Abstract
Reconstitution experiments have suggested that N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) proteins constitute a minimal membrane fusion machinery but have yielded contradictory results, and it is unclear whether the mechanism of membrane merger is related to the stalk mechanism that underlies physiological membrane fusion. Here we show that reconstitution of solubilized neuronal SNAREs into preformed 100 nm liposomes (direct method) yields proteoliposomes with more homogeneous sizes and protein densities than the standard reconstitution method involving detergent cosolubilization of proteins and lipids. Standard reconstitutions yield slow but efficient lipid mixing at high protein densities and variable amounts of lipid mixing at moderate protein densities. However, the larger, more homogenous proteoliposomes prepared by the direct method yield almost no lipid mixing at moderate protein densities. These results suggest that the lipid mixing observed for standard reconstitutions is dominated by the physical state of the membrane, perhaps due to populations of small vesicles (or micelles) with high protein densities and curvature stress created upon reconstitution. Accordingly, changing membrane spontaneous curvature by adding lysophospholipids inhibits the lipid mixing observed for standard reconstitutions. Our data indicate that the lipid mixing caused by high SNARE densities and/or curvature stress occurs by a stalk mechanism resembling the mechanism of fusion between biological membranes, but the neuronal SNAREs are largely unable to induce lipid mixing at physiological protein densities and limited curvature stress.
INTRODUCTION
Fusion of two membranes into a single membrane is a critical event for a wide variety of biological processes, including intracellular transport, fertilization, and viral entry into a host cell. Multiple types of functional and genetic evidence have shown that N-ethylmaleimide sensitive factor attachment protein receptors (SNAREs) play a key role in all steps of the secretory and endocytic pathways in eukaryotic cells (reviewed in (1–5)). Studies of the synaptic vesicle SNARE synaptobrevin/vesicle associated membrane protein and the presynaptic plasma membrane SNAREs syntaxin and SNAP-25 showed that they form a tight complex known as the SNARE complex through coiled-coil sequences called SNARE motifs (6–10). The discovery that this complex involves a parallel interaction between the SNARE motifs of synaptobrevin and syntaxin, which are adjacent to their transmembrane (TM) regions, showed that assembly of the SNARE complex must bring the synaptic vesicle and plasma membranes together and led to the hypothesis that SNARE complex formation may provide the energy for membrane fusion (11,12). The neuronal SNARE complex was later shown to consist of a bundle of four parallel α-helices (two from SNAP-25 and one each from syntaxin and synaptobrevin) (13,14), and characterization of SNAREs from diverse membrane compartments (e.g., Antonin et al. (15)) has indicated that all SNARE complexes adopt similar four-helix bundle structures. In addition, reconstitution experiments revealed lipid mixing between liposomes containing synaptobrevin and liposomes containing syntaxin/SNAP-25 (16), and similar results were obtained with yeast SNAREs involved in different membrane traffic processes (17).
These and other observations have led to a widespread model whereby the SNAREs constitute a minimal machinery for intracellular membrane fusion, but the validity of this model is still under intense debate (18–20). A key problem with this model is that it does not account for the strict requirement of Sec1/Munc18-1 homologs for all types of intracellular membrane fusion (reviewed in Rizo and Sudhof (3)). Moreover, additional proteins appear to act downstream of SNAREs in fusion of egg cortical vesicles (21). The debate over the precise function of the SNAREs also arises in part because of the intrinsic in vitro nature of the reconstitution experiments, which hinders conclusive demonstration of protein function in the absence of strict correlations with in vivo data. In addition, the initial reconstitution experiments with neuronal SNAREs (16) were performed with liposomes containing an exceedingly high amount of synaptobrevin, and the observed rate of lipid mixing (minutes-hours) was very slow compared to the timescale of neurotransmitter release (<0.5 ms). Whereas liposome fusion mediated by neuronal SNAREs was later observed at lower protein/lipid ratios (22), other experiments using different reconstitution schemes revealed no lipid mixing, which was attributed to membrane sequestration of part of the synaptobrevin SNARE motif (23,24). Note also that when SNARE-mediated lipid mixing is observed, the number of rounds of fusion calculated (see Parlati et al. (25)) is significantly lower than expected from the stoichiometry of the reaction mixture. It is also unclear why SNARE-induced fusion of individual vesicles with planar bilayers occurs at much faster timescales (<1 s) than the overall rate of liposome-liposome fusion and why only a small fraction of the vesicles fused in these experiments (26). Particularly worrisome was also the observation that significant lipid mixing was still observed when long linkers that can span 100 Å were introduced between the SNARE motifs and TM regions of syntaxin and synaptobrevin (27), since the linkers would have been expected to uncouple SNARE complex assembly from fusion and it seems highly unlikely that these mutants would be functional in vivo (28).
Despite these caveats and unanswered questions, the reconstitution experiments provide a powerful approach to study the roles of proteins involved in membrane traffic. Whether or not the SNAREs indeed constitute a minimal machinery for physiological membrane fusion, the extensive amount of work performed with these experiments (reviewed in Scott et al. (29)) has shown that SNAREs alone can induce lipid mixing and has laid a foundation to attempt to correlate the influence of these and other proteins in membrane merger with functional experiments performed in vivo. To increase the predictive value of this approach, it is important to understand the factors that determine the efficiency of fusion and the origin of the discrepancies between results obtained with different reconstitution methods. In addition, little is known about the mechanism of SNARE-induced bilayer merger. Since fusion appears to require relatively high densities of SNAREs on the vesicles, it is critical to assess whether the observed fusion occurs by a disordered process involving massive formation of SNARE complexes, which could force the membranes to collapse in a different way for each fusion event and perhaps could cause temporary membrane rupture, or by a more ordered, nonleaky mechanism as expected for physiological membrane fusion. In this context, studies with living cells have suggested that all physiological membrane fusion reactions proceed through a lipid-based mechanism involving a stalk intermediate resulting from initial merger of the outer leaflets ((18,30) but see also Frolov et al. (31) and Muller et al. (32)). The standard assay for vesicle leakiness during vesicle-vesicle fusion (33,34) has never been presented, and the only evidence for content mixing required unusually large probes (35), limiting their conclusiveness. On the other hand, reconstitution assays with low densities of the yeast plasma membrane SNAREs have revealed outer leaflet mixing without inner leaflet mixing (36). However, it is unknown whether SNARE-mediated liposome fusion is inhibited by inverted cone phospholipids such as lysophospholipids, which is a key hallmark of the stalk mechanism of membrane fusion and universal for biological membrane fusion (30).
Here we describe a comparative study of the ability of the neuronal SNAREs to induce lipid mixing upon reconstitution with the “standard” method that involves comicellization of the proteins and lipids with detergent (reviewed in Scott et al. (29)), or with a “direct” method involving incorporation of detergent solubilized SNAREs into preformed liposomes (24,37). We find that proteoliposomes prepared by the standard method at high SNARE densities yield slow but efficient lipid mixing that is inhibited by exogenously added lysophospholipids, indicating that in these experiments lipid mixing proceeds through a stalk mechanism analogous to that observed in physiological membrane fusion reactions. At moderate SNARE densities, proteoliposomes prepared by the standard method have a considerable dispersion in size and lipid/protein ratios and yield variable amounts of lipid mixing. In contrast, the direct method leads to more homogenous proteoliposomes but yields almost no lipid mixing at comparable average SNARE densities. Since the chemical composition of these two preparations of proteoliposomes is similar, these observations suggest that the ability of SNAREs to induce lipid mixing depends on the physical state of the reconstituted vesicles (i.e., protein density, size, tension, or curvature).
MATERIALS AND METHODS
Recombinant protein preparation
The construct to express a glutathione S-tranferase (GST) fusion of full-length human SNAP-25B (amino acids 1–206; abbreviated SN25) has been described previously (38). The construct to express His-tagged N-terminally truncated rat Syntaxin 1A (amino acids 183–288; abbreviated SyxH3) (22) was a kind gift from Dr. R. Jahn, and the construct to express a GST fusion of full-length rat synaptobrevin 2 (amino acids 1–116; abbreviated Syb) (24) was a kind gift from Dr. Y. K. Shin. The plasmids were transformed into Escherichia coli BL21(DE3) cells for protein expression. The GST tagged SN25 and Syb were isolated by affinity chromatography using glutathione-agarose beads (Sigma-Aldrich, St. Louis, MO) and cleaved with thrombin (Sigma-Aldrich). SN25 was further purified by ion exchange chromatography on MonoS (Pharmacia, Piscataway, NJ) and stored in 250 mM NaCl, 1 mM 1,4-dithiothreitol (DTT), 20 mM Tris buffer pH7.4. Syb was further purified by anion exchange chromatography using a Vivapure S column (Vivascience, Hanover, Germany) and stored in 25 mM Hepes buffer (pH 7.0), 1 mM DTT, and 1% octyl-β-D-glucopyranoside (OG) containing 500 mM NaCl. The His-tagged SyxH3 was isolated from inclusion bodies by solubilization with 6 M urea followed by affinity chromatography using Ni-NTA agarose (QIAGEN, Valencia, CA). The protein was cleaved from the His-tag with thrombin and further purified by ion exchange chromatography using a Vivapure Q column (Vivascience) and stored in 20 mM Tris buffer (pH 7.4), 1 mM DTT, and 1% OG containing 500 mM NaCl.
