7 and the data of Wang and Dohlman (9) was used to estimate the degradation rate δKK of Ste7 (see Table 1). Fig. 8 A is a bifurcation diagram for this system as a function of the Ste7 synthesis rate γKK. In this figure, the synthesis rate γP of Msg5 was chosen to produce molecular abundances similar to those measured experimentally. The bifurcation diagram is very similar to the one shown in Fig. 3, except that the model parameter being varied in this case is the Ste7 synthesis rate rather than the Ste7 abundance. We performed stochastic simulations to investigate if a binary response is possible in this case. Initially, the protein levels were taken to be at their steady-state values for a Ste7 synthesis rate of γKK = 0.224 s−1. The model does not consider upstream elements of the pathway such as the MAPK kinase kinase Ste11. Therefore, to simulate pathway activation the Ste7 synthesis rate was increased to γKK = 0.339 s−1 at t = 0. Because our model does not take into account the phosphorylation of Ste7, mathematically increasing the synthesis rate of Ste7 is similar to modeling the pheromone-induced activation of Ste7 by Ste11. Increasing the Ste7 synthesis rate moves the pathway just beyond the bistable regime (see Fig. 8 A). Fig. 8 B shows a simulated time-series for Fus3 activation. Because the total molecule numbers of Ste7 and Msg5 now fluctuate, there is considerably more noise in the system (compare with Fig. 5 A). This increase in fluctuations allows the system to transition between the two steady states more rapidly. This can be seen by comparing the rate equation result (solid line) and the result from averaging 500 realizations of the process (dashed line). The increased fluctuations are almost sufficient to eliminate the time lag. However, Fig. 8 C shows that the system still produces a transient binary response. Notice that the timescale for the pathway to respond is now considerably longer than that for the activation of Fus3 shown in Fig. 5 A. This is due to the fact that we have simulated pathway activation by increasing the synthesis rate as a surrogate for increasing the phosphorylation rate. Phosphorylation of Ste7 by Ste11 occurs on a faster timescale than protein synthesis. In fact, in Fig. 5 it was assumed to occur instantaneously. However, as long as Ste7 activation is not rate-limiting, we expect an extended model that includes Ste7 regulation to respond on a timescale similar to that observed experimentally. Case II represents an extreme scenario in which proteins can be degraded regardless of the chemical state. Therefore, this case maximizes the effects of biochemical fluctuations. One consequence of the large fluctuations is that the time lag produced by the rate equations is almost eliminated. In contrast, Case I minimizes the effects of fluctuations, because variability in concentration levels that arises from protein synthesis and degradation events is not present in this model.
are given by the ratio of their synthesis rate to their degradation rate (see Materials and Methods). The degradation rate δP of Msg5 was estimated from the data presented in Fig. 7 and the data of Wang and Dohlman (9) was used to estimate the degradation rate δKK of Ste7 (see Table 1). Fig. 8 A is a bifurcation diagram for this system as a function of the Ste7 synthesis rate γKK. In this figure, the synthesis rate γP of Msg5 was chosen to produce molecular abundances similar to those measured experimentally. The bifurcation diagram is very similar to the one shown in Fig. 3, except that the model parameter being varied in this case is the Ste7 synthesis rate rather than the Ste7 abundance. We performed stochastic simulations to investigate if a binary response is possible in this case. Initially, the protein levels were taken to be at their steady-state values for a Ste7 synthesis rate of γKK = 0.224 s−1. The model does not consider upstream elements of the pathway such as the MAPK kinase kinase Ste11. Therefore, to simulate pathway activation the Ste7 synthesis rate was increased to γKK = 0.339 s−1 at t = 0. Because our model does not take into account the phosphorylation of Ste7, mathematically increasing the synthesis rate of Ste7 is similar to modeling the pheromone-induced activation of Ste7 by Ste11. Increasing the Ste7 synthesis rate moves the pathway just beyond the bistable regime (see Fig. 8 A). Fig. 8 B shows a simulated time-series for Fus3 activation. Because the total molecule numbers of Ste7 and Msg5 now fluctuate, there is considerably more noise in the system (compare with Fig. 5 A). This increase in fluctuations allows the system to transition between the two steady states more rapidly. This can be seen by comparing the rate equation result (solid line) and the result from averaging 500 realizations of the process (dashed line). The increased fluctuations are almost sufficient to eliminate the time lag. However, Fig. 8 C shows that the system still produces a transient binary response. Notice that the timescale for the pathway to respond is now considerably longer than that for the activation of Fus3 shown in Fig. 5 A. This is due to the fact that we have simulated pathway activation by increasing the synthesis rate as a surrogate for increasing the phosphorylation rate. Phosphorylation of Ste7 by Ste11 occurs on a faster timescale than protein synthesis. In fact, in Fig. 5 it was assumed to occur instantaneously. However, as long as Ste7 activation is not rate-limiting, we expect an extended model that includes Ste7 regulation to respond on a timescale similar to that observed experimentally. Case II represents an extreme scenario in which proteins can be degraded regardless of the chemical state. Therefore, this case maximizes the effects of biochemical fluctuations. One consequence of the large fluctuations is that the time lag produced by the rate equations is almost eliminated. In contrast, Case I minimizes the effects of fluctuations, because variability in concentration levels that arises from protein synthesis and degradation events is not present in this model.
