Abstract
A putative pheromone precursor gene of Neurospora crassa, mfa-1 (which encodes mating factor a-1), was identified as the most abundant clone in starved mycelial and perithecial cDNA libraries. Northern analysis demonstrated high mfa-1 expression in all mating type a tissues and suggested low expression levels in mat A tissues. The mfa-1 gene was expressed as an approximately 1.2-kb transcript predicted to encode a 24-residue peptide, followed by a long 3′ untranslated region (3′ UTR). The predicted MFA1 sequence showed 100% sequence identity to PPG2 of Sordaria macrospora and structural similarity (a carboxy-terminal CAAX motif) to many hydrophobic fungal pheromone precursors. Mutants with a disrupted open reading frame (ORF) in which the critical cysteine residue had been changed to a nonprenylatable residue, tyrosine (YAAX mutants), were isolated, as were mfa-1 mutants with intact ORFs but multiple mutations in the 3′ noncoding region (CAAX mutants). The 3′ UTR is required for the full range of mfa-1 gene activity. Both classes of mutants showed delayed and reduced vegetative growth (which was suppressed by supplementation with a minute amount [30 μM] of ornithine, citrulline, or arginine), as well as aberrant sexual development. When crossed as female parents to wild-type males, the CAAX and YAAX mutants showed greatly reduced ascospore production. No ascospores were produced in homozygous mfa-1 crosses. As males, YAAX mat a mutants were unable to attract wild-type mat A trichogynes (female-specific hyphae) or to initiate sexual development, while CAAX mat a mutants were able to mate and produce sexual progeny despite their inability to attract mat A trichogynes. In the mat A background, both CAAX and YAAX mutants showed normal male fertility but defective vegetative growth and aberrant female sexual development. Thus, the mfa-1 gene appears to have multiple roles in N. crassa development: (i) it encodes a hydrophobic pheromone with a putative farnesylated and carboxymethylated C-terminal cysteine residue, required by mat a to attract trichogynes of mat A; (ii) it is involved in female sexual development and ascospore production in both mating types; and (iii) it functions in vegetative growth of both mating types.
In heterothallic fungi, mating occurs only between haploid cells of opposite mating types; the mating is initiated by cell-specific pheromone and receptor combinations (9, 36). The specific recognition launches a complex mitogen-activated protein kinase (MAPK) cascade, the pheromone response pathway. To date, two types of pheromone precursor genes have been identified in the fungi; the MFa and MFα genes of Saccharomyces cerevisiae are the best-studied examples (39). The MFa gene encodes a short peptide with a C-terminal CAAX motif (C = cysteine, A = aliphatic, and X = any amino acid residue); the mature a-factor mating pheromone is highly hydrophobic due to prenylation at the cysteine residue, which is also carboxymethylated. The MFα gene encodes a precursor containing multiple repeats of a pheromone sequence bordered by Kex2 protease processing sites; the mature α-factor is unmodified and hydrophilic. These critical features of pheromone precursor genes are conserved in other fungi.
Pheromone precursor genes have been identified throughout the fungal kingdom. In addition to S. cerevisiae, other heterothallic ascomycetes also contain pheromone precursor genes (19, 31, 58, 68), as do heterothallic basidiomycetes (4, 9, 14, 42, 61), homobasidiomycetes (36, 46), and a homothallic ascomycete (50). However, both classes of pheromone precursor genes have been identified in ascomycetes, while only one class of precursor genes (Mfa-like) has been identified in basidiomycetes. The presence of pheromone precursor genes in homothallic fungi suggests that their products may have functions beyond that of attracting a mate; this supposition has been supported by recent studies. In some fungi, pheromones play a role in postfusion events as well as cell-cell recognition and fusion. Specifically, pheromones have been implicated in the induction of meiosis in Schizosaccharomyces pombe, stimulation of filamentous growth in Ustilago maydis, and internuclear recognition in Schizophyllum commune and Podospora anserina (18, 22, 61). The suggested requirement of pheromones for postfertilization events such as karyogamy and meiosis may explain the presence of pheromones in homothallic species (9, 22, 50).
Neurospora crassa, a heterothallic filamentous fungus with two mating types, mat a and mat A, undergoes more complex sexual development than do the yeasts (38). Under conditions of nitrogen starvation, light, and low temperature N. crassa forms a spherical prefruiting body (protoperithecium) (48). This female reproductive structure extends a receptive hypha (trichogyne) toward a cell of the opposite mating type in a process mediated by pheromones (8). The trichogyne recruits a single fertilizing (or male) nucleus; the fertilized protoperithecium then develops into a perithecium, or fruiting body, from which the haploid meiotic products (ascospores) are eventually ejected.
In this study, a pheromone precursor gene of N. crassa, mfa-1 (previously called poi-1 [for plenty of it]), was analyzed at the molecular and functional levels. The results showed that the mfa-1 gene encodes a pheromone belonging to the CAAX family of hydrophobic peptide pheromones, and that the pheromone plays complex roles in N. crassa throughout sexual development (pre- and postfertilization) as well as in vegetative growth.
MATERIALS AND METHODS
Neurospora strains and plasmids.