SNARE reconstitutions
All lipids were obtained from Avanti Polar Lipids (Alabaster, AL). Proteoliposome reconstitutions by the standard comicellization scheme were performed essentially as described (16), except that the syntaxin construct did not have the N-terminal Habc domain and the t-SNARE complex was produced by mixing purified SN25 and SyxH3 on ice for 30 min before reconstitution. Briefly, the appropriate volumes of purified SNARE solutions were mixed with reconstitution buffer (25 mM Hepes, pH 7.5, 100 mM KCl, 1 mM DTT, 0.1 mM EGTA) containing 1% OG and added to dry lipids (100 μL final volume; 3 mM final total lipid concentration). The resulting solution were quickly diluted with 200 μL of reconstitution buffer and dialyzed overnight against reconstitution buffer, changing the buffer two times during the dialysis. Synaptobrevin vesicles contained 15% DOPS, 82% POPC, 1.5% N-NBD-1,2-dipalmitoyl phosphatidylethanolamine and 1.5% N-(lissamine rhodamine B sulfonyl)-1,2-dipalmitoyl phosphatidylethanolamine, and SNAP-25/syntaxin vesicles contained 15% DOPS and 85% POPC.
Reconstitutions by the direct method were performed essentially as described by Kweon et al. (24) except that the large unilamellar vesicles (LUVs) were prepared using 15 mM total lipids rather than 100 mM total lipids. Briefly, the LUVs were preformed by hydrating dry lipids with 200 μL reconstitution buffer, shaking vigorously for 5 min, freeze/thawing the samples five times to disrupt multilamellar vesicles, and extruding through an 80 nm polycarbonate filter (at least 19 times). Appropriate volumes of purified SNARE solutions were diluted with reconstitution buffer containing 1% OG to a final volume of 200 μL, and then they were mixed with 100 μL of the preformed liposomes. The resulting samples thus contained 0.66% OG and 5 mM final lipid concentration. These are optimal reconstitution conditions based on systematic studies of OG-mediated reconstitution (37). Samples were kept at room temperature for 30 min under gentle stirring, and the detergent was removed by dialysis against reconstitution buffer containing 1.0 g/L Biobeads SM2 beads (Bio-Rad, Hercules, CA) for 1 h at room temperature, followed by dialysis against fresh reconstitution buffer containing 1.0 g/L Biobeads for 2 h more at room temperature and final overnight dialysis 4°C against reconstitution buffer containing 1.0 g/L Biobeads. The resulting lipid/protein ratios for all reconstitutions were assessed from preparations used for the fine Nycodenz gradient analysis (see below). More than 80–90% of the proteins have the correct orientation as determined by chymotrypsin digestion followed by SDS-PAGE analysis.
Nycodenz gradient
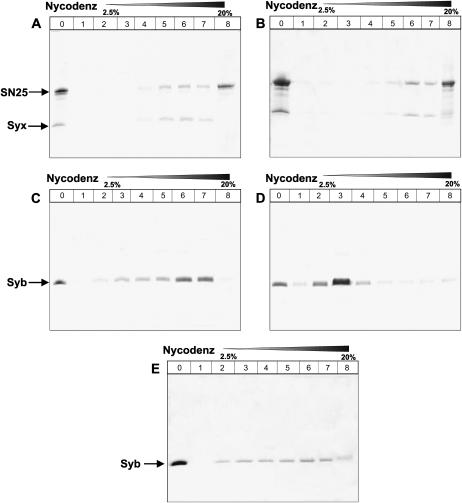
Nycodenz gradients were used to purify reconstituted proteoliposomes basically as described (39). Fine Nycodenz gradients were used to analyze the homogeneity of the proteoliposomes. All the proteoliposomes for this assay contained 1.5% (mol/mol) NBD-DPPE (N-(7-nitro-2,1,3-benzoxadiazole-4-yl)-1,2-dipalmitoyl-PE) and 1.5% (mol/mol) rhodamine-DPPE (N-(lissamine rhodamine B sulfonyl)-1,2-dipalmitoyl-PE) to facilitate lipid quantification by ultraviolet. A total of 250 μL of the proteoliposomes was mixed with an equal volume of 80% Nycodenz in an 11 × 60 mm ultraclear centrifuge tube (Beckman, Fullerton, CA) and overlaid sequentially with 500 ul of 30% Nycodenze, 500 μL of 20% Nycodenze, 500 μL of 10% Nycodenze, 500 μL of 5% Nycodenze, 500 μL of 2.5% Nycodenze, and 500 μL of reconstitution buffer. The gradient was then centrifuged in a SW60Ti rotor (Beckman) at 35,000 rpm for 4 h at 4°C. The fractions (250 μL) were collected from the top of the gradient. A total of 100 μL samples from each fraction were trichloroacetic acid (TCA) precipitated and resuspended in 10 μL loading buffer for SDS-PAGE analysis. For comparison, a 20 μL sample before the Nycodenz gradient was loaded also analyzed by SDS-PAGE (see Fig. 3, A–E, lane 0). The bottom fraction (∼1 ml) was TCA precipitated and resuspended in 10 μL loading buffer and also analyzed by SDS-PAGE (see Fig. 3, A–E, lane 8). The concentration of lipid in each fraction was estimated based on the absorption of rhodamine at 572 nm, and the protein content in each fraction was estimated using SDS-PAGE followed by Coomassie brilliant blue staining, by comparison with standard samples.
FIGURE 3.
Direct method yields synaptobrevin proteoliposomes with more homogeneous protein densities. (A–E) Nycodenz gradient flotation analyses of proteoliposomes containing neuronal SNAREs are shown. (A,C, and E) Proteoliposomes prepared by the standard method and containing syntaxin/SNAP-25 (A) or synaptobrevin (C and E) at 150:1 and 160:1 initial lipid/protein ratios, respectively. Panels C and E represent two experiments performed with different preparations under the same conditions. (B and D) Proteoliposomes prepared by the direct method and containing syntaxin/SNAP-25 (B) or synaptobrevin (D) at 185:1 and 200:1 initial lipid/protein ratios, respectively. Proteoliposomes were floated on a gradient containing Nycodenz concentrations that ranged from 2.5% to 40%. Fractions were pooled and analyzed by SDS-PAGE followed by Coomassie blue staining. Lane 0 corresponds to the starting material, and lanes 1–7 correspond to fractions containing 2.5–20% Nycodenz, where all proteoliposomes were found. Lane 8 corresponds to the bottom of the gradient (30–40% Nycodenz), which only contained unincorporated proteins. The volume of this layer used for SDS-PAGE analysis was 10-fold larger than those used for fractions 1–7 to facilitate detection of small amounts of unincorporated protein (see Materials and Methods).
Lipid mixing assays
The proteoliposomes (1 mM lipid concentration) were preincubated at 37°C before mixing. Typically, 5 μL Syb proteoliposomes were mixed with 45 ul SyxH3/SN25 proteoliposomes at 37°C in a 50 μL Quartz fluorometer cuvette (Nova Biotech, El Cajon, CA). Lipid mixing was followed by NBD fluorescence increase monitored with a Photon Technology Incorporated (PTI, Lawrenceville, NJ) spectrofluorometer (excitation 460 nm; emission 538 nm). At the end of the reaction, 1% OG was added to solubilize the proteoliposomes and the resulting NBD fluorescence was used as the maximal signal for normalization.
Dynamic light scattering tests
Dynamic light scattering (DLS) was performed on a DynaPro dynamic light scattering model 99D instrument (Wyatt Technology, Santa Barbara, CA) using 10 s acquisition time at 37°C. The liposomes or the proteoliposomes were diluted 100 times (10 μM final lipid concentration) and microfuged at 13,000 rpm for 10 min before the DLS measurement. The laser power was adjusted to keep the intensity between 500,000 counts and 2,000,000 counts. The results were then processed with the program Dynamics V6 (Wyatt Technology Corporation). The radii and the size distribution were calculated with the regularization algorithm provided by this software.