FIGURE 8.
(A) A bifurcation diagram showing the steady-state concentration of activated Fus3 concentration [Kpp] as a function of the Ste7 synthesis rate γKK for Case II (no protection from degradation). (B) Simulation time-series for the activated Fus3 molecule number NKpp. In this figure the Ste7 synthesis rate is 0.339 s−1. The solid line is the result from the rate equations and the dashed line is an average over 500 realizations of the process. The discrepancy between these two curves is due to the large effects of the fluctuations in the activated Fus3 concentration. These fluctuations eliminate the time lag in Fus3 activation. (C) Histograms of activated Fus3 molecule number from the stochastic simulations at times 150, 250, 300, and 350 min. Even though the system no longer shows a lag in Fus3 activation, the transient binary response is still produced.
We now move to an intermediate case in which protein-protein interactions (such as those leading to protein dimerization) and phosphorylation protect proteins from degradation. It is commonly observed that proteins in a functional complex are more stable than when expressed alone. One familiar example is the Gβ- and Gγ-subunits, neither of which can be stably expressed or purified in the absence of the other. For simplicity, we start with the case in which only Ste7 is degraded and synthesized, with the levels of total Fus3 and Msg5 remaining constant. That is, d[K]T/dt = d[P]T/dt = 0 and [KK]T satisfies Eq. 17, above. The rate equations for the rest of the chemical species remain unchanged except for the equation for the Ste7 concentration [KK], which now includes the same synthesis and degradation terms as in Eq. 17. Equation 17 implies that at steady state the free Ste7 concentration is given by the synthesis rate γKK divided by the degradation rate δKK. Fig. 9 A shows a plot of the total Ste7 concentration 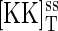 as function of the synthesis rate γKK. Surprisingly this plot is not monotonic, but goes through a local maximum when γKK is ∼0.7 s−1. This effect is a result of the protein complexes being protected from degradation and the dual phosphorylation reaction. At low levels of Ste7, most of the Fus3 is in the unphosphorylated or singly phosphorylated state. Ste7 interacts strongly with these two states and is then protected from degradation. As Ste7 levels increase, the reactions in Fig. 1 B favor the dually phosphorylated state. Therefore, there is less substrate for Ste7 to interact with and to protect from degradation. As the synthesis rate is increased further, the total amount of Ste7 again begins to increase. To illustrate the counterintuitive observation that increasing synthesis rate of Ste7 can lead to a decrease in its total concentration, the degradation rate of Ste7 was artificially increased to δKK = 4 × 10−3 s−1. If the experimentally measured value δKK = 3.2 × 10−4 s−1 is used, the results shown in Fig. 9 B are produced. As can be seen, the steady value of the total Ste7 concentration still goes through a local maximum. However, where the concentration declines, the steady state is unstable (dashed line). In this region, the system produces sustained oscillations. An example of these oscillations is shown in the inset. The circles shown in Fig. 9 B indicate the minimum and maximum values of the total Ste7 concentration. This model can also exhibit bistability (see Appendix B). However, the parameter range over which two stable steady states exist appears to be limited.
as function of the synthesis rate γKK. Surprisingly this plot is not monotonic, but goes through a local maximum when γKK is ∼0.7 s−1. This effect is a result of the protein complexes being protected from degradation and the dual phosphorylation reaction. At low levels of Ste7, most of the Fus3 is in the unphosphorylated or singly phosphorylated state. Ste7 interacts strongly with these two states and is then protected from degradation. As Ste7 levels increase, the reactions in Fig. 1 B favor the dually phosphorylated state. Therefore, there is less substrate for Ste7 to interact with and to protect from degradation. As the synthesis rate is increased further, the total amount of Ste7 again begins to increase. To illustrate the counterintuitive observation that increasing synthesis rate of Ste7 can lead to a decrease in its total concentration, the degradation rate of Ste7 was artificially increased to δKK = 4 × 10−3 s−1. If the experimentally measured value δKK = 3.2 × 10−4 s−1 is used, the results shown in Fig. 9 B are produced. As can be seen, the steady value of the total Ste7 concentration still goes through a local maximum. However, where the concentration declines, the steady state is unstable (dashed line). In this region, the system produces sustained oscillations. An example of these oscillations is shown in the inset. The circles shown in Fig. 9 B indicate the minimum and maximum values of the total Ste7 concentration. This model can also exhibit bistability (see Appendix B). However, the parameter range over which two stable steady states exist appears to be limited.