The following N. crassa strains were used (where a refers to mat a and A refers to mat A): the wild-type 74 A (74-OR23-IV A [FGSC 2489]) and ORS a (FGSC 2490) strains and the mutant strains fl A (FGSC 4317), fl a (FGSC 4347), lys-1 inl A (FGSC 209), fmf-1 pyr-3 A (FGSC 3108), qa-2 aro-9 inl al-2 a (RLM 63-01), Am44 (un-3 ad-3A nic-2 cyh-1 Am44 [FGSC 4570]) and am1 (ad-3B cyh-1 am1 [FGSC 4564]). A homothallic mat A idiomorph Neurospora species, N. africana (FGSC 1740) was also used. (Strains were from the Fungal Genetics Stock Center [FGSC], University of Kansas Medical Center, Kansas City, and the laboratory of R. L. Metzenberg [RLM]). FGSC strains 4411, 4416, and 4450 through 4487 were also used to carry out restriction fragment length polymorphism (RFLP) mapping.
A specialized plasmid, pMSN1 (43), containing the qa-2+ gene of N. crassa, was used as a selective marker for transformants. The pGEM3zf(+) vector (Promega, Madison, Wis.) was used in subcloning an approximately 2.9-kb XhoI fragment of mfa-1+ genomic DNA from a genomic cosmid clone (X23C8, the Orbach/Sachs pMOcosX library obtained from FGSC). The cosmid clone was used in RFLP mapping and complementation experiments as well. cDNA clones (NP2A8, NP2B4, NP2B5, NP2A12, NM3F9, and NM4D3) from the Neurospora Genome Project (44), containing the longest mfa-1 inserts, were used in sequence analyses, gene disruption experiments, and Southern and Northern analyses.
Media and culture conditions.
N. crassa cultures were maintained on solid Vogel's minimal medium N (66) with 1.5% sucrose plus any needed supplements. Strains were vegetatively grown either in liquid Vogel's medium, on solid medium in petri dishes (100 mm), or in race tubes (50 cm in length and 18 mm in diameter). All crosses were carried out on crossing medium containing 1% sucrose and any needed supplements (67). In growth of cultures for vegetative and perithecial RNA isolation, conidia were inoculated at 106/ml and strains were grown as described by Nelson and Metzenberg (43). For RNAs from mating conditions, strains were grown as floating mycelial mats on liquid crossing medium and incubated without agitation at 25°C in the light for 3 to 6 days. For preparation of perithecial RNAs, the fluffy strain fl a or fl A was grown on crossing plates covered with sterile Miracloth circles (Calbiochem) and fertilized with a heavy conidial suspension of the opposite mating type, 74 A or ORS a, respectively. Either 7 or 9 days after fertilization, the perithecia were scraped from the crossing plates, immediately frozen, and ground in liquid nitrogen, using a mortar and pestle. N. africana cultures were maintained on Vogel's slants with 1.5% sucrose. For preparation of N. africana perithecial RNAs, a fresh culture was finely crushed using a sterile mortar and pestle, evenly spread on crossing plates covered with sterile Miracloth circles, and grown for 7 or 9 days. The perithecia scraped from the plates were treated as described above.
DNA sequencing and sequence analyses.
The cDNA and genomic mfa-1 sequences were determined using the dideoxy chain termination method (55), using the Applied Biosystems (ABI) PRISM dye terminator kit (Perkin-Elmer) and the ABI model 377 DNA sequencer. To identify potential homologs, the DNA sequence of the mfa-1 gene and its derived protein sequence were compared with the DNA and protein databases available through the National Center for Biotechnology Information, using the BLAST algorithms (1, 2). The mfa-1 sequences were also analyzed using the DNAsis software package. Putative components of the pheromone response MAPK pathway were identified by analysis of the nearly complete N. crassa genome sequence available from the Whitehead Institute Center for Genome Research (http://www-genome.wi.mit.edu/annotation/fungi/neurospora/).
Preparation of genomic DNA and RNA.
Genomic N. crassa DNA was isolated as described previously (63). Total RNA was isolated using the PURESCRIPT RNA isolation kit (Gentra Systems, Minneapolis, Minn.), per the manufacturer's instructions.
Northern and Southern hybridization blots.
Northern and Southern blot procedures were performed as described previously (54). The constitutively expressed glutamate dehydrogenase (am) gene (34) was used as a control in all Northern blots. Probes were prepared using the random priming method (25), with the Pharmacia Biotech oligolabeling kit.
Heterologous Southern hybridizations (zoo blots).
In order to identify potential mfa-1 homologs in other fungi, low-stringency hybridization was performed. The ascomycetes used in the zoo blots were as follows: Gelasinospora reticulospora (FGSC 960), Gelasinospora tetrasperma (FGSC 966), Neurospora tetrasperma (FGSC 1270 and 1271), N. africana (FGSC 1740), Neurospora galapagosensis (FGSC 1739), Neurospora intermedia (FGSC 2316 and 1940), Neurospora sitophila (FGSC 2216 and 2217), Neurospora terricola (FGSC 1889), Sordaria fimicola (FGSC 2918), Sordaria macrospora (FGSC 4818), and Anixiella sublineolata (FGSC 5508). The N. crassa 74 A strain was included as a positive control. Genomic DNAs from the various fungal species were digested with XhoI, electrophoresed in 0.9% agarose gels, and then transferred to nylon membranes. The blots were probed overnight at 55°C and washed at low stringency (two 15-min washes in 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] at room temperature, followed by two 20-min washes at 50°C). These blots allowed the detection of sequences showing at least 55 to 65% nucleotide sequence identity (10).
