Abstract
The ability of cells to form tissues represents one of the most fundamental issues in biology. However, it is unclear what triggers cells to adhere to one another in tissues and to migrate once a piece of tissue is planted on culture surfaces. Using substrates of identical chemical composition but different flexibility, we show that this process is controlled by substrate rigidity: on stiff substrates, cells migrate away from one another and spread on surfaces, whereas on soft substrates they merge to form tissue-like structures. Similar behavior was observed not only with fibroblastic and epithelial cell lines but also explants from neonatal rat hearts. Cell compaction on soft substrates involves a combination of weakened adhesions to the substrate and myosin II-dependent contractile forces that drive cells toward one another. Our results suggest that tissue formation and maintenance is regulated by differential mechanical signals between cell-cell and cell-substrate interactions, which in turn elicit differential contractile forces and adhesions to determine the preferred direction of cell migration and association.
INTRODUCTION
The ability to form tissues represents one of the most fundamental behaviors of metazoan cells. Under physiological conditions, most cells in a metazoan remain associated with one another or with the extracellular matrix (ECM) and rarely venture out of the home tissue. However, once a piece of tissue is placed on culture surfaces such as polystyrene or glass, most adherent cells will migrate rapidly away from one another to cover the culture surface. Although this migration behavior has greatly facilitated the preparation of cell lines and the investigation of cellular functions in vitro, it is well recognized that most adherent cells in vivo show active migration only upon stimulations, e.g., during tissue morphogenesis (1), tumor metastasis (2), or wound healing (3). Despite the importance, the mechanism that regulates such association/dissociation behavior remains largely unknown.
Although chemotaxis has been under intensive investigations for decades (4–6), it seems difficult to explain cell migration from tissue explants, which occurs under a wide range of chemical conditions. We suspect that it is physical properties of the environment that is primarily responsible for regulating the formation and maintenance of tissues. Mechanical interactions have indeed been demonstrated to be a potent means for cell-cell and cell-substrate communications. Adherent cells exert strong traction forces at their anchorage sites to the matrix and/or neighboring cells (7). In turn, mechanical forces applied to surface receptors can cause stiffening of the cortex (8,9), enlargement of focal adhesions (10,11), and redirection of cell migration (12). Furthermore, fibroblasts are able to actively probe the stiffness of their environment and to turn toward substrates of high rigidity (13), a phenomenon referred to as durotaxis. Since the stiffness of artificial culture substrates is typically much higher than that of tissues, we hypothesize that substrate rigidity is a key factor in dictating the different behavior on artificial substrates versus in vivo.
Although this hypothesis of durotaxis-driven tissue formation appeared consistent with the spontaneous formation of tissue-like aggregates for cells grown in soft ECM (14–16), the interpretation of previous studies is complicated by changes in multiple chemical and physical parameters. In this study, we take advantage of the flexible polyacrylamide substrates developed for cell mechanical studies (17). The major advantage of the material is that its rigidity may be varied over a wide range while maintaining constant chemical properties by changing the ratio of acrylamide and bis-acrylamide during polymerization. Our observations indicate that substrate rigidity does play a pivotal role in tissue formation and maintenance for both cell lines and primary cultures. In addition, the response may be explained by myosin II-driven contractility combined with differential adhesions.
MATERIALS AND METHODS
Polyacrylamide substrates and tissue culture
Polyacrylamide substrates were prepared as described previously (18). The substrates contain 5% total acrylamide and 0.012% (referred to as soft substrates), 0.06%, or 0.1% bis-acrylamide (referred to as stiff substrates). Young's moduli of the substrates were determined by atomic force microscopy ((AFM); see Appendix). The uniformity of collagen coating on the substrate surface was examined as described previously (13,19) by immunofluorescence staining with monoclonal anti-collagen type I IgG (clone COL-1; Sigma-Aldrich, St. Louis, MO; 1:500 dilution in phosphate buffered saline/bovine serum albumin (PBS/BSA)) and Fluoresbrite carboxylate beads coated with secondary antibodies (1-μm diameter; Polysciences, Warrington, PA; 1:100 dilution in PBS/BSA).
Balb/c 3T3 mouse embryonic fibroblasts and NRK-52E normal rat kidney epithelial cells were obtained from the American Type Cell Collection (Manassas, VA) and maintained as described previously (13,17). Cell aggregates were prepared with cells detached from petri dishes with either trypsin or an enzyme-free solution (Specialty Media, Phillipsburg, NJ) by centrifugation for 2 min at 2000 rpm. To obtain pieces of heart tissues, heart was removed from neonatal rat, quickly dipped in 70% ethanol, and washed with PBS until the solution was clear. The tissue was cut into small pieces ∼1 mm3 in size, washed with PBS, and planted on substrates.