FIGURE 9.
(A) The steady-state total Ste7 concentration  as a function of its synthesis rate γKK. The total concentration goes through a local maximum near γKK = 0.7 s−1. This results from protection from degradation and the dual phosphorylation reactions. In this figure the degradation rate was artificially increased to δKK = 4 × 10−3 s−1 to illustrate the nonmonotonic behavior. (B) When the experimentally determined value δKK = 3.2 × 10−4 s−1 is used, the steady-state total Ste7 concentration
as a function of its synthesis rate γKK. The total concentration goes through a local maximum near γKK = 0.7 s−1. This results from protection from degradation and the dual phosphorylation reactions. In this figure the degradation rate was artificially increased to δKK = 4 × 10−3 s−1 to illustrate the nonmonotonic behavior. (B) When the experimentally determined value δKK = 3.2 × 10−4 s−1 is used, the steady-state total Ste7 concentration  again shows nonmonotonic behavior. However, in the case the steady-state is not stable for the part of the curve with negative slope (dashed line). In this region the system undergoes sustained oscillations. The minimum and maximum values of [KK]T are plotted as circles. (Inset) A time-series [KK]T of illustrating the periodic behavior. In this figure, γKK = 0.1 s−1.
again shows nonmonotonic behavior. However, in the case the steady-state is not stable for the part of the curve with negative slope (dashed line). In this region the system undergoes sustained oscillations. The minimum and maximum values of [KK]T are plotted as circles. (Inset) A time-series [KK]T of illustrating the periodic behavior. In this figure, γKK = 0.1 s−1.
We now consider the case in which the uncomplexed/free forms of Fus3 and Msg5 also are degraded. In this case, the total concentrations satisfy Eqs. 16–18. To estimate the degradation rate δK of Fus3, we used the experimental results in absence of pheromone presented in Fig. 7. The rationale for this decision is that, in the absence of pheromone, most Fus3 molecules are in the inactive state and available for degradation, whereas, upon pheromone stimulation, phosphorylation of Fus3 leads to kinase/substrate interactions that protect the protein from degradation. Therefore, the data in the absence of pheromone provide a good estimate for the degradation rate of free inactive Fus3. If this reasoning is correct, then it implies that the pheromone-induced increase in the degradation rate of Ste7 is actually larger than that reported in Table 1, because these estimates did not take into account protection against degradation through protein/protein interactions. However, for simplicity we assume that the rates estimated from the data for these two proteins are the rates at which free protein is degraded. The synthesis rates were chosen to produce protein numbers consistent with experimental measurements. In this case, the steady-state concentrations of free Fus3, Ste7, and Msg5 are given by the ratio of their synthesis rates and degradation rates (i.e., [K]ss = γK/δK, [KK]ss = γKK/δKK, and [P]ss = γP/δP), and it is straightforward to show that the system has a unique steady state. That is, this system cannot be bistable. Fig. 10 A is a bifurcation diagram for the system as a function of the Ste7 synthesis rate γKK. At small values of γKK, the system always approaches steady state. However, at values greater than γKK = 0.013 s−1, the system undergoes sustained oscillations in concentration. The circles again indicate the maximum and minimum values of the activated Fus3 concentration [Kpp]. The periodic behavior is illustrated in Fig. 10 B, which shows time-series of the total Fus3 concentration [K]T (solid line), the activated Fus3 concentration [Kpp] (shaded line), and the unphosphorylated Fus3 concentration [K] (dashed line).
FIGURE 10.
(A) A bifurcation diagram showing the activated Fus3 concentration [Kpp] as a function of its synthesis rate γKK. For small values of γKK there is a single stable steady state (solid line). At γKK = 0.013 s−1, this steady state becomes unstable (dashed line) and the system undergoes sustained oscillations (circles). (B) Time-series of the total Fus3 concentration [K]T (solid line), the activated Fus3 concentration [Kpp] (shaded line), and the inactive Fus3 concentration [K] (dashed line). In this figure, γKK = 0.016 s−1. (C) A plot of the activated the free Ste7 concentration [KK] versus the total Ste7 concentration [KK]T (dotted line) using the same value of γKK as in B. Also drawn on this figure is the bifurcation diagram generated by using the time-averaged values of [K]T and [P]T. The periodic trajectory closely follows the upper and lower branches of this curve and rapidly transition between the two branches near the bifurcation points.