RFLP mapping of the mfa-1 gene.
The map location of a cosmid encoding mfa-1 was determined using the RFLP technique (41, 45). This method uses RFLPs as genetic markers and examines the ordered progeny from a cross of a multiply marked laboratory strain (multicent-2 a, in Oak Ridge genetic background) with a wild-collected strain (Mauriceville-1c A). For this analysis, the cosmid encoding mfa-1 was linearized with a restriction enzyme, labeled, and hybridized with Southern blots of digested genomic DNAs from the mapping strains.
Gene disruption experiments (repeat-induced point [RIP] mutation method).
The pMSN1 plasmid and mfa-1 cDNA were cotransformed into freshly harvested conidia of the qa-2 aro-9 inl al-2 a (RLM 63-01) strain via electroporation (15). The qa-2+ transformants were selected based on their ability to grow without the aromatic amino acid supplement required by qa-2 aro-9 double-mutant strains. Single-colony isolates of the primary transformants were judged to be homokaryotic if they failed to produce conidia requiring the aromatic amino acid supplement (43). Transformants containing only two copies of mfa-1 (an endogenous mfa-1+ and one extra copy introduced by transformation) were identified by Southern hybridization. The selected transformants were then crossed with lys-1 inl A (FGSC 209), which has an auxotrophic mutation (lys-1) mapping near the mfa-1 gene and contains no duplicated mfa-1 sequences. The progeny of these crosses were plated on medium lacking the lysine required by the auxotrophic normal sequence strain, consequently enriching for progeny containing the potentially disrupted mfa-1 gene (43).
Sequence analysis of mfa-1 mutants.
The sequences of meiotic progeny with putative disrupted mfa-1 genes were examined. A 2.5-kbp fragment including the endogenous mfa-1 gene of selected progeny was amplified by PCR. Approximately 0.5 μg of genomic DNA was amplified with primers 1 and 2 (5′ TGAGAGAATAGGATAGCCCG 3′ and 5′ ACCTAACCTGGCCGAAACAG 3′). The PCR product was verified by agarose gel electrophoresis, purified using the QIAquick spin PCR purification kit (Qiagen), and sequenced using nested primers 3, 4, 5, and 6 (5′ TAAGAACGTCCGTCTCCTCC 3′, 5′ GTCTTACGAAGAGGCATACG 3′, 5′ GGGAAACGATGCACAACTTC 3′, and 5′ AAGGAAGATCCTCACCGCTC 3′, respectively) with the ABI PRISM dye terminator kit (Perkin-Elmer) and ABI model 377 DNA sequencer. Primers 1 to 4 were designed from the flanking sequences of the mfa-1 gene, and primers 5 and 6 were from the mfa-1 cDNA sequence. The mfa-1 sequences were compared with the wild-type mfa-1 sequence to identify specific mutations.
Linear growth tests.
Growth rates of the mfa-1 mutant strains were tested on minimal, complex (supplemented with 0.5% yeast extract), and low-nitrogen (Westergaard) media at either 25 or 30°C, using race tubes (21).
Fertility tests.
To examine the role of the mfa-1 gene in sexual development, mfa-1 mutant strains were crossed to wild type as either the female (protoperithecial) or male (fertilizing) parent to detect dominant mating-specific defects (43). When used as a male, a small drop of mfa-1 conidial suspension was spotted onto fluffy (fl A and fl a) strains grown on plates with crossing medium. The fluffy mutants feature high fertility and the inability to produce macroconidia (40). These crosses also served to identify the mating type of the mutants, as perithecia were formed on the fl A or the fl a plates. When used as females, the mutants were grown in crossing slants and fertilized with conidia of the opposite mating type. The mfa-1 strains were crossed to sibling strains to detect recessive mutations affecting sexual development (43).
Mating response tests.
Attraction between trichogynes of one mating type and conidia of the opposite mating type was assayed to investigate pheromone function in the mfa-1 mutants. An agar strip covered by dense growth of fl a or fl A was placed next to a 2% agar strip, and a heavy suspension of wild-type or mfa-1 conidia was streaked on the 2% agar approximately 5 mm from the fluffy strain. Mating responses were monitored to determine if the conidia could stimulate the directed growth of trichogynes at this distance and to localize the sites of perithecial production.
Complementation of the mfa-1 defect.
Transformation of an mfa-1 YAAX mat a mutant with genomic mfa-1+ DNA (X23C8 genomic cosmid clone) was carried out via electroporation (15). Freshly harvested conidia of a YAAX mat a mutant were transformed with the genomic cosmid clone containing the mfa-1+ gene in the pMOcosX vector (obtained from the FGSC). The pMOcosX vector contains an ampicillin resistance gene as a selective marker in Escherichia coli and a hygromycin B resistance gene (Hyr) as a selective marker in N. crassa (47). Transformants were selected based on their resistance to the fungicide hygromycin B (220 μg/ml) and tested for complementation of vegetative and sexual defects of the mfa-1 mutant.