Time-lapse microscopy, immunofluorescence staining, and drug application
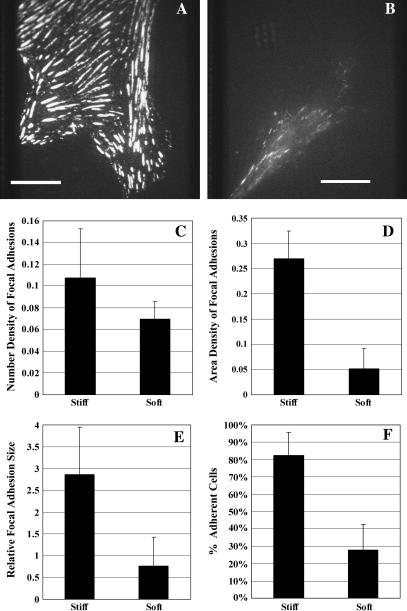
Phase-contrast images were recorded every 5 min with a cooled slow-scan charge-coupled device (CCD) camera (NTE/CCD-512-EBFT; Princeton Instruments, Trenton, NJ) or a video-rate CCD camera (12V1E-EX, Mintron, Taipei, Taiwan) attached to a Zeiss (Jena, Germany) Axiovert 100S microscope equipped with a 10×, CP-Achromat phase objective lens or a 40×, Plan-Neofluar phase objective lens and a stage incubator for time-lapse imaging. Immunofluorescence staining of focal adhesion was performed using primary antibodies against paxillin (ICN Biomedicals, Aurora, OH; 1:200 dilution in PBS/BSA), after fixing cells with 4% paraformaldehyde (EMS, Hatfield, PA). Images were collected with a 100× Plan-Neofluar numerical aperture 1.3 oil lens, a QLC100 spinning disk confocal head (Solamere Technology, Salt Lake City, UT), and a low light electron-multiplying CCD (EMCCD) camera (iXon DV887DCS-BV; Andor Technology, South Windsor, CT). Total focal adhesion area was calculated after intensity thresholding using custom software. Area density of focal adhesions was calculated as the total area of focal adhesions divided by the total cell area, and number density as the number of focal adhesions divided by the total cell area. Relative average focal adhesion size was calculated by dividing the total area of focal adhesions with the number of focal adhesions.
Cells were treated with 50 μM of Y-27632 (diluted from 10 mM stock solution in PBS; Calbiochem, San Diego, CA) or 100 μM of blebbistatin (diluted from 100 mM stock solution in dimethylsulfoxide; Calbiochem, San Diego, CA) after plating on substrates and incubating for 3–4 h.
Cell-substrate adhesion assay
Cell-substrate adhesiveness was measured with a centrifugation assay. Cells were plated on a modified 60 mm petri dish with polyacrylamide substrate attached to a large coverslip that forms the bottom surface and allowed to adhere for 25 min at room temperature. To avoid air bubbles that may interfere with the assay, a sealed subchamber was formed by placing a large glass slide in the chamber, separated from the lid and the substrate by two O-rings above and below. The chamber was clamped together with work clips and inverted and centrifuged for 10 min at 1,495 × g in a Beckman (Fullerton, CA) TJ-6 centrifuge with a TH-4 rotor. The force was determined empirically as the force required to detach ∼10–20% of the cells from stiff substrates. The number of cells on the substrate before and after centrifugation was counted, and the strength of adhesion was determined as the percentage of cells remaining on the substrate after centrifugation. Design of the chamber assembly is shown in Supplemental Fig. 1.
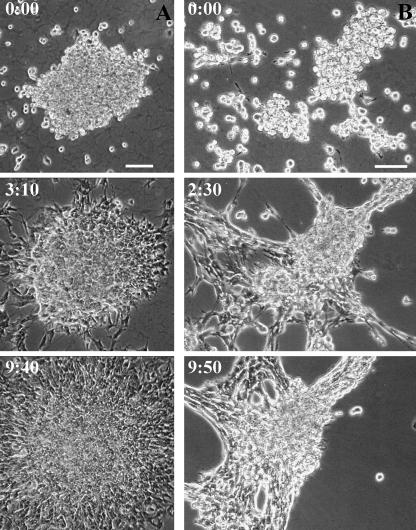
FIGURE 1.
Response of cell-cell associations of cultured fibroblasts to substrate flexibility. 3T3 fibroblast cell aggregates are plated on either stiff (A) or soft (B) substrates. On stiff substrates, cells show the typical scattering behavior as seen on conventional culture dishes (A). In contrast, cells form tissue-like aggregates when plated on soft substrates (B). Time in hours and minutes is indicated. Bar, 100 μm.