Electron microscopy
Reaction mixtures such as those used for lipid mixing assays were prepared at 37°C, and 4 μL of the samples were applied to carbon-coated glow-discharged holey grids. The grids were blotted and fast-plunged into liquid ethane. Electron microscopy (EM) images were obtained with a JEM-2200SE transmission electron microscope at 200 kV. Electron micrographs were taken with a charge-coupled device digital camera with a 10,000–20,000 Å defocus.
Leakage assay
For leakage assays, the proteoliposomes were prepared by the standard method as described above but including 100 mM 5(6)-carboxyfluorescein (CF, Sigma-Aldrich) in initial protein-lipid-detergent mixture and lowering the KCl concentration to match the tonicity to that of the standard reconstitution buffer, which was later used for dialysis. The release of CF from its self-quenched concentration in liposomes or proteoliposomes was monitored with a PTI spectrofluorometer with a 50 μL Quartz fluorometer cuvette (Nova Biotech, El Cajon, CA). The emission spectra from 505 to 535 nm were acquired for 2 h at 37°C, with an excitation wavelength of 490 nm. After 2 h, 1% OG was added to solubilize the proteoliposomes and the emission spectrum was then recorded as 100% release of CF.
RESULTS
SNARE-mediated lipid mixing using two different reconstitution schemes
Multiple studies of the ability of reconstituted neuronal SNAREs to induce lipid mixing have been described (e.g., (16,22–25,40)). A range of lipid mixing efficiencies were observed in these experiments, which probably arose from differences in the protein densities and lipid composition of the proteoliposomes and perhaps in the reconstitution method. At one end of the spectrum, slow but efficient lipid mixing was observed in experiments performed with proteoliposomes reconstituted by the standard comicellization method and using very high synaptobrevin densities (20:1 lipid/protein ratio) (16,25). On the opposite end, no significant lipid mixing was observed in assays carried out with proteoliposomes reconstituted by the direct method and using lower synaptobrevin densities (300:1 lipid/protein ratio) (24), which more closely approximate those present on synaptic vesicles (41,42).
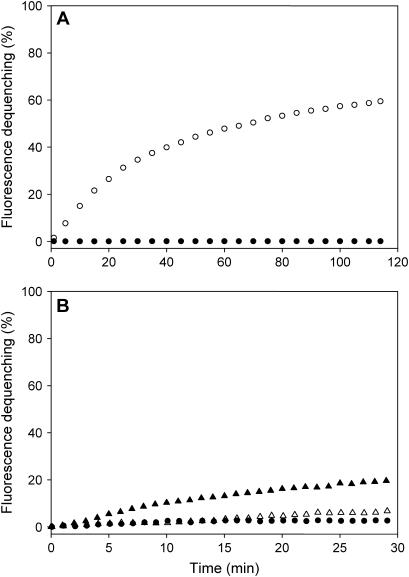
To compare the potential advantages and disadvantages of these two reconstitution methods, we first tested whether in our hands we could reproduce these results using the same fluorescence dequenching assay that has been employed in most of these studies. For this purpose, we used both reconstitution methods to prepare separate populations of “donor” proteoliposomes containing full-length synaptobrevin and a quenched mixture of NBD- and rhodamine-labeled lipids (1.5% each), and “acceptor” proteoliposomes containing full-length SNAP-25 and a syntaxin fragment spanning the SNARE motif and TM region but lacking the N-terminal regulatory Habc domain, which binds intramolecularly to the SNARE motif and inhibits SNARE complex formation (43). For simplicity, we will refer to this fragment as syntaxin. The lipid composition of the vesicles consisted of 15% DOPS and 85% POPC (82% for the donor vesicles), which has been extensively used in previous studies (e.g., Weber et al. (16) and Kweon et al. (24)). We did not coexpress syntaxin and SNAP-25, as described by Weber et al. (16), but rather expressed them separately and mixed them before reconstitution with 1.2 equivalents of SNAP-25 to favor quantitative binding to syntaxin. Note that syntaxin and SNAP-25 are expected to form a 2:1 heterodimer, and hence the actual molar excess of SNAP-25 is substantially higher. This procedure resulted in efficient incorporation of Syntaxin/SNAP-25 heterodimers into the liposomes (see below). The final lipid/synaptobrevin and lipid/syntaxin ratios in vesicles prepared by the standard reconstitution method were 20:1 and 150:1, respectively, and for vesicles prepared by the direct method they were 185:1 and 200:1, respectively.
As described previously (25), efficient lipid mixing was observed for the proteoliposomes prepared by the standard comicellization method and using very high synaptobrevin densities on the donor vesicles, whereas no significant lipid mixing was observed in control experiments where the acceptor vesicles were replaced by protein-free liposomes (Fig. 1 A). In contrast, assays performed with proteoliposomes prepared by the direct reconstitution method and using lower synaptobrevin densities on the donor vesicles yielded almost no lipid mixing (Fig. 1 B), also in agreement with previous results (24). To investigate whether lipid mixing can still be observed with proteoliposomes prepared by the standard method but using lower synaptobrevin densities, we also prepared donor vesicles with a 160:1 lipid/synaptobrevin ratio by this method and performed analogous lipid mixing assays with syntaxin/SNAP-25 vesicles. Some degree of lipid mixing was observed in these experiments, as described previously (40), but there was significant variability in the results obtained with different preparations (Fig. 1 B). Hence, although only small amounts of fluorescence dequenching were generally observed, some preparations yielded substantially higher dequenching (Fig. 1 B). These results confirm that the lipid mixing efficiency of the neuronal SNAREs depends to a large extent on the protein densities on the vesicles and that lipid mixing is very inefficient when protein densities comparable to those present in biological membranes are used. In addition, these results suggest that proteoliposomes prepared by the standard comicellization method have a higher fusion propensity than those prepared by the direct method. It is worth noting that in a very recent study (44) where neuronal SNAREs were reconstituted by the direct method into proteoliposomes with similar protein densities to those used in our experiments, some lipid mixing was observed (∼4% dequenching in 30 min compared to <2% in our experiments). This higher efficiency likely arises from the use of a higher amount of DOPS in the vesicles (35%) and/or the absence of a divalent cation chelator in the buffer, since phosphatidylserine (PS) and Ca2+ are known to facilitate hemifusion.
FIGURE 1.
Comparison of lipid mixing between synaptobrevin “donor” proteoliposomes and syntaxin/SNAP-25 “acceptor” proteoliposomes prepared by different methods. (A) Lipid mixing measured by fluorescence dequenching using donor proteoliposomes containing 1.5% NBD-DPPE and 1.5% rhodamine-DPPE and a 20:1 lipid/synaptobrevin ratio, and acceptor proteoliposomes with a 150:1 lipid/syntaxin/SNAP-25 ratio (open circles). Both proteoliposome populations were prepared by the standard reconstitution method. NBD fluorescence was normalized using the starting point as 0 and the fluorescence observed after addition of 1% OG as 100. The solid circles represent an analogous experiment performed with protein-free liposomes as acceptor vesicles. (B) Analogous experiments performed with synaptobrevin and syntaxin/SNAP-25 proteoliposomes prepared by the standard method with 160:1 and 150:1 lipid/protein ratios, respectively (solid and open triangles, which represent experiments performed under the same conditions but with different preparations), or with synaptobrevin and syntaxin/SNAP-25 proteoliposomes prepared by the direct method with 185:1 and 200:1 lipid/protein ratios, respectively (solid circles). All experiments were performed with a 1 mM total lipid concentration.
Comparison of proteoliposome homogeneity
The direct method whereby detergent solubilized proteins are incorporated into preformed liposomes has been extensively optimized by Rigaud and co-workers to reconstitute proteins of widely diverse sizes and biological activities and has been shown to commonly lead to more homogeneous proteoliposome preparations than the standard comicellization method whereby lipids and proteins are initially cosolubilized with detergent (37). To examine whether this is also the case for reconstitutions of neuronal SNARE proteins, we analyzed the size and protein density distributions of the synaptobrevin and syntaxin/SNAP-25 proteoliposomes prepared by the standard and direct methods.