The qualitative explanation for the origin of the oscillations is that protein degradation acts as a negative feedback on the bistable system. The combination of bistability and negative feedback often leads to periodic behavior referred to as hysteresis oscillations (6). This effect is illustrated in Fig. 10 C, which is a plot of the free Ste7 concentration [KK] versus the total Ste7 concentration [KK]T. Also drawn is the bifurcation curve computed under the assumption of constant enzyme concentrations (i.e., no synthesis or degradation). Therefore, to draw the bifurcation curve the time-averaged values of the total Fus3 and Msg5 concentrations were used. As can be seen, the periodic trajectory closely follows the upper and lower branches of the curve in the region of bistability, rapidly jumping between the two branches at the bifurcation points.
Stochastic modeling and oscillations
We next performed stochastic simulations to determine how random fluctuations in protein abundance affect the oscillatory dynamics described above. One important result of these investigations is that the fluctuations increase the parameter range over which the system exhibits oscillatory behavior. Fig. 11 A shows a time-series from a stochastic simulation. In this figure, the Ste7 synthesis rate is γKK = 0.01 s−1. For this value, the rate equations predict a stable steady state (see Fig. 10 A). However, the biochemical fluctuations are sufficiently strong to destabilize the steady state and generate behavior that appears periodic. Such behavior is typical of noisy systems near a bifurcation and is referred to as noise-induced oscillations. To investigate the periodicity of the oscillations, we computed the power spectrum of the time-series generated from the stochastic model (Fig. 11 B). A clear peak is seen in the spectrum at a frequency of 8 × 10−4 min−1, indicating that the noise-induced oscillations are indeed periodic. The inset in Fig. 11 B shows a histogram of the interbeat interval (i.e., time between successive peaks in concentration) and provides a measure of the amount of variability in the period of the oscillations. The fact that stochastic effects enlarge the parameter range over which the protein concentrations oscillate makes it more likely that this periodic behavior has biological significance.
FIGURE 11.
Noise-induced oscillations. (A) A time-series from a stochastic simulation with γKK = 0.01 s−1. For this value of the Ste7 synthesis rate the rate equations predict a stable steady state. However, biochemical fluctuations are sufficient to destabilize the steady state leading to sustained oscillations. (B) The power spectrum computed from stochastic simulations using the same parameter values as in A. A clear peak is seen at a frequency of 8 × 10−4 min−1. (Inset) A histogram of the in interbeat interval.
The explanation of the transient binary response required that the pathway operate near a bistable region. However, when synthesis and degradation of all three enzymes are included in the model, the system can no longer exhibit bistability. To investigate if a binary response is possible in this scenario, we performed stochastic simulations of pathway activation. Histograms from the simulations are shown in Fig. 12. To generate the histograms, the stochastic simulations were started near the stable steady state that exists for a Ste7 synthesis rate of γKK = 0.005 s−1. Once again, to model pathway activation, the Ste7 synthesis rate was increased at t = 0. In this case, the increase in synthesis rate to γKK = 0.016 s−1 moves the system into a regime where oscillations occur (see Fig. 10 A). The results presented in Fig. 12 illustrate that the strength of the biochemical fluctuations is sufficient to generate significant variability in the activation time of individual cells and produce a clear binary response. The slow response of the pathway again results from simulating pathway activation by increasing the Ste7 synthesis rate rather than modeling the phosphorylation of Ste7. If larger values of the synthesis rate are used, the response becomes graded (data not shown).
FIGURE 12.
Histograms from the stochastic model when protein synthesis and degradation is considered. These results illustrate that a binary response is possible when the system is undergoing sustained oscillations.
The oscillatory behavior presented above represents a macroscopic phenomenon in the sense that it emerges from the stochastic dynamics only if sufficiently large volumes and protein abundances are considered. To investigate the onset of oscillations, we performed stochastic simulations at various volumes. The synthesis rate and second-order rate constant were scaled appropriately to ensure that the rate equations remained unchanged as the volume was increased (see Materials and Methods). The results are summarized in Figs. 13 and 14. Fig. 13 shows time-series of the active Fus3 molecule number for various volumes. Fig. 13 A corresponds to a volume of 0.6 μm3. For this volume and protein abundance, the time-series is dominated by fluctuations and no periodic behavior is discernable. The steady-state distribution (two-dimensional histogram) for the activated Fus3 and total Ste7 molecule numbers is shown in Fig. 14 A. In this figure, red indicates regions where the system spends large amounts of time and blue indicates regions that are visited infrequently. For this small volume, the steady-state distribution has very little structure. Figs. 13 B and 14 B correspond to a volume of 6.0 μm3. Periodic behavior is beginning to appear in the time-series. However, the steady-state distribution indicates that the system is still dominated by fluctuations. Figs. 13 C and 14 C are for a volume of 30 μm3, which is similar to the volume of a yeast cell. Here, the periodic behavior is clear. The steady-state distribution is clearly structured and lies mostly along the deterministic trajectory (compare Fig. 14, C and D). These investigations reveal that at volumes and protein abundances typical of yeast cells, the qualitative behavior of the system is accurately captured by the rate equations. However, these findings highlight the importance of stochastic simulations to account for the variability observed in single cell measurements.