RESULTS
Flexible substrates induce tissue-like cell compaction
To explore if substrate rigidity plays a role in regulating cell-cell interactions during tissue formation, we prepared a series of polyacrylamide substrates with different rigidity but identical coating of type I collagen. Substrates prepared with 5% acrylamide and 0.1% or 0.06% bis-acrylamide will be referred to as stiff substrates, whereas those with 5% acrylamide and 0.012% bis-acrylamide will be referred to as soft substrates. Measurements with AFM yielded values of Young's modulus of 7.69 ± 2.85 kPa for stiff substrates with 0.1% bis-acrylamide and 2.68 ± 0.99 kPa for soft substrates, where the range of values represents the relative uncertainty. There is less uncertainty in values for indentation moduli, which are 12.40 ± 1.61 kPa and 4.41 kPa ± 0.57 kPa for stiff and soft samples, respectively (see Appendix). Immunostaining with anti-collagen antibodies and bead-conjugated secondary antibodies (to limit the detection to surface-bound collagen on the porous substrate) confirmed that the substrates were coated with a similar concentration of collagen irrespective of rigidity (13,19; and data not shown).
Cell migration on these substrates was tested primarily with Balb/c 3T3 fibroblasts, although similar results were obtained with NRK epithelial cells. Cells were plated either directly from a dispersed suspension or as aggregates after centrifugation of the suspension. Their behavior on stiff substrates was indistinguishable from that on glass or plastic surfaces. Individual cells spread and migrated on the substrate, showing the typical “contact inhibition” upon collision with other cells—cells moved away from one another and were never able to form sizeable three-dimensional aggregates even at a high density (Fig. 1 A; Supplementary Video 1). In contrast, cells plated on soft substrates readily aggregated with one another to form tight, tissue-like spheroids. The aggregates continuously incorporated cells and merged with one another until the culture turned into one or few very large aggregates (Fig. 1 B; Supplementary Video 2).
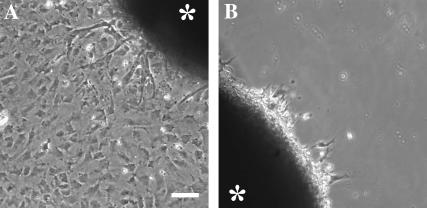
To determine if substrate rigidity is also responsible for the migration of cells from tissue explants in primary cultures, we performed similar experiments with small pieces of neonatal rat heart tissues. Consistent with the observations of cell lines, the cells migrated away from the tissue explants on stiff substrate (Fig. 2 A) but failed to do so on soft substrates (Fig. 2 B).
FIGURE 2.
Response of explanted neonatal cardiac tissues to substrate flexibility. Heart tissue explants from newborn rat are plated on either stiff (A) or soft (B) substrates for 3 days. Many cells have migrated out of the tissue explants onto stiff substrates (A), but few cells are able to migrate onto soft substrate (B). Asterisks indicate explanted tissues. Bar, 100 μm.
Physical contacts with soft substrates and with other cells directly activate cell compaction
To determine if cell-cell aggregation on soft substrates was signaled directly by physical contacts with the substrate or indirectly by chemical factors released into the medium, we cultured 3T3 cells on soft or stiff substrates placed within the same chamber in close proximity for 6 h. The behavior of cells was similar to that found in separate chambers (data not shown), indicating that cells responded directly to local physical stimuli rather than released chemical factors.
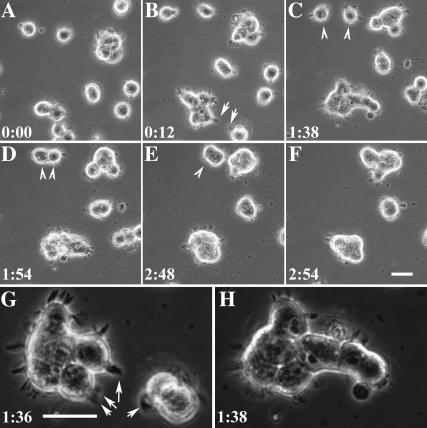
Time-lapse observations at a high magnification provided further clues as to the mechanism of cell compaction. Cells on soft substrates spread out poorly but were highly motile (17). They form dynamic, filopodia-like extensions, which appeared to contract upon cell-cell contact to bring cells into an aggregate (Fig. 3, arrowhead; Supplementary Video 3). Although cells in aggregates were unable to migrate out, those along the periphery maintained dynamic filopodia that extended for 5–7 μm from the edge of the aggregate (Fig. 3, arrow). Interactions of these filopodia upon contact with single cells or other aggregates again lead to the incorporation of additional cells and enlargement of the aggregates, possibly through contractions of the filopodia structure.
FIGURE 3.
Involvement of cell migration and contractions in cell compaction. Time-lapse images (A–F) show that 3T3 cells on soft substrates contracted toward each other upon establishment of contact through their extensions (arrowheads). Cells along the periphery of the aggregates maintain their dynamic extensions (arrows), which mediate interactions and incorporations of additional cells or aggregates. Enlarged views show that filopodia-like extensions formed by aggregates (G) were responsible for the incorporation of additional cells and enlargement of the aggregates (H). Bar, 25 μm.