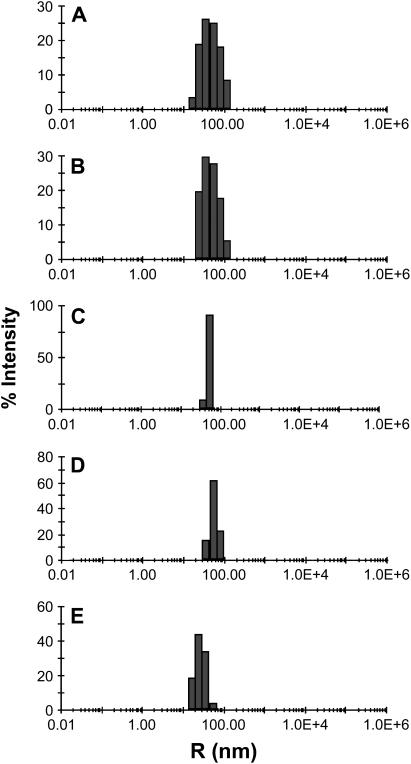
Fig. 2, A and B, shows size profiles measured by DLS for synaptobrevin and syntaxin/SNAP-25 proteoliposomes obtained using the standard comicellization method (both with 150–160:1 lipid/protein ratios). These intensity distributions are biased toward larger particles, which scatter light more efficiently, and conversion of intensities to mass distributions for different particle sizes is complicated by the proximity of the particle diameters to the laser wavelength. However, it is clear from these profiles that the proteoliposomes obtained by the standard method exhibit a broad size distribution centered around 30 nm radii, with substantial populations of small vesicles (i.e., 5–20 nm radius). These results are consistent with size distributions that have been previously measured by EM (16,29). In contrast, DLS analysis of proteoliposomes prepared by the direct method with comparable protein/lipid ratios (Fig. 2, C and D) revealed substantially narrower size distributions. These distributions were similar to those obtained for the original liposomes before protein addition (not shown), which were obtained by extrusion through filters with 80 nm pores. The average radii of the liposomes only increased slightly upon protein incorporation (from 50 nm to 55–60 nm), which can be attributed to the presence of protein at the surface. These observations indicate that insertion of the detergent solubilized SNAREs into the liposomes does not result in liposome lysis or fusion. We also analyzed the size distribution for synaptobrevin-containing vesicles prepared by the standard method with a 20:1 lipid/protein ratio. Interestingly, the size distribution of these vesicles was centered around smaller radii (∼20 nm; compare Fig. 2, A and E). This observation suggests that larger synaptobrevin densities lead to formation of smaller proteoliposomes. This result is not surprising since the SNARE motif of synaptobrevin is unstructured before SNARE complex assembly (45) and is expected to occupy a larger area of the vesicle surface than the cross section of the helical TM region. Hence, the increased steric hindrance between the synaptobrevin SNARE motifs expected at high protein densities should favor the higher curvature of small vesicles.
FIGURE 2.
The direct method yields proteoliposomes with more homogeneous size distributions. The diagrams show DLS analyses of the proteoliposomes used for the lipid mixing assays shown in Fig. 1 but diluted 100-fold (10 μM final lipid concentration). (A and E) Synaptobrevin proteoliposomes prepared by the standard method with a 160:1 lipid/protein ratio (A) or a 20:1 lipid/protein ratio (E). (B) Syntaxin/SNAP-25 proteoliposomes prepared by the standard method with a 150:1 lipid/protein ratio. (C and D) Synaptobrevin (C) and syntaxin/SNAP-25 (D) proteoliposomes prepared by the direct method with lipid/protein ratios of 185:1 and 200:1, respectively.
To investigate the distribution of protein densities, we performed flotation experiments with a fine Nycodenz gradient (0–40%), using proteoliposomes prepared under analogous conditions to those used for the lipid mixing and DLS experiments described above. In these experiments, proteoliposomes of different protein densities are expected to distribute over the fine Nycodenz gradient, whereas unincorporated proteins are expected to remain at the bottom, highest density layer. The syntaxin/SNAP-25 proteoliposomes prepared by either the standard or the direct method were found largely at the fractions containing 10–20% Nycodenz (Fig. 3, A and B) and had similar lipid/protein ratios as the average values calculated before the gradient. Note that part of SNAP-25 coeluted with syntaxin and that the bottom, high density layer contained excess SNAP-25 but little syntaxin (lane 8 of Fig. 3, A and B), showing that syntaxin/SNAP-25 heterodimers incorporated efficiently into the liposomes (note also that lane 8 was loaded 10-fold over the other lanes to increase the sensitivity of protein detection).
Synaptobrevin-containing liposomes obtained by the direct method floated over a narrow density range that corresponds to the expected protein density (Fig. 3 D). The starting average lipid/protein ratio was 185:1, and the lipid/protein ratio of the fraction containing most of the proteoliposomes (lane 3) was 180:1. Only small amounts of synaptobrevin were found in the bottom layer, showing that the protein was also incorporated efficiently into the liposomes (Fig. 3 D). Flotation gradients of synaptobrevin proteoliposomes prepared by the standard method with an average lipid/protein ratio of 160:1 also revealed efficient protein incorporation (Fig. 3, C and E). However, the proteoliposomes were found in multiple fractions over the fine Nycodenz gradient, which correspond to lipid/protein ratios ranging from 250:1 (lane 3) to 100:1 (lane 7). Importantly, we found significant variability in the protein distribution observed in separate experiments performed under analogous conditions. Hence, a large percentage of the synaptobrevin was found at the higher density proteoliposomes in the experiment shown in Fig. 3 C, whereas the protein was more evenly distributed over the different fractions in the experiment shown in Fig. 3 E. This variability is likely to underlie the variability observed in the lipid mixing efficiency observed for these preparations (Fig. 1 B), since it seems clear that higher protein densities strongly increase this efficiency and a larger percentage of vesicles with high protein densities would thus lead to an increase in the observed lipid mixing.
It is important to note that heterogeneous sizes and lipid/protein ratios are common in proteoliposomes prepared by the standard comicellization method since the ternary lipid-protein-detergent mixtures are inherently heterogeneous and unstable; such heterogeneity translates to the proteoliposomes obtained upon detergent removal, which is a nonequilibrium process (37). On the other hand, in preliminary experiments using the direct method where we incorporated synaptobrevin to preformed vesicles with a lipid composition that approximates that present in synaptic vesicles (phosphatidylcholine/phosphatidylethanolalmine/PS/cholesterol 48:20:12:20; see Hu et al. (23)), we found that 10% of the proteoliposomes contain 90% of the protein and the remaining synaptobrevin incorporates into the remaining 90% of vesicles (X. Chen and J. Rizo, unpublished results). These preparations yield small but significant lipid mixing in contrast to those with the lipid composition of 15% DOPS and 85% POPC, which have a homogeneous protein density. These observations reinforce the notion that fractions of proteoliposomes with high protein densities may account for the lipid mixing observed for heterogeneous preparations and further emphasize the need to characterize the distribution of protein densities in the proteoliposomes used for fusion assays regardless of the reconstitution method employed.
SNARE-mediated lipid mixing is inhibited by lysophospholipids
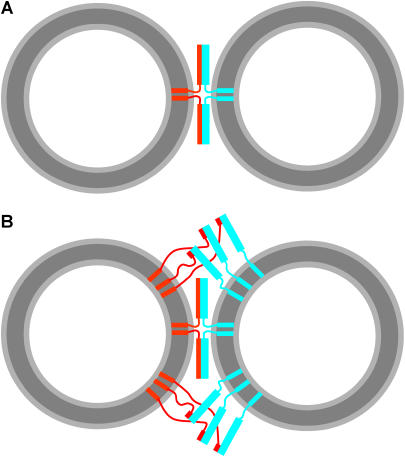
The above results indicate that the physical state of the reconstituted vesicles determines the success or failure of SNARE-mediated lipid mixing between vesicles. On this basis alone, we would like to test if SNARE-mediated fusion in vitro follows the physical principles established for both phospholipid bilayer membrane fusion and biological membrane fusion (18). In addition, our data and those of multiple studies described in the literature (e.g., (16,17,22,25)) indicate that a threshold protein density is necessary to observe some degree of SNARE-mediated lipid mixing (at least dozens of SNARE molecules per vesicle, based on the lipid/protein ratios used and assuming a vesicle radius of 20 nm). Although these threshold protein densities may resemble those of biological membranes, it is natural to expect that, if the SNAREs are indeed primarily responsible for membrane fusion in vivo, only a few SNARE complexes likely assembled in a ring-like fashion would mediate the fusion reaction. The question that arises is why then is a much larger number of SNAREs necessary to observe lipid mixing in the reconstitution assays? In addition, SNARE complex assembly is expected to bring two opposing membranes within 20–30 Å, based on the crystal structure of the neuronal SNARE complex (14), but it is unclear how the SNARE complex can bend the membranes to induce membrane fusion. One possibility to explain these observations is that formation of a few SNARE complexes between two membranes may not cause membrane fusion (Fig. 4 A), but massive formation of SNARE complexes between more distal parts of the two membranes (Fig. 4 B) may force collapse of the two membranes in a disordered (and perhaps leaky) fashion that may be completely unrelated to physiological membrane fusion. An alternative possibility is that the SNAREs induce liposome fusion by a more ordered process related to the stalk mechanism that is believed to underlie all types of physiological membrane fusion (30), but the high entropic cost of arranging a few SNARE complexes in a very restrictive proper orientation to induce membrane merger underlies the need for a sufficiently high protein density and also the slow rate of fusion observed. To obtain experimental data that could help to distinguish between these possibilities, we performed cryo-EM and leakage experiments and examined whether lysophospholipids inhibit SNARE-induced lipid mixing. All these experiments were performed with proteoliposomes prepared with the standard method and using high synaptobrevin densities (20:1 lipid/protein ratio) to favor efficient lipid mixing.