RESULTS
Representation of the mfa-1 cDNA in cDNA libraries.
The mfa-1 gene was identified by the Neurospora Genome Project at the University of New Mexico as the gene that is the most highly expressed under starved conditions (44). In the project, three cDNA libraries were initially constructed using mRNA isolated from conidial (germinating asexual spores), starved mycelial (branching hyphae), and perithecial (fruiting body) tissues (44). An additional cDNA library, the Westergaard library, was subsequently constructed to represent unfertilized sexual or protoperithecial tissue (24). More recently, two “time-of-day-specific” cDNA libraries (morning and evening libraries) were constructed and analyzed to address the expression of genes in mycelial tissues of N. crassa over the course of the circadian day (69). The four developmental cycle-specific cDNA libraries were constructed using RNAs extracted from the mat A wild-type strain 74 A (24, 44), and the two time-of-day-specific cDNA libraries were constructed using RNAs from selected circadian rhythm cycle mutant strains in a mat a background (bd; a and bd; a; frq7 [69]).
Analysis of the unsubtracted libraries showed that mfa-1 clones account for 4.6% of the starved mycelial library (43 of 937 isolates) and 5.5% of the perithecial library (121 of 2,196 isolates), both of which represent starved conditions. Neither cDNA has been identified in the conidial, Westergaard, or morning or evening libraries.
Sequence analyses.
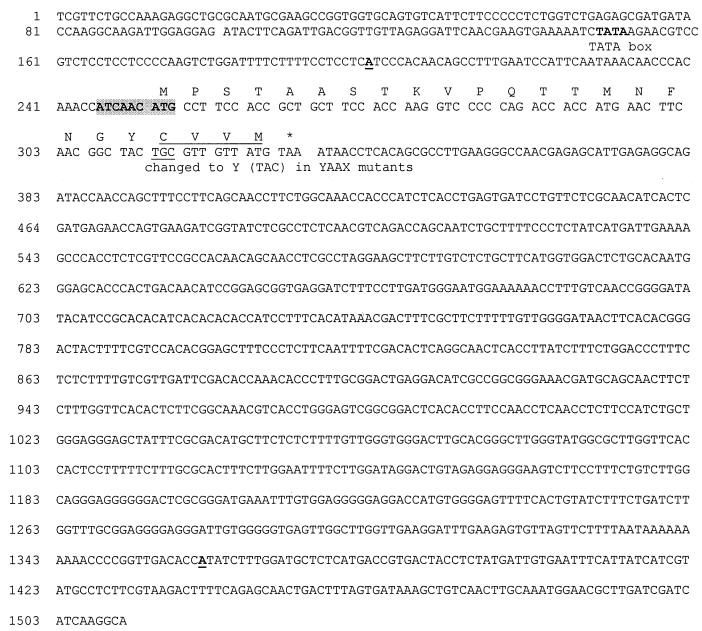
Selected mfa-1 cDNA inserts and subcloned genomic mfa-1 DNA were sequenced. The genomic mfa-1 sequence and the derived amino acid sequence of the MFA1 protein are shown in Fig. 1. The mfa-1 gene encodes a transcript of approximately 1.2 kb, as deduced by the longest cDNA sequences; it is not interrupted by an intron. A potential TATA box in the promoter region of the mfa-1 gene is noted in Fig. 1. A good Kozak sequence (37) for the initiation of translation is present at the proposed mfa-1 start codon (in italics): ATCAACATG. The 24 codon mfa-1 open reading frame (ORF) shows typical Neurospora codon bias (52).
FIG. 1.
Genomic nucleotide and predicted amino acid sequences of the putative N. crassa pheromone precursor gene, mfa-1. The potential TATA box is noted. The 5′ end of the longest cDNAs and the polyadenylation sites are underlined in boldface type; the site of polyadenylation was identical in all mfa-1 cDNA clones. A good consensus ATG start site is highlighted, and the translation termination codon is indicated by an asterisk. The deduced amino acids are shown above the sequence of the coding region. The prenylation signal (CAAX) is underlined.