Cell compaction on soft substrates is driven by a combination of myosin II-dependent contractile forces and differential adhesions
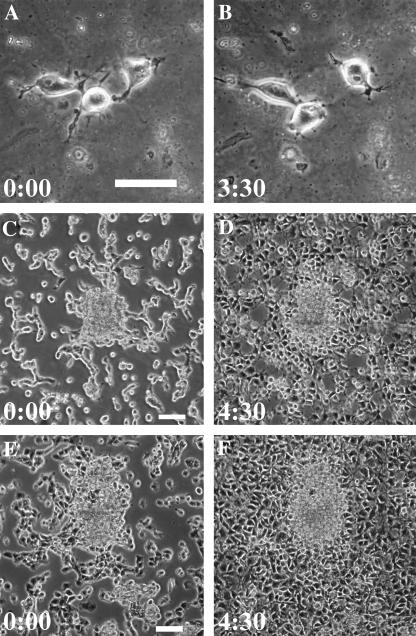
The above observations strongly suggest that cell compaction on soft substrates involves contractility. Since myosin II is the primary motor protein for the generation of traction forces (20,21), we asked if blebbistatin, a potent and reversible inhibitor of nonmuscle myosin II ATPase (22), is able to inhibit the compaction process. Dispersed cells maintained the ability to migrate randomly in the presence of blebbistatin for 3 h but were unable to form tissue-like aggregates on soft substrates (Fig. 4, A and B; Supplementary Video 4). In addition, when blebbistatin was added to cells that were plated on soft substrates for 3–4 h to allow the initiation of compaction, cells started random migration within 20 min, which eventually lead to the scattering of the aggregates (Fig. 4, C and D; Supplementary Video 5).
FIGURE 4.
Involvement of myosin II and Rho kinase activity in cell compaction. 3T3 cells are plated on soft substrates and treated with either blebbistatin (A–D) or Y-27632 (E and F). Single cells are able to migrate randomly, although the movement appears highly disorganized and cells fail to form aggregates upon contact (A and B). Application of blebbistatin to cells that have initiated compaction for 3–4 h (C) causes cells that have entered the aggregates to scatter (D). Application of Y-27632 causes a similar effect (E and F). Time in hours and minutes is indicated. Bar, 50 μm.
Since the Rho small GTPases and the downstream Rho-dependent kinase play a pivotal role in cell rounding and in regulating myosin II-dependent contractility (23,24), we asked if Y-27632, an inhibitor of Rho-dependent kinase, has a similar effect as does blebbistatin. Y-27632 applied after the initiation of compaction caused cells to disperse as did blebbistatin (Fig. 4, E and F; Supplementary Video 6), supporting a Rho-regulated, myosin II-dependent contractile mechanism that drove cells into aggregates.
In addition to contraction, cell aggregation/dispersion may be controlled by the strength of cell-substrate adhesions. From the poorly spread morphology on soft substrates (17), we suspected that cells may not adhere as strongly to soft substrate as they did to stiff substrates. Immunofluorescence staining of paxillin showed numerous large, elongated focal adhesions on stiff substrates (Fig. 5 A) and small, dot-like focal adhesions on soft substrates (Fig. 5 B). Striking differences were observed with the area density (p = 3.23 E-06; Fig. 5 D) and average size (p = 0.0014; Fig. 5 E) of focal adhesions. However, the number density of focal adhesions was not significantly higher on stiff substrates than on soft substrates (p = 0.0567; Fig. 5 C). Using a centrifugation assay to compare the strength of cell-substrate adhesions, we found that only ∼30% of cells remained on soft substrates after centrifugation, whereas more than 80% of the cells remained adherent to stiff substrates under the same condition (Fig. 5 F). A similar result was recently reported by using a micropipette peeling method (25). Together, these results suggest that substrate stiffness regulates the strength of adhesions by promoting the formation of large focal adhesions.
FIGURE 5.
Effects of substrate flexibility on the size of focal adhesions and the strength of adhesion. 3T3 cells are plated on stiff (A) or soft (B) substrates for 1 day, fixed and stained for focal adhesions with anti-paxillin monoclonal antibody. Bar, 20 microns. The number density of focal adhesions shows no significant difference between cells on stiff or soft substrates (p = 0.0567; C). However, both the area density (D; p < 0.01) and average size (E; relative size in pixels; p < 0.01) of focal adhesions are significantly higher on stiff substrates than on soft substrates. The strength of cell-substrate adhesion is measured with an inversion centrifugation assay. The percentage of cells remaining on the substrate after centrifugation is significantly higher for stiff substrates than for soft substrates (F; p < 0.01). Error bars represent standard deviations.