FIGURE 4.
How many SNARE complexes induce lipid mixing? (A) Model of two fully assembled SNARE complexes located between the most proximal regions of two vesicles. The model was drawn approximately to scale to represent the relative sizes of the SNARE complexes and 40 nm vesicles and to illustrate that SNARE complex assembly should bring the opposing membranes within 2–3 nm, but it is unclear whether this is sufficient for membrane merger. (B) Model analogous to A but including additional SNARE complexes that are initiating assembly at their N-termini with the rest of their SNARE motifs unstructured. The model is intended to illustrate the possibility that massive formation of SNARE complexes between more distal parts of the two membranes might actually induce lipid mixing rather than formation of a few SNARE complexes in the proximal intermembrane space. In panels A and B, synaptobrevin is colored in red and syntaxin/SNAP-25 heterodimers in cyan. Curves with arbitrary shapes indicate unstructured regions (i.e., the SNARE motif of free synaptobrevin and the linker sequences between the TM regions and the SNARE motifs of syntaxin and synaptobrevin). Thin rectangles represent individual helices (i.e., TM regions and the synaptobrevin SNARE motif upon partial or full assembly of SNARE complexes), and wide rectangles represent the helix bundle formed by the syntaxin and SNAP-25 SNARE motifs.
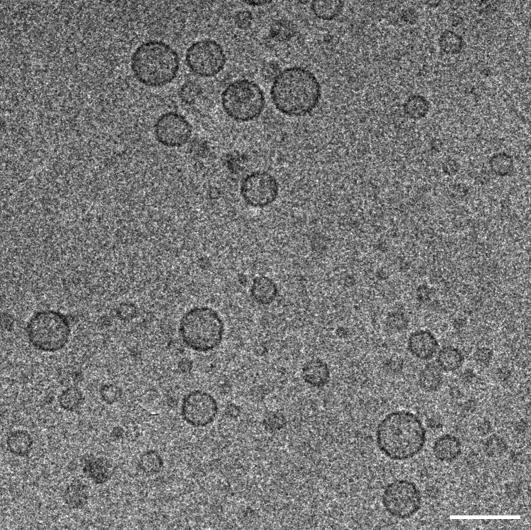
Cryo-EM experiments were performed in an attempt to examine the appearance of vesicle-vesicle interfaces during fusion. Samples were fast-frozen a few minutes after mixing donor and acceptor proteoliposomes, when the kinetics of lipid mixing is fastest (Fig. 1 A), to increase the probability of “catching” two vesicles in the act of fusion. A representative image obtained in these experiments is shown in Fig. 5. Surprisingly, no vesicle-vesicle contacts, let alone fusion intermediates, were observed in this and multiple regions of the samples that were examined (note that some vesicle pairs appear to be close in the plane shown but are more distant in the perpendicular axis as assessed from a tomographic series of images). Although these experiments did not allow us to observe membrane fusion interfaces, they showed that vesicle clustering or pairwise vesicle docking must be short lived during the course of these fusion reactions. This result correlates with DLS experiments, where we failed to observe significant increases in particle size during these early stages of the reaction, and with results obtained previously by cryo-EM using proteoliposomes with lower protein densities (22). These observations indicate that collisions between donor and acceptor vesicles are usually unproductive and that lipid mixing occurs very fast when a productive collision is established, which could be explained by the following model. Syntaxin and SNAP-25 form a heterodimer that consists of a four-helix bundle with two copies of syntaxin and one of SNAP-25; one of the syntaxin helices occupies the synaptobrevin binding site and thus needs to be replaced by synaptobrevin (46). Since the synaptobrevin SNARE motifs are unstructured (45) and partially sequestered by the membrane (23,24), the probability that an individual synaptobrevin SNARE motif from one vesicle approaches a syntaxin/SNAP-25 heterodimer from another vesicle in the proper orientation to replace one of the syntaxin SNARE motifs and initiate formation of a SNARE complex is probably very low. Even if one SNARE complex starts forming, steric hindrance between the majority of synaptobrevin SNARE motifs and syntaxin/SNAP-25 heterodimers could cause vesicle-vesicle repulsion and dissociation of the single nascent SNARE complex. Hence, most collisions would be unproductive. In this model, simultaneous initiation of assembly of several SNARE complexes in a given collision would occur with very low probability but, when a minimum number of such complexes is formed, their combined stabilization energy could overcome the steric hindrance of the unassembled SNAREs and facilitate assembly of additional SNARE complexes, leading quickly to membrane merger (this would constitute a productive collision).
FIGURE 5.
No significant docking is observed during lipid mixing reactions mediated by the neuronal SNAREs. The figure shows a cryo-EM image of a mixture of donor and acceptor vesicles prepared under the same conditions to those used for the lipid mixing assays shown in Fig. 1 A. The sample was fast-frozen a few minutes after mixing donor and acceptor proteoliposomes. The scale bar corresponds to 100 nm.
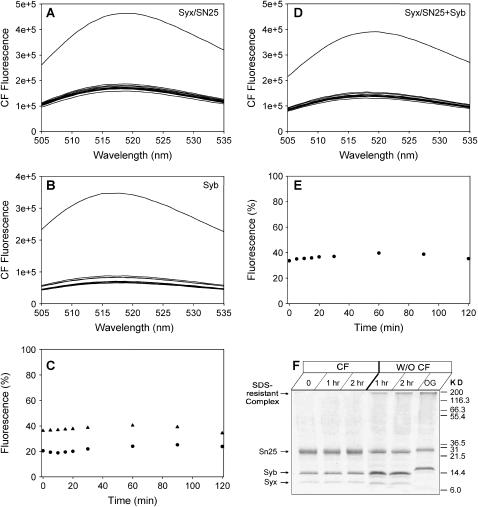
We next attempted to study whether SNARE-mediated lipid mixing occurs without membrane rupture. Previous experiments using oligonucleotides trapped inside proteoliposomes suggested that SNARE-mediated membrane fusion occurs without leakage of its contents (35), but some leakage cannot be ruled out based on these data due to the large size of these probes and their tendency to adhere to membranes. Hence, in our studies we employed a fluorescein dequenching assay that has been widely used in the literature to analyze liposome leakage (47). Separate synaptobrevin and syntaxin/SNAP-25 vesicle populations were prepared by the standard method in the presence of high fluorescein concentrations, and untrapped fluorescein was removed by dialysis. Both proteoliposome populations exhibited a small amount of leakage that decayed over time (Fig. 6, A–C). This result suggests that at least a fraction of the vesicles are relatively stable, although we cannot rule out the possibility that a substantial vesicle population lost most of its contents during dialysis. A similar amount of leakage was observed when synaptobrevin and syntaxin/SNAP-25 liposomes were mixed (Fig. 6, D and E). However, analysis of these mixtures by SDS-PAGE showed that no SDS-resistant SNAREs complexes were formed during the course of these experiments, in contrast to parallel experiments performed without fluorescein trapped inside the vesicles (Fig. 6 F). This observation suggests that the trapped fluorescein inhibits lipid mixing, perhaps due to inhibition of membrane contacts arising from the higher membrane tension in vesicles containing the osmotically active fluorescein (48). This may also explain why the vesicle to planar bilayer reconstitution system gives higher kinetics of lipid mixing, since the planar membrane does not have the tension that inhibits contacts and can wrap around the tense vesicle. Thus, although these data are inconclusive, they emphasize the dependence of SNARE-mediated lipid mixing on the physical state of the vesicles.
FIGURE 6.