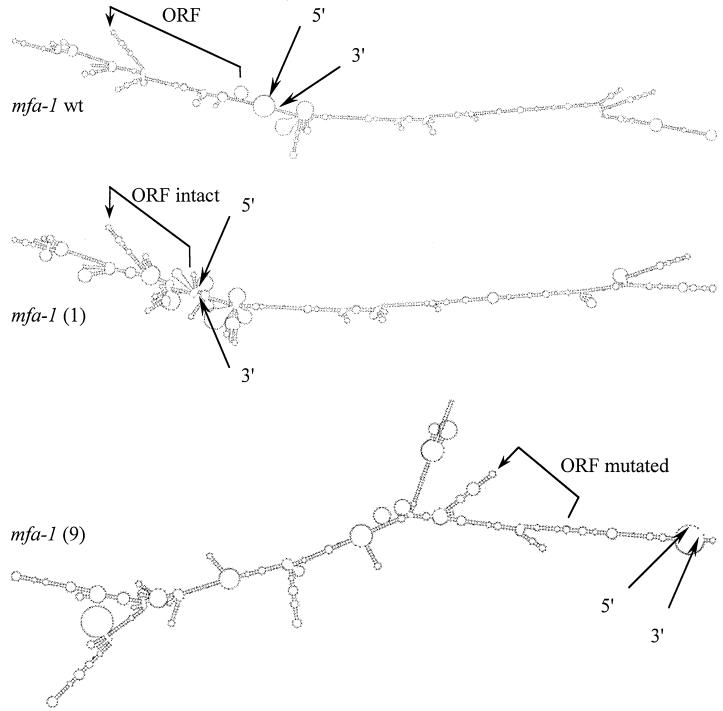
The mfa-1 mRNA has two unusual features: (i) it is mostly noncoding and (ii) it has an unusual predicted secondary structure. The predicted 24-residue ORF begins with an AUG at position 53 and ends with a UAA codon at position 125 in the 1.2-kb transcript, resulting in a 3′ untranslated region (3′ UTR) of more than 1 kb. The predicted mfa-1 mRNA secondary structure, obtained using the Vienna-RNA-Package-1.3 (with help from Peter Stadler at the University of Vienna, Vienna, Austria) (29, 30) is shown in Fig. 2. The long stable hairpins are atypical of mRNA (P. Stadler, personal communication).
FIG. 2.
Predicted secondary structure of mRNA of the mfa-1+ (wild type), mfa-1(1), and mfa-1(9) alleles. The mfa-1(1) strain is a CAAX mutant (with an intact ORF), and mfa-1(9) is a YAAX strain (with a mutated ORF). Vienna-RNA-Package-1.3 was used with parameters for 37°C (29, 30). Courtesy of Peter Stadler at the University of Vienna.
To identify potential mfa-1 homologs, the DNA sequences were compared with the NCBI nucleotide and protein databases, using BLASTN and BLASTX, respectively. The protein sequences from putative ORFs were compared with the NCBI protein database (using BLASTP and PSI-BLAST). The results of these sequence analyses showed more than 80% sequence identity with the ppg2 gene of S. macrospora (50) and 100% nucleotide identity within its predicted coding region. No other significant homologies were observed. An unusually long 3′ UTR is also present in some other fungal pheromone precursor genes, including MF1-1 of Magnaporthe grisea, mfa2 of Ustilago hordei, and ppg2 of S. macrospora (4, 50, 58).
The predicted MFA1 protein contained a CAAX motif at its carboxy terminus that is common in fungal hydrophobic pheromone precursors (Fig. 3). In characterized mature pheromones, the C-terminal cysteine is prenylated and carboxymethylated during posttranslational processing; these modifications are essential for their biological activity (12, 19, 35, 61).
FIG. 3.
Derived amino acid sequences of the MFA1 pheromone precursor from N. crassa (Nc) and its homologs in other fungal species: S. macrospora (Sm) (50), S. cerevisiae (Sc) (12), S. pombe (Sp) (19), C. parasitica (Cp) (68), M. grisea (Mg) (58), U. maydis (Um) (61), and U. hordei (Uh) (4). The CAAX motifs are indicated in boldface type, and the known mature pheromone sequences are underlined.
Presence of mfa-1 homologs in other filamentous ascomycetes.
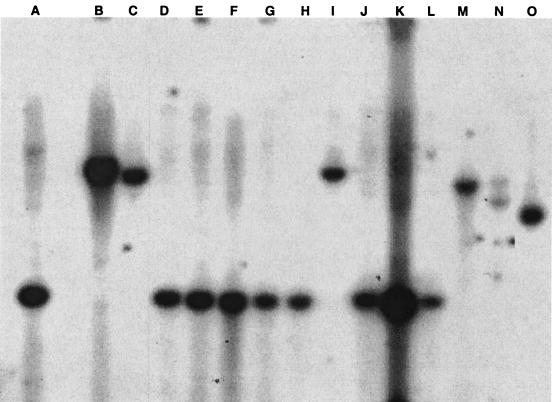
Heterologous hybridizations of the mfa-1 probe with digested ascomycete genomic DNAs showed that there is a single copy of the mfa-1 gene in the N. crassa genome, and that many other Neurospora species and their closely related ascomycete species also have an mfa-1 homolog in their genomes (Fig. 4). Striking homogeneity (or constancy) was observed within Neurospora species, in that bands of the same size were recognized in both mating types of N. tetrasperma and N. sitophila. However, in N. intermedia, fragments containing the mfa-1 homolog were of different sizes in the mat A (FGSC 2316) and mat a (FGSC 1940) strains; although both strains were isolated from Florida (Clewiston and LaBelle, respectively) on the same day, they are thought to be of independent genetic background (David Perkins, personal communication). Interestingly, the mfa-1 pheromone precursor gene was maintained in the true homothallic species G. reticulospora, N. africana, N. galapagosensis, S. fimicola, S. macrospora, and A. sublineolata.
FIG. 4.