FIGURE 13.
Simulation time-series of the activated Fus3 molecule number NKpp. (A) At small volumes (0.6 μm3) fluctuations dominate the dynamics, and no periodic behavior is observable. (B) As the volume increases (6 μm3), the fluctuations become less significant and periodic behavior is starting to emerge. (C) At biological realistic volumes (30 μm3) periodic behavior is clearly observable. (D) The result from the rate equations.
FIGURE 14.
(A–C) The corresponding steady-state distribution for the activated Fus3 molecule number NKpp and total Ste7 molecule number NKKT for the cases shown in Fig. 13. In these figures, red corresponds to regions where the system spends large amounts of time and blue regions are visited infrequently. (D) Plot of NKpp versus NKKT computed from the rate equations.
DISCUSSION
The mating response system in yeast is arguably the best-characterized signaling pathway of any eukaryote, and it has long served as a prototype for hormone, neurotransmitter, and sensory response systems in humans. At the receptor level, signal transduction occurs in a graded fashion, with pathway activation proportional to the agonist concentration. However, downstream components of the pathway can convert the graded response to a binary one. A common mechanism for achieving this conversion is through positive feedback (6). For example, we recently demonstrated that pheromone promotes transcriptional induction as well as ubiquitin-mediated degradation of Sst2, a negative regulator of the pheromone pathway. These pheromone-regulated changes in expression are likely to be functionally important, since twofold overexpression of Sst2 converts the normally graded response into a binary response (19). Moreover, induction of Sst2 expression by pheromone is delayed when Sst2 is overexpressed (2XSst2 strain). These results led us to suggest a model in which pheromone-induced degradation of Sst2 acts as a positive feedback mechanism to counteract the negative effects of Sst2 on G-protein signaling. The model of Sst2 regulation also suggested that the binary response exists only transiently, with all cells eventually becoming activated.
Although Sst2 acts at the G-protein level, other steps of the pathway may likewise regulate the graded-to-binary transformation. Poritz et al. (21) demonstrated, using sst2Δ mutants, that inhibiting the pathway downstream of the G-protein also converts a graded response to a binary one. In this case, the binary response was long-lived, existing for several hours. To further investigate pathway activation and attenuation, we performed single cell fluorescence-based transcription measurements on sst2Δ, wild-type, and 2XSst2 strains. The wild-type and 2XSst2 cells were both found to exhibit a transient binary response, with all the cells becoming activated within 2 h. The sst2Δ mutant strain, in contrast, showed a graded response. These new findings coupled with our previous work and the results of Poritz et al. (21) led us to investigate the regulation of the protein kinase Fus3 as a potential mediator of the graded-to-binary response. We focused on Fus3, because previous theoretical results of Markevich et al. (7) demonstrated that a distributive kinetic mechanism for the dual phosphorylation of protein kinases is sufficient to generate bistability. Our stochastic modeling revealed that this mechanism, applied to Fus3 regulation, can account for all the experimental observations outlined above. To test the validity of the model, we genetically altered expression of proteins that normally regulate Fus3. As predicted by the model, increasing expression of Msg5, a negative regulator of the pathway, counteracted the positive effects of deleting Sst2 and restored the binary response. Conversely, to counteract the increased negative effects of Sst2 in the 2XSst2 strain, we engineered cells to overexpress Ste7. In full agreement with model predictions, increased levels of Ste7 had the effect of removing the time delay in Sst2 induction.
Our computational and experimental analysis suggests that Fus3 regulation is responsible for the graded-to-binary conversion in the yeast pheromone response. However, the possibility that this conversion takes place downstream of Fus3 cannot be ruled out. For example, Blake et al. (26) used a stochastic model of transcription initiation to show that pulsatile mRNA production, through reinitiation of transcription, could account for the binary transcriptional response observed for the yeast GAL1 promoter. However, our ability to predict the results of altering the expression of proteins that regulate Fus3 provides strong evidence that this step of the pathway mediates the graded-to-binary conversion in the pheromone response pathway.