Analysis of fluorescein leakage of neuronal SNARE proteoliposomes. (A and B) Fluorescence spectra at 37°C as a function of time of samples of synaptobrevin (A) and syntaxin/SNAP-25 (B) proteoliposomes obtained by the standard method with analogous protein densities to those used for the lipid mixing assays of Fig. 1 A and with trapped fluorescein (100 mM). The top curve represents the fluorescence spectrum obtained upon addition of 1% OG after 2 h of incubation. (C) Time dependence of the fluorescence intensity at 518 nm observed in the experiments of panels A and B, expressed as percentage of the fluorescence intensity obtained after liposome lysis with 1% OG (circles, synaptobrevin liposomes; triangles, syntaxin/SNAP-25 liposomes). (D) Fluorescence spectra at 37°C as a function of time of a mixture of synaptobrevin vesicles and syntaxin/SNAP-25 vesicles analogous to those used in panels A and B. (E) Time dependence of the fluorescence intensity at 518 nm for the experiment shown in panel D. (F) SDS-PAGE analysis of the mixture used for the experiments in (D and E) at 0, 1, and 2 h (left lanes) and of a comparable mixture with proteoliposomes prepared without trapped fluorescein. Note the appearance of SDS-resistant SNARE complexes in the latter but not in the former experiment.
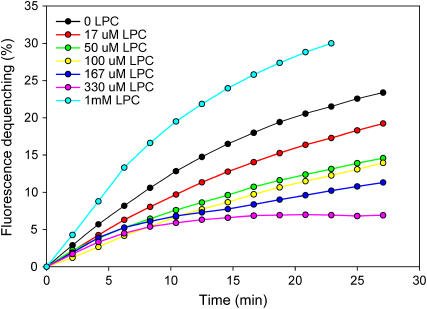
In a final set of experiments, we studied whether SNARE-mediated lipid mixing is inhibited by lysophospholipids, which is a key hallmark of the stalk mechanism of membrane fusion that is believed to mediate all types of membrane fusion in vivo (18). Importantly, addition of increasing amounts of oleoyl lysophosphatidyl choline (LPC) up to a concentration of 330 μM lead to a progressive decrease in the efficiency of SNARE-induced lipid mixing (Fig. 7). At 1 mM concentrations, LPC increased lipid mixing efficiency, most likely because of liposome lysis induced by the detergent nature of LPC. Note that a small degree of lipid mixing was still observed even at the highest inhibitory concentrations of LPC, but this may arise from unavoidable, partial liposome lysis caused by high local concentrations of LPC upon mixing the vesicles with the highly concentrated detergent. The inhibition of lipid mixing by LPC was not a specific effect of the headgroup, since oleoyl lysophosphatidylserine was equally efficient in inhibiting lipid mixing (data not shown). Overall, these results strongly support the notion that SNARE-mediated lipid mixing occurs largely by a stalk mechanism.
FIGURE 7.
Lysophosphatidylcholine inhibits SNARE-mediated lipid mixing. Lipid mixing assays were performed and monitored as in Fig. 1 A but with different additions of oleoyl LPC at the start of the reaction. The final concentrations of oleoyl LPC are color coded and indicated in the inset.
DISCUSSION
Extensive studies of membrane traffic in different membrane compartments of eukaryotic organisms from yeast to humans have established that SNARE proteins are key components of a conserved membrane fusion machinery that mediates most types of intracellular membrane traffic, and detailed structural characterization of the complexes formed by SNARE proteins have left little doubt that these complexes should bring two opposing membranes into close proximity (reviewed in Chen and Scheller (2) and Jahn et al. (4)). The major debate about the function of SNARE proteins centers around their exact role in membrane fusion: do the SNAREs set up fusion or are they the primary fusion proteins whose fusogenic activity is regulated by other proteins involved in membrane traffic? The observation that reconstituted SNAREs can induce slow lipid mixing (16,17) provided support for the latter possibility, but the biological relevance of this finding can be questioned based on the slow kinetics of lipid mixing, the requirement for high protein densities, the contradictory results obtained in different studies, and the lack of studies establishing rigorous correlations with in vivo data. In addition, it was extremely surprising that significant lipid mixing was still observed when long linkers that can span 100 Å were introduced between the SNARE motif and the TM region of syntaxin and synaptobrevin (27), since these linkers would be expected to uncouple the energy of SNARE complex formation from approximation of the two membranes to the short distances required for membrane fusion (<3 nm; see Burger (49)). On the other hand, the observation that inverted SNAREs exposed on a cell surface can induce intercellular fusion (50) further supported the notion of a fusogenic role for the SNAREs, but it is also uncertain whether this result arose from expression of massive amounts of SNAREs on the surface of the cells that fused. Moreover, a variety of data, particularly those obtained in studies of yeast vacuolar fusion (51,52) and fusion of egg cortical vesicles (21,53), have suggested that additional events after SNARE complex assembly lead to membrane fusion.
With all its caveats, the reconstitution approach provides a powerful means to attempt to correlate the influence of proteins on membrane fusion observed in vitro with their functional activities observed in vivo, and the extensive work that has been carried out to optimize the reconstitution experiments (reviewed in Scott et al. (29)) has been fundamental to pursue this goal. The experiments described here were designed to further understand the factors that influence the lipid mixing induced by reconstituted neuronal SNAREs and to shed light on the mechanism of lipid mixing. Our data show that the direct method involving incorporation of detergent solubilized proteins into preformed liposomes yields considerably more homogenous proteoliposomes than the standard comicellization method that has been most widely used in reconstitution experiments with SNARE proteins. Moreover, our data further emphasize the notion that the protein density is a critical determinant of lipid mixing efficiency (Fig. 1) and suggest that higher protein densities lead to the formation of smaller vesicles (Fig. 2 E), smaller than the sizes of many secretory vesicles such as the dense core vesicles of chromaffin cells. Since the lipids used for these assays favor planar or slightly negative membrane curvature, the high positive geometric curvature of small vesicles should result in substantial negative curvature stress. Consequently, these small vesicles are expected to have a higher tendency to form stalks and fuse to reduce such stress. Note for instance that small sonicated vesicles are unstable and fuse spontaneously (54), which is also likely to arise from curvature stress. On the other hand, small SNARE-containing proteoliposomes may not easily fuse with each other or with protein-free liposomes such as those used in controls because of steric hindrance generated by the SNAREs on the surface, but they may be able to fuse with proteoliposomes containing the cognate SNAREs because SNARE complex formation overcomes the steric hindrance and facilitates release of the curvature stress.
The heterogeneity in both the size and the protein density distribution in the proteoliposomes obtained by the standard method is of particular concern since the limited amount of SNARE-induced lipid mixing commonly observed in these reconstitution experiments may actually arise from the existence of substantial populations of small vesicles with high protein densities that are more prone to fusion. Note also that our DLS and cryo-EM data (Figs. 2 and 5) revealed the presence of numerous particles of very small size (<10 nm radius) that are more likely to be micelles rather than vesicles. Altogether, these observations indicate that the neuronal SNAREs may be even less efficient in inducing lipid mixing than previously thought. On the other hand, it is worth noting that the ability of the neuronal SNAREs to induce lipid mixing is likely hindered by the interaction of the SNARE motif of synaptobrevin with the membrane (23,24) and that other SNAREs such as those from the yeast plasma membrane appear to be considerably more efficient in causing lipid mixing (55).
The partial sequestration of the synaptobrevin SNARE motif by the membrane, a slow kinetics of SNARE complex formation between proteins in two membranes, and inhibition by one of the two syntaxin SNARE motifs that form the syntaxin/SNAP-25 heterodimers, likely determine that most collisions between donor and acceptor vesicles are not productive, as suggested by the absence of vesicle clusters in our cryo-EM images (Fig. 5). However, since lipid mixing does occur under the conditions of these experiments (Fig. 1 A), it appears that when a sufficient number of SNAREs engage across the intermembrane space, lipid mixing ensues on a relatively fast timescale. This conclusion correlates with the fast timescale of individual SNARE-mediated lipid mixing events observed between vesicles and planar bilayers by total internal reflection fluorescence spectroscopy (26). However, it should be noted that only a fraction of the vesicles yielded lipid mixing during the course of these experiments. Given the heterogeneity in the synaptobrevin vesicles prepared by the standard method, it is plausible that the observed lipid mixing arose only from small vesicles with high protein densities. Although the relatively fast speed of individual lipid mixing events partially mitigates the concerns regarding the slow overall rate of SNARE-mediated lipid mixing, it is still clear that high protein densities are required for efficient lipid mixing. Hence, it was crucial to assess whether lipid mixing is caused by a “brute force” mechanism involving massive SNARE complex formation that collapses the membranes, perhaps even involving many vesicles with a concomitant increase in surface tension with each newly adherent vesicle to a cluster. Our cryo-EM data clearly rule out any such clustering in this system, even at the time of peak lipid mixing.