Zoo blot probed with mfa-1. (A) N. crassa (2489); (B) G. reticulospora (960); (C) G. tetrasperma (966); (D and E) N. tetrasperma (1270 and 1271); (F) N. africana (1740); (G) N. galapagosensis (1739); (H and I) N. intermedia (2316 and 1940); (J and K) N. sitophila (2216 and 2217); (L) N. terricola (1889); (M) S. fimicola (2918); (N) S. macrospora (4818); (O) A. sublineolata (5508). Numbers in parentheses indicate the FGSC strain number.
Mapping of mfa-1.
To determine whether mfa-1 represents a newly identified gene or one previously characterized, the chromosomal location of mfa-1 was determined using the RFLP method (41, 45). The mfa-1 gene was mapped to the right arm of linkage group V, with an RFLP pattern identical to that of cyh-2, sod-2, and vma-1. The region did not include any previously identified sexual developmental mutants (49).
Expression of the mfa-1 gene.
The mfa-1 cDNA was the most abundant clone in starved mycelial and perithecial cDNA libraries of N. crassa (as discussed above). Northern analysis was carried out to examine the expression of mfa-1 in other conditions. A forced heterokaryon containing both the mat a and mat A loci was used to mimic mating conditions (43). Also, since protoperithecia form efficiently only on the surface, only the stationary cultures (floating mycelial mats) grown on crossing medium would be expected to contain significant levels of transcripts specific to sexual development. Expression in the Am44 sterile mutant (56), the fmf-1 (for female and male fertility-1) mutant (33), and the sterile am1 mutant (27) was also examined.
Northern analysis showed high levels of mfa-1 expression in the heterokaryotic (mat a/mat A) strain grown under nitrogen starvation conditions (Fig. 5A, lane 2) and in vegetative and sexual tissues of the wild-type mat a strain (Fig. 5B, lanes 2 to 4 and Fig. 5C, lane 1). In the wild-type ORS a strain, mfa-1 expression in unfertilized sexual tissue (protoperithecia, Fig. 5B, lane 4) was higher than that in exponentially growing vegetative tissue (Fig. 5B, lane 3) but similar to that in germinating conidia (Fig. 5B, lane 2). Interestingly, while the mfa-1 mRNA was abundant in protoperithecia, its level was greatly increased in 7-day-old perithecia and even more so in 9-day-old perithecia (Fig. 5B, lanes 5 and 6, respectively). No expression was detected in the mat am1 sterile mutant (Fig. 5B, lane 1) (27), and little or no expression was observed in the wild-type mat A strain (Fig. 5A, lane 6, and Fig. 5C, lane 2). However, the expression of the mfa-1 homolog was high in sexual tissues of the true homothallic species N. africana, which contains only the mat A idiomorph (Fig. 5C, lane 4) (26).
FIG. 5.
Expression of the mfa-1gene and the constitutively expressed am (glutamate dehydrogenase) gene. (A) Expression of mfa-1 in mat A strains and mat a/mat A heterokaryons. RNAs in lanes 1, 2, and 3 were prepared from a mat a tol/mat A tol heterokaryon, and RNAs in lanes 4, 5, and 6, were from Am44, fmf-1 mat A, and 74 A (wild type), respectively. The strains were grown with or without agitation under these conditions: with shaking in vegetative medium (lane 1), with shaking in crossing medium (lane 2), and without agitation in crossing medium (lanes 3 to 6). (B) Expression of mfa-1 in mat a strains and in perithecia. RNA in lane 1 was prepared from the sterile am1 mutant, grown with agitation in vegetative medium for 14 h. RNAs in lanes 2 to 4 were prepared from ORS a (wild type), grown under these conditions: with agitation in vegetative medium for 5 h (lane 2), with agitation in vegetative medium for 14 h (lane 3), and without agitation in crossing medium for 5 days (lane 4). RNAs in lanes 5 and 6 were prepared from a cross of fl a with 74 A, prepared from perithecia harvested 7 days (lane 5) or 9 days (lane 6) after fertilization. Ethidium bromide staining of the separated rRNA is shown. (C) Expression of mfa-1 in sexual tissues of N. crassa (lanes 1 and 2) and N. africana (lanes 3 and 4). RNAs in lanes 1 and 2 were prepared from the N. crassa ORS a and 74 A wild-type strains, respectively, grown without agitation in crossing medium for 5 days. RNAs in lanes 3 and 4 were prepared from N. africana grown without agitation in crossing medium for 3 or 5 days, respectively.
The mat a locus of N. crassa contains a single gene, mat a-1, that encodes a putative transcriptional regulator (62). The MATa1 protein contains the high mobility group DNA binding domain (32) and is thought to regulate expression of genes required for mating interactions, including the production of pheromones (59). This protein specifically binds CAAAG sequences in DNA (38), as do other high mobility group box proteins such as STE11 of S. pombe and Prf1 of U. maydis (28, 64). The mfa-1 gene contains the CAAAG sequence in its promoter region (from position 9 to position 13 in Fig. 1), suggesting that it may be directly controlled by MATa1.
Isolation of mfa-1 mutants.