Protein kinase cascades are a reoccurring feature of signal transduction pathways and are found in all eukaryotic cells. For this reason, many recent theoretical investigations have focused on understanding their behavior (7,27–33). Protein kinase cascades have been shown to exhibit ultrasensitivity (28) and to lead to bistability and sustained oscillations of concentration levels in the presence of feedback regulation (30,31,33). Here we have demonstrated that a MAPK cascade can generate sustained oscillations in the absence of feedback regulation. This result builds on the work of Markevich et al. (7), who demonstrated that multisite phosphorylation in protein kinase cascades is sufficient to generate bistability. We have also shown that periodic behavior occurs when protein synthesis and degradation are included in the model of MAPK regulation. Thus, protein turnover, which is often overlooked in MAPK signaling, can profoundly influence the response of a signaling pathway and may provide an important regulatory mechanism. In our model, protein degradation acts as a negative feedback on a bistable system and produces periodic behavior through hysteresis oscillations. The oscillations occur when physical modifications such as protein oligomerization and phosphorylation protect enzymes against degradation. The period of the oscillations is determined mainly by the protein degradation rate. Taken together, these findings highlight the importance of considering protein stability and degradation in generating models of biological processes.
One measure of our understanding of biological systems is our ability to predict their behavior in detail. Intracellular signaling pathways are highly nonlinear and often regulated by multiple feedback loops. Additionally, these networks are subject to stochastic fluctuations that arise from the random nature of biochemical reactions. These features make predicting a pathway's response to an external stimulus difficult, if not impossible, without the aid of mathematical modeling. Our stochastic modeling of the biochemical steps that regulate the MAPK Fus3 reveal that very small changes in the abundance of the MAPKK Ste7 have a significant effect on pathway activation. Such a high sensitivity to Ste7 levels might underlie the cell's ability to convert a small external signal into a strongly amplified response. Any computational model necessarily includes biological assumptions and mathematical simplifications. For example, many models of signaling pathways ignore protein synthesis and degradation. However, our analysis reveals that including protein turnover can lead to sustained oscillations in protein concentrations that are likely to have biological significance. These findings demonstrate that mathematical modeling, when combined with experimental analysis, provides a powerful tool for understanding the complex behavior of signaling pathways.
Acknowledgments
We thank Yuqi Wang and Christine Fraser for providing the protein turnover data.
This work was supported by National Institutes of Health grants No. GM059167 (to H.G.D.) and No. GM073180 (to H.G.D. and T.C.E.), and DARPA grant No. F30602-01-2-0579 (to T.C.E.).
APPENDIX A: BIOCHEMICAL REACTIONS
In this Appendix, we list the biochemical reactions used in the stochastic simulations and rate equations. Case I, in which proteins are not synthesized or degraded, consists of the following reactions:
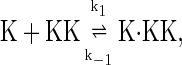 |
(19) |
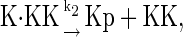 |
(20) |
 |
(21) |
 |
(22) |
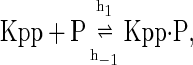 |
(23) |
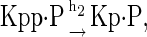 |
(24) |
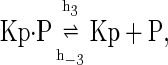 |
(25) |
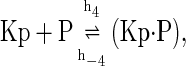 |
(26) |
 |
(27) |
 |
(28) |
The above reactions are identical to the ones considered by Markevich et al. (7). Both phosphorylation events follow standard Michaelis-Menten kinetics, Eqs. 19–22. Dephosphorylation occurs in two chemical steps. First the phosphate is released, Eqs. 24 and 27, and then the kinase and phosphatase dissociate, Eqs. 25 and 28. The dissociation of the kinase and phosphatase is assumed to be reversible. The backward rate constants, h−3 and h−6, for this process can be taken to be zero without significantly changing the results. In this case, the kinetics is essentially Michaelis-Menten. The chemical species (Kp·P) in Eqs. 26 and 27 results from the assumption that the phosphotyrosine is dephosphorylated first (7,17). That is, the chemical species (Kp·P) indicates the phosphatase attacking the phosphothreonine, whereas Kp·P is the product after phosphotyrosine has been dephosphorylated.In addition to Eqs. 19–28, Case II consists of the following reactions:
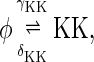 |
(29) |
 |
(30) |
 |
(31) |
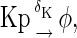 |
(32) |
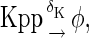 |
(33) |
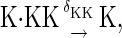 |
(34) |
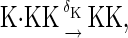 |
(35) |
 |
(36) |
 |
(37) |
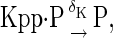 |
(38) |
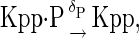 |
(39) |
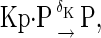 |
(40) |
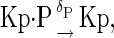 |
(41) |
 |
(42) |
 |
(43) |
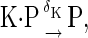 |
(44) |
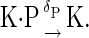 |
(45) |
APPENDIX B: DISCUSSION ON BISTABILITY FOR CASE II
It was shown that the inclusion of protein synthesis and degradation of Ste7 in the mathematical description of Fus3 regulation destroyed bistability in this model for the parameter values listed in Table 1. The addition of protein degradation and synthesis forces the steady-state Ste7 concentration to take the value γKK/δKK, and a plot of the active Fus3 concentration [Kpp]ss versus the synthesis rate γKK, does not have an S-shape similar to Fig. 3. However, if [Kpp]ss is plotted against the total Ste7 concentration 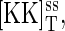 the resulting curve is identical to Fig. 3. The multiple values of [Kpp]ss for a single value of