Does SNARE-mediated lipid mixing in reconstituted systems go through a nonlamellar intermediate of negative geometric monolayer curvature, such as the stalk? Our finding that lysophospholipids inhibit SNARE-mediated lipid mixing (Fig. 7) shows that changing lipid composition to make the membrane curvature more positive decreases lipid mixing, strongly suggesting that a stalk-like intermediate is required for the action of neuronal SNAREs. A study of the effects of mutations in the TM region of syntaxin on fusion pore conductance during Ca2+-triggered exocytosis led to the conclusion that the TM regions line the fusion pore (56), in contradiction with the lipidic nature of the fusion pore assumed by the stalk model. However, the results of this study were not inconsistent with a lipidic fusion pore, as was pointed out later (57). Moreover, it would be surprising if Ca2+-triggered exocytosis does not occur by the stalk mechanism of membrane fusion since this mechanism generally mediates fusion between viruses and target cells, between cell membranes, between organelles, and between organelles and plasma membranes (30). In particular, Ca2+-triggered exocytosis in the egg cortical exocytosis is similarly inhibited by lysolipids. Since the pathway of purely phospholipid membrane fusion also proceeds through a stalk-like intermediate of negative curvature and is inhibited by lysolipids (18,58), it is simplest to imagine that the intermembrane proximity caused by formation of multiple SNARE complexes, together with the existing curvature stress, lower the energy barrier for formation of a stalk intermediate in this in vitro system.
Can the SNAREs be considered a “minimal membrane fusion machinery” as is often stated in the literature? This question arises from a common tendency to simplify complex biological systems, but it may not have a simple valid answer. For instance, the lack of lipid mixing in the experiments performed with the homogeneous proteoliposomes obtained with the direct method (Fig. 1 B and (24)) could be used to argue that the neuronal SNAREs do not constitute a minimal fusion machinery, but it is plausible that SNARE complex formation does provide the primary driving force for membrane fusion in vivo with the help of additional proteins that mediate assembly of multiple SNARE complexes in a proper orientation (e.g., in a ring-like fashion). A fusogenic role for the SNAREs is indeed attractive because of their key importance for all types of intracellular membrane fusion and because of the similar structural characteristics of SNARE complexes with viral fusion proteins (11), which use the energy of interactions between coiled coils to induce membrane fusion. On the other hand, it is still unclear how SNARE complex formation can bend the membranes to initiate fusion (see Fig. 4 A), and abundant data have suggested that additional proteins function downstream of the SNAREs (see above). Furthermore, a key issue that needs to be addressed is the role of Sec1/Munc18 homologs which, like the SNAREs, are crucial for most types of intracellular membrane fusion (reviewed in Rizo and Sudhof (3)). Although Sec1/Munc18 homologs have been suggested to assist in SNARE complex formation, some evidence has indicated that they may act after SNARE complex assembly (59), and their function is currently still a mystery. Hence, a true understanding of the exact roles of SNAREs in membrane fusion will require elucidation of the roles of other key proteins involved in membrane traffic. The reconstitution approach can help to address these questions, but the relevance of any results obtained with these in vitro experiments needs to be validated by testing whether they correlate with functional experiments performed in vivo. The homogeneity of the SNARE-containing proteoliposomes obtained by the direct reconstitution method shows that this method provides an improved tool for rigorous studies directed at pursuing this goal. Their lack of activity instructs us that more than SNAREs alone is needed for even the first stage of fusion: the physical state of the vesicle in which they are reconstituted is a regulatory partner.
Acknowledgments
We thank Thomas C. Südhof, Yeon-Kyun Shin, Reinhard Jahn, and Masahide Kikkawa for fruitful discussions.
This work was supported by a grant for the Welch Foundation (I-1304) and by National Institutes of Health grant NS37200 (to J.R.).
References
- 1.Pelham, H. R. 1999. SNAREs and the secretory pathway—lessons from yeast. Exp. Cell Res. 247:1–8. [DOI] [PubMed] [Google Scholar]
- 2.Chen, Y. A., and R. H. Scheller. 2001. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2:98–106. [DOI] [PubMed] [Google Scholar]
- 3.Rizo, J., and T. C. Sudhof. 2002. Snares and munc18 in synaptic vesicle fusion. Nat. Rev. Neurosci. 3:641–653. [DOI] [PubMed] [Google Scholar]
- 4.Jahn, R., T. Lang, and T. C. Sudhof. 2003. Membrane fusion. Cell. 112:519–533. [DOI] [PubMed] [Google Scholar]
- 5.Schiavo, G., M. Matteoli, and C. Montecucco. 2000. Neurotoxins affecting neuroexocytosis. Physiol. Rev. 80:717–766. [DOI] [PubMed] [Google Scholar]
- 6.Sollner, T., M. K. Bennett, S. W. Whiteheart, R. H. Scheller, and J. E. Rothman. 1993. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 75:409–418. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi, T., H. McMahon, S. Yamasaki, T. Binz, Y. Hata, T. C. Sudhof, and H. Niemann. 1994. Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J. 13:5051–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poirier, M. A., J. C. Hao, P. N. Malkus, C. Chan, M. F. Moore, D. S. King, and M. K. Bennett. 1998. Protease resistance of syntaxin.SNAP-25.VAMP complexes. Implications for assembly and structure. J. Biol. Chem. 273:11370–11377. [DOI] [PubMed] [Google Scholar]
- 9.Fasshauer, D., W. K. Eliason, A. T. Brunger, and R. Jahn. 1998. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry. 37:10354–10362. [DOI] [PubMed] [Google Scholar]
- 10.Calakos, N., M. K. Bennett, K. E. Peterson, and R. H. Scheller. 1994. Protein-protein interactions contributing to the specificity of intracellular vesicular trafficking. Science. 263:1146–1149. [DOI] [PubMed] [Google Scholar]
- 11.Hanson, P. I., R. Roth, H. Morisaki, R. Jahn, and J. E. Heuser. 1997. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 90:523–535. [DOI] [PubMed] [Google Scholar]
- 12.Lin, R. C., and R. H. Scheller. 1997. Structural organization of the synaptic exocytosis core complex. Neuron. 19:1087–1094. [DOI] [PubMed] [Google Scholar]
- 13.Poirier, M. A., W. Xiao, J. C. Macosko, C. Chan, Y. K. Shin, and M. K. Bennett. 1998. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat. Struct. Biol. 5:765–769. [DOI] [PubMed] [Google Scholar]
- 14.Sutton, R. B., D. Fasshauer, R. Jahn, and A. T. Brunger. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 395:347–353. [DOI] [PubMed] [Google Scholar]
- 15.Antonin, W., D. Fasshauer, S. Becker, R. Jahn, and T. R. Schneider. 2002. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 9:107–111. [DOI] [PubMed] [Google Scholar]
- 16.Weber, T., B. V. Zemelman, J. A. McNew, B. Westermann, M. Gmachl, F. Parlati, T. H. Sollner, and J. E. Rothman. 1998. SNAREpins: minimal machinery for membrane fusion. Cell. 92:759–772. [DOI] [PubMed] [Google Scholar]
- 17.McNew, J. A., F. Parlati, R. Fukuda, R. J. Johnston, K. Paz, F. Paumet, T. H. Sollner, and J. E. Rothman. 2000. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 407:153–159. [DOI] [PubMed] [Google Scholar]
- 18.Zimmerberg, J., and L. V. Chernomordik. 1999. Membrane fusion. Adv. Drug Deliv. Rev. 38:197–205. [DOI] [PubMed] [Google Scholar]
- 19.Rizo, J. 2003. SNARE function revisited. Nat. Struct. Biol. 10:417–419. [DOI] [PubMed] [Google Scholar]
- 20.Szule, J. A., and J. R. Coorssen. 2003. Revisiting the role of SNAREs in exocytosis and membrane fusion. Biochim. Biophys. Acta. 1641:121–135. [DOI] [PubMed] [Google Scholar]
- 21.Coorssen, J. R., P. S. Blank, F. Albertorio, L. Bezrukov, I. Kolosova, X. Chen, P. S. Backlund Jr., and J. Zimmerberg. 2003. Regulated secretion: SNARE density, vesicle fusion and calcium dependence. J. Cell Sci. 116:2087–2097. [DOI] [PubMed] [Google Scholar]