DISCUSSION
To maintain the stability of a multicellular organism, it is critical that cells remain stably associated with one another within specific tissues and migrate out only upon receiving signals such as after wounding. Although modern cell biology is largely built upon the ability of single cells to migrate away from explanted tissues to form primary and secondary cultures in vitro, there was no clear answer as to why cells show the paradoxically opposite behavior in vivo and in vitro. It was noted that many types of cells form aggregates when cultured in soft gels of basement membrane proteins and that in vitro angiogenesis took place only when endothelial cells were plated on “malleable” materials (14). However it was never clear if it is the rigidity or the chemical composition of the substrate that is primarily responsible for the tissue-forming behavior.
By allowing the variation of flexibility over a wide range without changing surface chemical properties, polyacrylamide substrates represent a powerful tool for testing the hypothesis that substrate flexibility plays a pivotal role in tissue formation. We demonstrated that on stiff substrates, cells dispersed from tissues or aggregates to cover the surface of the substrates. Conversely, on soft substrates, dispersed cells or small aggregates coalesce to form multicellular tissue-like structures. These observations may be understood if cells compare mechanical signals mediated by substrate adhesion with those by interactions with neighboring cells. They migrate away from one another if physical signals from the substrates are stronger than those from cell-cell interactions and toward one another if cell-cell interactions provide a stronger mechanical input.
This behavior of aggregation mimicking tissue formation may be driven at least partially by durotaxis, the preferential migration of cells toward stiff substrates or away from soft substrates (13). Closely related phenomena guide cells toward maximal adhesiveness (haptotaxis; (26)) or maximal substrate tension (13). The same process may also explain why, during tissue formation, cells tend to maximize their net area of adhesion to one another while minimizing the area exposed to the surrounding environment. The general principle underlying these observations appears to be a preference of cells to maximize mechanical input from the environment. This surface tension-like mechanism is believed to also drive cell sorting as well as morphogenetic movements during gastrulation and neurulation (27).
Our results further suggest that cell migration toward maximal mechanical input is driven by differential traction forces and substrate adhesions. We reported previously that cells exert stronger traction forces on stiff than on soft substrates (19) and that active traction forces are concentrated at the leading edge where nascent focal adhesions are assembled (28,29). Substrate stiffness also regulates the strength of adhesion, as seen in these experiments and in a recent micromanipulation study (25). Furthermore, a recent report indicated that β5 integrin was expressed at a higher level on stiff substrates than on soft substrates (30), which may contribute to the enhancement of adhesion. A closely related phenomenon is referred to as “cortical reinforcement,” where mechanical forces exerted through adhesive microbeads induce strengthening of cortical forces and rigidity (8,9). Therefore, a plausible mechanism for cell compaction would involve a tug of war, possibly combined with a negative feedback loop, among protrusions and adhesions in different regions. If those interacting with neighboring cells receive a stronger mechanical stimulation than those adhering to substrates, localized increase in adhesion strength and traction forces would drive cells toward each other into an aggregate. Implied in this model is that, at the mechanical level, cell-ECM adhesions compete directly with cell-cell adhesions in determining the interactions between cells and the environment. In addition, cadherins at cell-cell adhesions may show similar responses to mechanical signals as does paxillin at focal adhesions, as supported by the similar interactions of cell-cell and cell-ECM adhesion structures with the actin cytoskeleton and by a recent report that cells in myosin II-A-deficient mice are defective in cell-cell adhesions (31). However, our attempts to image and quantify cadherin in tissue-like aggregates were hampered by limited optical qualities.
Consistent with this idea, pharmacological studies suggest that myosin II plays a major role in the cell compaction process, likely under the regulation of the small GTPase Rho and Rho-dependent kinase, which phosphorylates myosin light chain and inhibits myosin phosphatase (32). Myosin II may be involved in tissue formation in three ways. First, myosin II is known to generate tractions forces (20,21) and is likely to be responsible for the contraction of cellular processes during the compaction process. Second, myosin II may provide forces for probing the flexibility of the environment, which cannot otherwise be detected in a chemically homogeneous environment. This is supported by the defects of myosin II-deficient cells in establishing polarity and undergoing durotaxis (33). Third, myosin II, together with Rho, is known to regulate the formation of focal adhesions, the affinity of integrins for the ECM, and cadherin-mediated cell-cell adhesions (31), a phenomenon referred to as inside-out signaling (34).
Substrate stiffness regulates not only cell migration and adhesion but also cell growth and apoptosis (19), which are closely related to cell shape and adhesion and are equally important in tissue formation. It is also important to note that stiffness varies over a wide range among different tissues and different types of cells also show quantitative and/or qualitative differences in their responses to substrate stiffness. For example, myocytes prefer moderately stiff substrates (25), whereas neurons grow more branching on soft than on stiff substrates (35). Therefore, although we designated soft and stiff in this report based on the behavior of fibroblasts and epithelial cells, the same physical signal may elicit cell type-specific responses.