the resulting curve is identical to Fig. 3. The multiple values of [Kpp]ss for a single value of 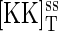 results from the nonmonotonic behavior of
results from the nonmonotonic behavior of 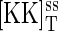 shown in Fig. 9, A and B. The reason why the functional dependence of [Kpp]ss on
shown in Fig. 9, A and B. The reason why the functional dependence of [Kpp]ss on  is the same in this case as it is in Case I (no synthesis or degradation) is as follows. The concentrations of all the chemical species satisfy exactly the same equations as in Case I, except for the equation for the Ste7 concentration [KK], which contains the two additional terms γKK – δKK [KK]. In Case I, when the system is in the bistable region, the steady-state equations have three solutions. However, Eq. 17 for the total Ste7 concentration imposes an additional constraint on the steady state of the system, [KK]ss = γKK/δKK. This constraint uniquely selects one of the three possible solutions. This argument does not rule out the possibility of bistability in this model for other parameter values. In fact, it can be shown that the system can be bistable. For this to occur, the steady-state equations for the concentrations possess three solutions with identical values of [KK]ss. We have assumed that only the free form of Ste7 is degraded. If we relax this assumption and allow other chemical forms of Ste7 to be degraded, then the constraint [KK]ss = γKK/δKK no longer applies, and it seems likely that the region of parameter values that show bistability would increase.
is the same in this case as it is in Case I (no synthesis or degradation) is as follows. The concentrations of all the chemical species satisfy exactly the same equations as in Case I, except for the equation for the Ste7 concentration [KK], which contains the two additional terms γKK – δKK [KK]. In Case I, when the system is in the bistable region, the steady-state equations have three solutions. However, Eq. 17 for the total Ste7 concentration imposes an additional constraint on the steady state of the system, [KK]ss = γKK/δKK. This constraint uniquely selects one of the three possible solutions. This argument does not rule out the possibility of bistability in this model for other parameter values. In fact, it can be shown that the system can be bistable. For this to occur, the steady-state equations for the concentrations possess three solutions with identical values of [KK]ss. We have assumed that only the free form of Ste7 is degraded. If we relax this assumption and allow other chemical forms of Ste7 to be degraded, then the constraint [KK]ss = γKK/δKK no longer applies, and it seems likely that the region of parameter values that show bistability would increase.
References
- 1.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G-protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703–754. [DOI] [PubMed] [Google Scholar]
- 2.Wang, Y., and H. G. Dohlman. 2004. Pheromone signaling mechanisms in yeast: a prototypical sex machine. Science. 306:1508–1509. [DOI] [PubMed] [Google Scholar]
- 3.Hume, D. A. 2000. Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood. 96:2323–2328. [PubMed] [Google Scholar]
- 4.Ferrell, J. E., Jr. 2002. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14:140–148. [DOI] [PubMed] [Google Scholar]
- 5.Kaern, M., T. C. Elston, W. J. Blake, and J. J. Collins. 2005. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 6:451–464. [DOI] [PubMed] [Google Scholar]
- 6.Tyson, J. J., K. C. Chen, and B. Novak. 2003. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 15:221–231. [DOI] [PubMed] [Google Scholar]
- 7.Markevich, N. I., J. B. Hoek, and B. N. Kholodenko. 2004. Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. J. Cell Biol. 164:353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hicke, L. 1999. Gettin' down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 9:107–112. [DOI] [PubMed] [Google Scholar]
- 9.Wang, Y., and H. G. Dohlman. 2002. Pheromone-dependent ubiquitination of the mitogen-activated protein kinase kinase Ste7. J. Biol. Chem. 277:15766–15772. [DOI] [PubMed] [Google Scholar]
- 10.Esch, R. K., and B. Errede. 2002. Pheromone induction promotes Ste11 degradation through a MAPK feedback and ubiquitin-dependent mechanism. Proc. Natl. Acad. Sci. USA. 99:9160–9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, Y., Q. Ge, D. Houston, J. Thorner, B. Errede, and H. G. Dohlman. 2003. Regulation of Ste7 ubiquitination by Ste11 phosphorylation and the Skp1-Cullin-F-box complex. J. Biol. Chem. 278:22284–22289. [DOI] [PubMed] [Google Scholar]
- 12.Nelson, C., S. Goto, K. Lund, W. Hung, and I. Sadowski. 2003. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 421:187–190. [DOI] [PubMed] [Google Scholar]
- 13.Ferrell, J. E., Jr., and R. R. Bhatt. 1997. Mechanistic studies of the dual phosphorylation of mitogen-activated protein kinase. J. Biol. Chem. 272:19008–19016. [DOI] [PubMed] [Google Scholar]
- 14.Gillespie, D. 1977. Exact stochastic simulation of coupled chemical reactions. J. Chem. Phys. 81:2340–2361. [DOI] [PubMed] [Google Scholar]
- 15.Adalsteinsson, D., D. McMillen, and T. C. Elston. 2004. Biochemical Network Stochastic Simulator (BioNetS): software for stochastic modeling of biochemical networks. BMC Bioinformatics. 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ermentrout, B. 2002. Simulating, Analyzing, and Animating Dynamical Systems: A Guide to XPPAUT for Researchers and Students. Society for Industrial and Applied Mathematics, Philadelphia, PA.