- 22.Schuette, C. G., K. Hatsuzawa, M. Margittai, A. Stein, D. Riedel, P. Kuster, M. Konig, C. Seidel, and R. Jahn. 2004. Determinants of liposome fusion mediated by synaptic SNARE proteins. Proc. Natl. Acad. Sci. USA. 101:2858–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, K., J. Carroll, S. Fedorovich, C. Rickman, A. Sukhodub, and B. Davletov. 2002. Vesicular restriction of synaptobrevin suggests a role for calcium in membrane fusion. Nature. 415:646–650. [DOI] [PubMed] [Google Scholar]
- 24.Kweon, D. H., C. S. Kim, and Y. K. Shin. 2003. Regulation of neuronal SNARE assembly by the membrane. Nat. Struct. Biol. 10:440–447. [DOI] [PubMed] [Google Scholar]
- 25.Parlati, F., T. Weber, J. A. McNew, B. Westermann, T. H. Sollner, and J. E. Rothman. 1999. Rapid and efficient fusion of phospholipid vesicles by the alpha-helical core of a SNARE complex in the absence of an N-terminal regulatory domain. Proc. Natl. Acad. Sci. USA. 96:12565–12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fix, M., T. J. Melia, J. K. Jaiswal, J. Z. Rappoport, D. You, T. H. Sollner, J. E. Rothman, and S. M. Simon. 2004. Imaging single membrane fusion events mediated by SNARE proteins. Proc. Natl. Acad. Sci. USA. 101:7311–7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNew, J. A., T. Weber, D. M. Engelman, T. H. Sollner, and J. E. Rothman. 1999. The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol. Cell. 4:415–421. [DOI] [PubMed] [Google Scholar]
- 28.Wang, Y., I. Dulubova, J. Rizo, and T. C. Sudhof. 2001. Functional analysis of conserved structural elements in yeast syntaxin Vam3p. J. Biol. Chem. 276:28598–28605. [DOI] [PubMed] [Google Scholar]
- 29.Scott, B. L., J. S. Van Komen, S. Liu, T. Weber, T. J. Melia, and J. A. McNew. 2003. Liposome fusion assay to monitor intracellular membrane fusion machines. Methods Enzymol. 372:274–300. [DOI] [PubMed] [Google Scholar]
- 30.Chernomordik, L. V., S. S. Vogel, A. Sokoloff, H. O. Onaran, E. A. Leikina, and J. Zimmerberg. 1993. Lysolipids reversibly inhibit Ca(2+)-, GTP- and pH-dependent fusion of biological membranes. FEBS Lett. 318:71–76. [DOI] [PubMed] [Google Scholar]
- 31.Frolov, V. A., A. Y. Dunina-Barkovskaya, A. V. Samsonov, and J. Zimmerberg. 2003. Membrane permeability changes at early stages of influenza hemagglutinin-mediated fusion. Biophys. J. 85:1725–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller, M., K. Katsov, and M. Schick. 2003. A new mechanism of model membrane fusion determined from Monte Carlo simulation. Biophys. J. 85:1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duzgunes, N., T. M. Allen, J. Fedor, and D. Papahadjopoulos. 1987. Lipid mixing during membrane aggregation and fusion: why fusion assays disagree. Biochemistry. 26:8435–8442. [DOI] [PubMed] [Google Scholar]
- 34.Bentz, J., and N. Duzgunes. 1985. Fusogenic capacities of divalent cations and effect of liposome size. Biochemistry. 24:5436–5443. [DOI] [PubMed] [Google Scholar]
- 35.Nickel, W., T. Weber, J. A. McNew, F. Parlati, T. H. Sollner, and J. E. Rothman. 1999. Content mixing and membrane integrity during membrane fusion driven by pairing of isolated v-SNAREs and t-SNAREs. Proc. Natl. Acad. Sci. USA. 96:12571–12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, Y., F. Zhang, Z. Su, J. A. McNew, and Y. K. Shin. 2005. Hemifusion in SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 12:417–422. [DOI] [PubMed] [Google Scholar]
- 37.Rigaud, J. L., and D. Levy. 2003. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 372:65–86. [DOI] [PubMed] [Google Scholar]
- 38.Chen, X., J. Tang, T. C. Sudhof, and J. Rizo. 2005. Are neuronal SNARE proteins Ca2+ sensors? J. Mol. Biol. 347:145–158. [DOI] [PubMed] [Google Scholar]
- 39.Weber, T., F. Parlati, J. A. McNew, R. J. Johnston, B. Westermann, T. H. Sollner, and J. E. Rothman. 2000. SNAREpins are functionally resistant to disruption by NSF and alphaSNAP. J. Cell Biol. 149:1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tucker, W. C., T. Weber, and E. R. Chapman. 2004. Reconstitution of Ca2+-regulated membrane fusion by synaptotagmin and SNAREs. Science. 304:435–438. [DOI] [PubMed] [Google Scholar]
- 41.Jahn, R., and T. C. Sudhof. 1994. Synaptic vesicles and exocytosis. Annu. Rev. Neurosci. 17:219–246. [DOI] [PubMed] [Google Scholar]
- 42.Walch-Solimena, C., J. Blasi, L. Edelmann, E. R. Chapman, G. F. Von Mollard, and R. Jahn. 1995. The t-SNAREs syntaxin 1 and SNAP-25 are present on organelles that participate in synaptic vesicle recycling. J. Cell Biol. 128:637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dulubova, I., S. Sugita, S. Hill, M. Hosaka, I. Fernandez, T. C. Sudhof, and J. Rizo. 1999. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 18:4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu, X., F. Zhang, J. A. McNew, and Y. K. Shin. 2005. Membrane fusion induced by neuronal SNAREs transits through hemifusion. J. Biol. Chem. 280:30538–30541. [DOI] [PubMed] [Google Scholar]
- 45.Hazzard, J., T. C. Sudhof, and J. Rizo. 1999. NMR analysis of the structure of synaptobrevin and of its interaction with syntaxin. J. Biomol. NMR. 14:203–207. [DOI] [PubMed] [Google Scholar]
- 46.Xiao, W., M. A. Poirier, M. K. Bennett, and Y. K. Shin. 2001. The neuronal t-SNARE complex is a parallel four-helix bundle. Nat. Struct. Biol. 8:308–311. [DOI] [PubMed] [Google Scholar]
- 47.Wilschut, J., N. Duzgunes, R. Fraley, and D. Papahadjopoulos. 1980. Studies on the mechanism of membrane fusion: kinetics of calcium ion induced fusion of phosphatidylserine vesicles followed by a new assay for mixing of aqueous vesicle contents. Biochemistry. 19:6011–6021. [DOI] [PubMed] [Google Scholar]
- 48.Kwok, R., and E. Evans. 1981. Thermoelasticity of large lecithin bilayer vesicles. Biophys. J. 35:637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burger, K. N. 2000. Greasing membrane fusion and fission machineries. Traffic. 1:605–613. [DOI] [PubMed] [Google Scholar]
- 50.Hu, C., M. Ahmed, T. J. Melia, T. H. Sollner, T. Mayer, and J. E. Rothman. 2003. Fusion of cells by flipped SNAREs. Science. 300:1745–1749. [DOI] [PubMed] [Google Scholar]
- 51.Ungermann, C., K. Sato, and W. Wickner. 1998. Defining the functions of trans-SNARE pairs. Nature. 396:543–548. [DOI] [PubMed] [Google Scholar]
- 52.Reese, C., F. Heise, and A. Mayer. 2005. Trans-SNARE pairing can precede a hemifusion intermediate in intracellular membrane fusion. Nature. 436:410–414. [DOI] [PubMed] [Google Scholar]
- 53.Tahara, M., J. R. Coorssen, K. Timmers, P. S. Blank, T. Whalley, R. Scheller, and J. Zimmerberg. 1998. Calcium can disrupt the SNARE protein complex on sea urchin egg secretory vesicles without irreversibly blocking fusion. J. Biol. Chem. 273:33667–33673. [DOI] [PubMed] [Google Scholar]
- 54.Suurkuusk, J., B. R. Lentz, Y. Barenholz, R. L. Biltonen, and T. E. Thompson. 1976. A calorimetric and fluorescent probe study of the gel-liquid crystalline phase transition in small, single-lamellar dipalmitoylphosphatidylcholine vesicles. Biochemistry. 15:1393–1401. [DOI] [PubMed] [Google Scholar]
- 55.Chen, Y., Y. Xu, F. Zhang, and Y. K. Shin. 2004. Constitutive versus regulated SNARE assembly: a structural basis. EMBO J. 23:681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han, X., C. T. Wang, J. Bai, E. R. Chapman, and M. B. Jackson. 2004. Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis. Science. 304:289–292. [DOI] [PubMed] [Google Scholar]
- 57.Szule, J. A., and J. R. Coorssen. 2004. Comment on “Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis”. Science. 306:813. [DOI] [PubMed] [Google Scholar]
- 58.Chernomordik, L., A. Chanturiya, J. Green, and J. Zimmerberg. 1995. The hemifusion intermediate and its conversion to complete fusion: regulation by membrane composition. Biophys. J. 69:922–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grote, E., C. M. Carr, and P. J. Novick. 2000. Ordering the final events in yeast exocytosis. J. Cell Biol. 151:439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]