These observations are relevant not only to tissue formation in development but also a number of important physiological and pathological processes. For example, cell migration during wound healing may be guided by the contractile forces at the wound, whereas cancer metastasis may be caused by cells losing the ability to respond to the differential mechanical signals between the home tissue and the basement membrane. Cancerous invasion may be further stimulated by the increased stiffness of the connective tissue referred to as desmoplasia, which occurs in some tumors such as breast cancer and pancreatic adenocarcinoma (36,37). Finally, given the profound effects of mechanical signals on cell migration and growth, engineering of artificial tissues as well as design of cell-based therapies must take into account physical parameters, particularly rigidity of the materials and the environment.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Barbara Walker for providing neonatal rats and Stephen Lambert for helpful discussion. We also thank Adam Engler and Dennis Discher at the University of Pennsylvania for advice on atomic force microscopy.
This work was funded by the National Institutes of Health grant GM-32476 to Yu-li Wang.
APPENDIX: CHARACTERIZATION OF POLYACRYLAMIDE GELS WITH ATOMIC FORCE MICROSCOPY
Materials and Methods
Hertz contact mechanics and atomic force microscopy
Hertz contact mechanics relates force, F, to indentation depth, δ, based on the geometry of an indenter for two elastic bodies. The theoretical indentation of a planar sample with a spherical indenter of radius, R, is also related to the Poisson ratio, ν, and Young's modulus, E, of the sample (Eq. 1) (38). An AFM can be used as a nano-indenter to obtain force-indentation profiles of a sample for a given probe geometry; in this case, the force is related to the spring constant, k, of the indenter and the resulting deflection, d (Eq. 2).
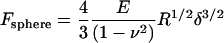 |
(1) |
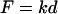 |
(2) |
The force-indentation profile obtained with the AFM (Eq. 2) is then fit to the theoretical model of contact (Eq. 1) to estimate the Young's modulus of the sample.
AFM measurements of polyacrylamide gels
Polyacrylamide substrates of 5% total acrylamide and 0.10% or 0.012% bis-acrylamide were prepared as described previously (18) and were stored at 4°C in PBS for no more than 3 days before testing. Testing was performed using an Autoprobe M4 AFM (Veeco, Santa Barbara, CA) equipped with ProScan V1.51b software (Veeco) and a MicroCell Kit (APMC-0001; Veeco) for testing hydrated samples.
Measurements were performed with two different types of AFM probes: particle tip contact mode probes (BioForce Nanosciences, Ames, IA) with a 5-μm spherical borosilicate tip on a cantilever with a nominal spring constant of 0.06 N/m, and probes with 20 μm polystyrene beads (Polysciences; Warrington, PA) glued onto CSC12 tipless cantilevers (MikroMasch, Wilsonville, OR), using Norland Optical Adhesive No. 71 (Norland Products, Cranbury, NJ) cured for 30 min under ultraviolet light, with a nominal spring constant of either 0.05 or 0.03 N/m. These probes gave comparable results. Before measurements were made on the substrates, the stiffness of each cantilever was accurately determined using a thermal calibration method (39) and tip geometry was imaged using a TGZ03 step grating (MikroMasch). The spherical cap portions of the tip-imaging profiles were fit to a spherical model to yield the tip radius (40). Force-indentation profiles were obtained at a rate of 1 Hz on multiple samples (n = 3 and 4 for stiff and soft samples, respectively) at various locations. Each measurement (n = 15 and 13 for the stiff and soft samples, respectively) is the average of either 16 or 10 force curves at that location.
Results and Discussion
The retraction portion of the force-indentation profiles obtained by AFM was used for analysis (41). These profiles and the values from the theoretical Hertz sphere model at the corresponding indentation depths were first converted to logarithmic form. These data were then fit using a linear least squares method and Excel Solver to yield Young's moduli of 7.69 ± 2.85 kPa and 2.68 ± 0.99 kPa for the stiff and soft samples, respectively, where the range of values represents the relative uncertainty. Results for a depth up to 100 nm did not vary appreciably using either a fixed range of indentation or a range optimized for each test based on the logarithmic fit. At the sampling frequency of 1 Hz, the indentation interaction may not have been quasistatic; this could yield moduli values somewhat greater than those obtained with lower frequencies (42). However we found that lower sampling frequencies increase the uncertainty, at least with the present device.
Error analysis
The estimated uncertainty of the value for E (Eq. 3) is mainly affected by the uncertainty of the value for ν (43); values between 0.3 and 0.5 have been used, though we chose a value of 0.45 in our estimates (44).
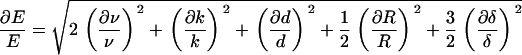 |
(3) |
For uncertainties of 0.10, 0.02, 0.09, and 0.04 for k, d, R, and δ, respectively (39,40), the uncertainty of E was calculated to be 37%. Indentation modulus, Kind (Eq. 4), can describe the material properties without the dependence on ν and thus has an uncertainty of only 13%.