Mutants with disrupted or intact mfa-1 coding regions were isolated by the RIP mutation approach (13, 57). A transformant containing two copies of the mfa-1 gene (the endogenous copy plus one extra copy introduced by transformation) was crossed with a strain having an auxotrophic mutation (lys-1) near the mfa-1 gene. The progeny of the crosses were plated on medium lacking lysine supplement in order to enrich for progeny in which this region was inherited from the transformed strain, with potential disruptions of the endogenous mfa-1 gene. Seven potential mfa-1 mutants were initially recognized based on their extremely poor vegetative growth, and their mfa-1 alleles were examined for the presence of typical RIP mutations (GC-to-AT transitions). One mutant allele containing 33 typical RIP-generated transitions, all 3′ to the coding region, was identified and designated mfa-1(1) a [where (1) a indicates allele 1, mat a]. Since mfa-1 mutants with disrupted ORFs were desired, the mfa-1(1) a strain (still containing the ectopic copy of mfa-1) was used as the male parent in a second RIP cross. Eight more mfa-1 mutants, in strains of both mating types, were isolated from the second RIP cross; they were designated mfa-1(2) through mfa-1(9). Subsequently, the mfa-1(9) mutant (which contained the ectopic copy of mfa-1 and was mat a) was used for a third round of RIP, and nine additional mutants were isolated [mfa-1(10) through mfa-1(18)].
Analysis of mfa-1 mutant sequences.
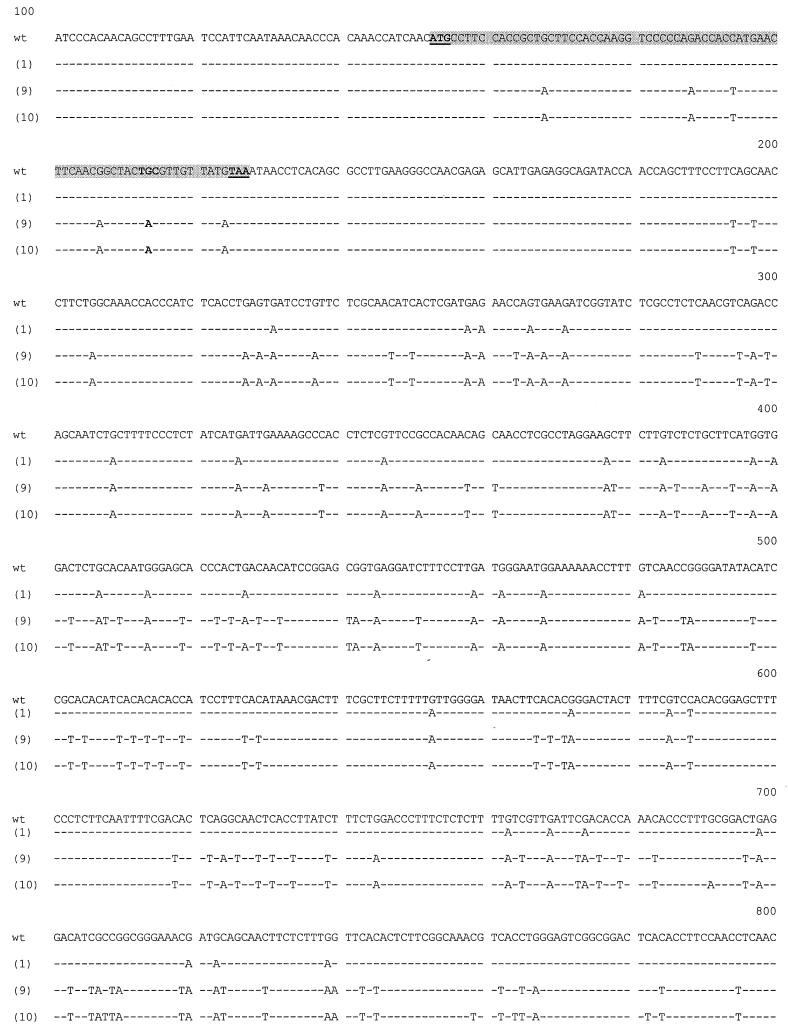
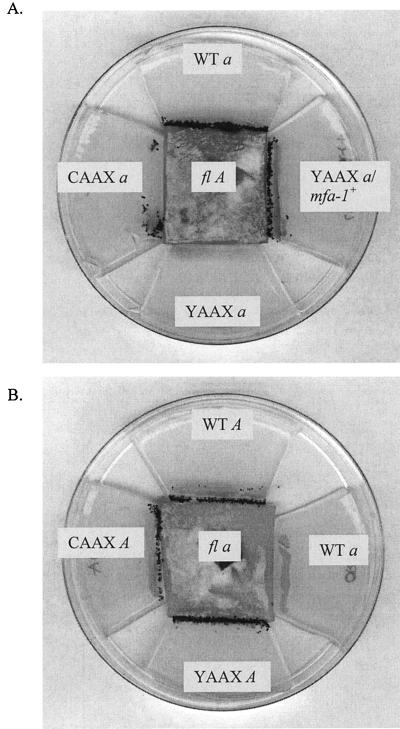
The nucleotide sequence of the mfa-1 gene was determined for all 18 mutant strains, and two distinct classes of mfa-1 mutants were identified. CAAX mutants [e.g., mfa-1(1)] had intact ORFs but multiple mutations in the 3′ UTRs (Fig. 6). However, in crosses of mutants of mating type a acting as males (CAAX a and YAAX a), both mutants failed to attract trichogynes. CAAX mutants were able to mate and undergo sexual development, but only if they were placed directly onto the wild-type female or only after the wild-type female had grown over the streak of CAAX a conidia (Fig. 7), whereas YAAX mutants of mating type a (YAAX a) were unable to either attract or mate with fl A trichogynes, although occasionally a few perithecia were formed when 74 A was presented as the female.