- 17.Zhao, Y., and Z. Y. Zhang. 2001. The mechanism of dephosphorylation of extracellular signal-regulated kinase 2 by mitogen-activated protein kinase phosphatase 3. J. Biol. Chem. 276:32382–32391. [DOI] [PubMed] [Google Scholar]
- 18.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741. [DOI] [PubMed] [Google Scholar]
- 19.Hao, N., N. Yildirim, Y. Wang, T. C. Elston, and H. G. Dohlman. 2003. Regulators of G-protein signaling and transient activation of signaling: experimental and computational analysis reveals negative and positive feedback controls on G-protein activity. J. Biol. Chem. 278:46506–46515. [DOI] [PubMed] [Google Scholar]
- 20.Siekhaus, D. E., and D. G. Drubin. 2003. Spontaneous receptor-independent heterotrimeric G-protein signalling in an RGS mutant. Nat. Cell Biol. 5:231–235. [DOI] [PubMed] [Google Scholar]
- 21.Poritz, M. A., S. Malmstrom, M. K. Kim, P. J. Rossmeissl, and A. Kamb. 2001. Graded mode of transcriptional induction in yeast pheromone signalling revealed by single-cell analysis. Yeast. 18:1331–1338. [DOI] [PubMed] [Google Scholar]
- 22.Bardwell, L., J. G. Cook, E. C. Chang, B. R. Cairns, and J. Thorner. 1996. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol. Cell. Biol. 16:3637–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strogatz, S. H. 1994. Nonlinear Dynamics and Chaos: With Applications to Physics, Biology, Chemistry, and Engineering. Addison-Wesley, Reading, MA.
- 24.Dietzel, C., and J. Kurjan. 1987. Pheromonal regulation and sequence of the Saccharomyces cerevisiae SST2 gene: a model for desensitization to pheromone. Mol. Cell. Biol. 7:4169–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elion, E. A., B. Satterberg, and J. E. Kranz. 1993. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol. Biol. Cell. 4:495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blake, W. J., M. Kaern, C. R. Cantor, and J. J. Collins. 2003. Noise in eukaryotic gene expression. Nature. 422:633–637. [DOI] [PubMed] [Google Scholar]
- 27.Heinrich, R., B. G. Neel, and T. A. Rapoport. 2002. Mathematical models of protein kinase signal transduction. Mol. Cell. 9:957–970. [DOI] [PubMed] [Google Scholar]
- 28.Huang, C. Y., and J. E. Ferrell, Jr. 1996. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. USA. 93:10078–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shvartsman, S. Y., M. P. Hagan, A. Yacoub, P. Dent, H. S. Wiley, and D. A. Lauffenburger. 2002. Autocrine loops with positive feedback enable context-dependent cell signaling. Am. J. Physiol. Cell Physiol. 282:C545–C559. [DOI] [PubMed] [Google Scholar]
- 30.Bhalla, U. S., P. T. Ram, and R. Iyengar. 2002. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science. 297:1018–1023. [DOI] [PubMed] [Google Scholar]
- 31.Kholodenko, B. N. 2000. Negative feedback and ultrasensitivity can bring about oscillations in the mitogen-activated protein kinase cascades. Eur. J. Biochem. 267:1583–1588. [DOI] [PubMed] [Google Scholar]
- 32.Lewis, T. S., P. S. Shapiro, and N. G. Ahn. 1998. Signal transduction through MAP kinase cascades. Adv. Cancer Res. 74:49–139. [DOI] [PubMed] [Google Scholar]
- 33.Yang, L., W. R. MacLellan, Z. Han, J. N. Weiss, and Z. Qu. 2004. Multisite phosphorylation and network dynamics of cyclin-dependent kinase signaling in the eukaryotic cell cycle. Biophys. J. 86:3432–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]

