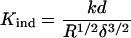 |
(4) |
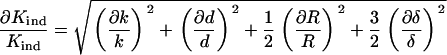 |
(5) |
Therefore, it is preferable to describe material properties of the substrates using the indentation modulus. Indentation moduli are 12.40 ± 1.61 kPa and 4.41 ± 0.57 kPa for the stiff and soft samples, respectively.
References
- 1.Juliano, R. L., and S. Haskill. 1993. Signal transduction from the extracellular matrix. J. Cell Biol. 120:577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein, L. R., and L. A. Liotta. 1994. Molecular mediators of interactions with extracellular matrix components in metastasis and angiogenesis. Curr. Opin. Oncol. 6:106–113. [DOI] [PubMed] [Google Scholar]
- 3.Martin, P. 1997. Wound healing: aiming for perfect skin regeneration. Science. 276:75–81. [DOI] [PubMed] [Google Scholar]
- 4.Parent, C. A., and P. N. Devreotes. 1999. A cell's sense of direction. Science. 284:765–770. [DOI] [PubMed] [Google Scholar]
- 5.Chung, C. Y., S. Funamoto, and R. A. Firtel. 2001. Signaling pathways controlling cell polarity and chemotaxis. Trends Biochem. Sci. 26:557–566. [DOI] [PubMed] [Google Scholar]
- 6.Postma, M., L. Bosgraaf, H. M. Loovers, and P. J. Van Haastert. 2004. Chemotaxis: signalling modules join hands at front and tail. EMBO Rep. 5:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dembo, M., and Y.-L. Wang. 1999. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 76:2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang, N., J. P. Butler, and D. E. Ingber. 1993. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 260:1124–1127. [DOI] [PubMed] [Google Scholar]
- 9.Choquet, D., D. P. Felsenfeld, and M. P. Sheetz. 1997. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell. 88:39–48. [DOI] [PubMed] [Google Scholar]
- 10.Riveline, D., E. Zamir, N. Q. Balaban, U. S. Schwarz, T. Ishizaki, S. Narumiya, Z. Kam, B. Geiger, and A. D. Bershadsky. 2001. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J. Cell Biol. 153:1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, H.-B., M. Dembo, S. K. Hanks, and Y.-L. Wang. 2001. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc. Natl. Acad. Sci. USA. 98:11295–11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, S., P. Butler, Y. Wang, Y. Hu, D. Han, S. Usami, J. Guan, and S. Chien. 2002. The role of the dynamics of focal adhesion kinase in the mechanotaxis of endothelial cells. Proc. Natl. Acad. Sci. USA. 99:3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo, C.-M., H.-B. Wang, M. Dembo, and Y.-L. Wang. 2000. Cell movement is guided by the rigidity of the substrates. Biophys. J. 79:144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingber, D. E., and J. Folkman. 1989. Mechanochemical switching between growth and differentiation during fibroblast growth factor-stimulated angiogenesis in vitro: role of extracellular matrix. J. Cell Biol. 109:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keely, P., A. Fong, M. Zutter, and S. Santoro. 1995. Alteration of collagen-dependent adhesion, motility, and morphogenesis by the expression of antisense α 2 integrin mRNA in mammary cells. J. Cell Sci. 108:595–607. [DOI] [PubMed] [Google Scholar]
- 16.Wozniak, M. A., D. Radhika, A. S. Patricia, J. D. Channing, and P. J. Keely. 2003. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J. Cell Biol. 163:583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pelham, R. J. Jr., and Y.-L. Wang. 1997. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 94:13661–13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, Y.-L., and R. J. Pelham Jr. 1998. Preparation of a flexible, porous polyacrylamide substrate for mechanical studies of cultured cells. Methods Enzymol. 298:489–496. [DOI] [PubMed] [Google Scholar]
- 19.Wang, H.-B., M. Dembo, and Y.-L. Wang. 2000. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am. J. Physiol. Cell Physiol. 279:C1345–C1350. [DOI] [PubMed] [Google Scholar]
- 20.Jay, P. Y., P. A. Pham, S. A. Wong, and E. L. Elson. 1995. A mechanical function of myosin II in cell motility. J. Cell Sci. 108:387–393. [DOI] [PubMed] [Google Scholar]
- 21.Pelham, R. J. Jr., and Y.-L. Wang. 1999. High resolution detection of mechanical forces exerted by locomoting fibroblasts on the substrate. Mol. Biol. Cell. 10:935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Straight, A. F., A. Cheung, J. Limouze, I. Chen, N. J. Westwood, J. R. Sellers, and T. J. Mitchison. 2003. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 299:1743–1747. [DOI] [PubMed] [Google Scholar]
- 23.Raftopoulou, M., and A. Hall. 2004. Cell migration: Rho GTPases lead the way. Dev. Biol. 265:23–32. [DOI] [PubMed] [Google Scholar]
- 24.Maekawa, M., T. Ishizaki, S. Boku, N. Watanabe, A. Fujita, A. Iwamatsu, T. Obinata, K. Ohashi, K. Mizuno, and S. Narumiya. 1999. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 285:895–898. [DOI] [PubMed] [Google Scholar]
- 25.Engler, A. J., M. A. Griffin, S. Sen, C. G. Bonnemann, H. L. Sweeney, and D. E. Discher. 2004. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J. Cell Biol. 166:877–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carter, S. B. 1967. Haptotaxis and the mechanism of cell motility. Nature. 213:256–260. [DOI] [PubMed] [Google Scholar]
- 27.Foty, R. A., C. M. Pfleger, G. Forgacs, and M. S. Steinberg. 1996. Surface tensions of embryonic cells predict their mutual envelopment behavior. Development. 122:1611–1620. [DOI] [PubMed] [Google Scholar]
- 28.Munevar, S., M. Dembo, and Y.-L. Wang. 2001. Traction force microscopy of normal and transformed fibroblasts. Biophys. J. 80:1744–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beningo, K. A., M. Dembo, I. Kaverina, J. V. Small, and Y.-L. Wang. 2001. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 153:881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeung, T., P. C. Georges, L. A. Flanagan, B. Marg, M. Ortiz, M. Funaki, N. Zahir, W. Ming, V. Weaver, and P. A. Janmey. 2005. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 60:24–34. [DOI] [PubMed] [Google Scholar]
- 31.Conti, M. A., S. Even-Ram, C. Liu, K. M. Yamada, and R. S. Adelstein. 2004. Defects in cell adhesion and the visceral endoderm following ablation of nonmuscle myosin heavy chain II-A in mice. J. Biol. Chem. 279:41263–41266. [DOI] [PubMed] [Google Scholar]
- 32.Kimura, K., M. Ito, M. Amano, K. Chihara, Y. Fukata, M. Nakafuku, B. Yamamori, J. Feng, T. Nakano, K. Okawa, A. Iwamatsu, and K. Kaibuchi. 1996. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science. 273:245–248. [DOI] [PubMed] [Google Scholar]
- 33.Lo, C.-M., D. B. Buxton, G. C. Chua, M. Dembo, R. S. Adelstein, and Y.-L. Wang. 2004. Nonmuscle myosin IIB is involved in the guidance of fibroblast migration. Mol. Biol. Cell. 15:982–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz, M. A., M. D. Schaller, and M. H. Ginsberg. 1995. Integrins: emerging paradigms of signal transduction. Annu. Rev. Cell Dev. Biol. 11:549–599. [DOI] [PubMed] [Google Scholar]
- 35.Flanagan, L. A., Y. E. Ju, B. Marg, M. Osterfield, and P. A. Janmey. 2002. Neurite branching on deformable substrates. Neuroreport. 13:2411–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller, M. M., and N. E. Fusenig. 2004. Friends or foes—bipolar effects of the tumour stroma in cancer. Nat. Rev. Cancer. 4:839–849. [DOI] [PubMed] [Google Scholar]
- 37.Paszek, J., N. Zahir, K. R. Johnson, J. N. Lakins, G. I. Rozenberg, A. Gefen, C. A. Reinhart-King, S. S. Margulies, M. Dembo, D. Boettiger, D. A. Hammer, and V. M. Weaver. 2005. Tensional homeostasis and the malignant phenotype. Cancer Cell. 8:241–254. [DOI] [PubMed] [Google Scholar]
- 38.Hertz, H. 1882. Uber die Beruhrung fester elastischer Korper. J. Reine Angew. Mathematik. [in German]. 92:156–171. [Google Scholar]
- 39.Burnham, N. A., X. Chen, C. S. Hodges, G. A. Matei, E. J. Thoreson, C. J. Roberts, M. C. Davies, and S. J. B. Tendler. 2003. Comparison of calibration methods for atomic-force microscopy cantilevers. Nanotechnology. 14:1–6. [Google Scholar]
- 40.Thoreson, E. J., and N. A. Burnham. 2004. Standard-deviation minimization for calibrating the radii of spheres attached to AFM cantilevers. Rev. Sci. Instrum. 75:1359–1362. [Google Scholar]
- 41.Burnham, N. A., and R. J. Colton. 1989. Measuring the nanomechanical properties and surface forces of materials using an atomic force microscope. J. Vac. Sci. Technol. A. 7:2906–2913. [Google Scholar]
- 42.Mahaffy, R. E., S. Park, E. Gerde, J. Kas, and C. K. Shih. 2004. Quantitative analysis of the viscoelastic properties of thin regions of fibroblasts using atomic force microscopy. Biophys. J. 86:1777–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor, J. R. 1997. An Introduction to Error Analysis: The Study of Uncertainties in Physical Measurements. University Science Books, Sausalito, CA.
- 44.Engler, A., L. Bacakova, C. Newman, A. Hategan, M. Griffin, and D. Discher. 2004. Substrate compliance versus ligand density in cell on gel responses. Biophys. J. 86:617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.