FIG. 6.
Sequence comparison of mfa-1+ (wild type) and the mfa-1(1), mfa-1(9), and mfa-1(10) alleles. The wild-type sequence is shown, and the changes in the mfa-1 mutants are noted below (dashes indicate identical nucleotides). The highlighted region indicates the putative ORF with its initiation and termination codons underlined; the asterisk indicates the polyadenylation site.
FIG. 7.
Pheromone responses. Conidia of wild-type, mfa-1 CAAX a, YAAX a, and a YAAX a/mfa-1+ transformant (A) and CAAX A and YAAX A (B) were placed on 2% agar in close proximity to an fl female of opposite mating type (at center). (A) Many perithecia formed along the fl A border when ORS a or YAAX a/mfa-1+ was streaked a slight distance away. Few perithecia formed when the fertilizing parent was CAAX a, and those formed only after the fl A strain had grown over the streak of CAAX conidia. No perithecia formed when YAAX a was the male parent. (B) No defective mating response was observed when mfa-1 mat A strains were used as males.
Complementation of a YAAX a mutant by transformation with genomic mfa-1+ was tested. A male-sterile (YAAX a) strain, mfa-1(18) a, was transformed with a genomic mfa-1+ clone, and 100 transformants were selected based on their hygromycin resistance. Fertility and mating responses were assayed as described above, and 30 transformants with complementing mfa-1+ genes were identified. Southern analysis verified acquisition of the mfa-1+ DNA (not shown). The transformants, used as male parents, were able to attract and mate with fl A strains, and ascospores were produced. While the male sexual defects were fully reversed, the vegetative defects were not suppressed by the complementation; perhaps a higher level of mfa-1+ activity is required for normal vegetative growth than is needed by the male parent in the sexual cycle. When CAAX a, CAAX A, or YAAX A mutants were used as male parents to fertilize CAAX or YAAX mutants of the opposite mating type, a few perithecia developed after some delay. However, development in those perithecia aborted at an early stage before ascogenous hyphae developed, resulting in very small, empty perithecia that never formed beaks or produced ascospores. When the YAAX a mutant was used as the male parent to fertilize CAAX A or YAAX A strains, the crosses were sterile; on those rare occasions when fertilization occurred, sexual development ceased very early.
DISCUSSION
mfa-1 is a gene encoding the precursor of a presumptive hydrophobic hormone.
Sequence analysis of the mfa-1 gene showed 100% sequence identity with a pheromone precursor gene, ppg2, of S. macrospora (50), which encodes a 24-amino-acid S. cerevisiae a-factor-like peptide pheromone (3). In S. cerevisiae, a-factor precursors undergo complex amino- and carboxy-terminal posttranslational processing (20): S-farnesylation at the cysteine residue by Ram2p and Ram1p (farnesyltransferase α and β, respectively), proteolysis of AAX residues by Rce1p (or Afc1p; AAX protease), carboxymethylation of the exposed cysteine by Ste14p (methyltransferase), and two-step proteolysis of the amino terminus by Ste24p (aminopeptidase 1) and Axl1p or Ste23p (both aminopeptidase 2). The mature pheromone is exported directly across the membrane via Ste6p, an ATP-dependent transporter. BLAST searches through NCBI and the Whitehead Institute Center for Genome Research indicated that the N. crassa genome contains homologs of all known genes for the processing and transport of the budding yeast a-pheromone (Table 1).
TABLE 1.
N. crassa homologs of S. cerevisiae and S. pombe genes that encode components of the pheromone response MAPK pathway and the posttranslational processing of a-factora
| Function | S. cerevisiae | S. pombe | N. crassa | E value |
|---|---|---|---|---|
| Mating type regulatory protein | A/ALPHA | matM/matP | mat a/mat A | ≫1 |
| Mating factor | MFA1 & 2 | mfm1, 2 & 3 | mfa-1 | ≫1 |
| Mating factor | MFα1 & 2 | map2 | ccg-4 | ≫1 |
| Mating factor receptor | STE2 | mam2 | Supercontig 9 | 1e-05 |
| Mating factor receptor | STE3 | map3 | Supercontig 147 | 8e-03 |
| G protein α subunit | GPA1 | gpa1 | gna-1 | 3e-62 |
| G protein β subunit | STE4 | git5 | Supercontig 59 | 1e-49 |
| G protein γ subunit | STE18 | git11 | Supercontig 128 | 6e-07 |
| PAK type kinase | STE20 | pak1 | Supercontig 59 | e-107 |
| Scaffold protein | STE5 | * | Supercontig 39 | 7e-05 |
| MAPKKK | STE11 | byr2 | Supercontig 8 | 4e-88 |
| MAPKK | STE7 | byr1 | Supercontig 109 | 9e-57 |
| MAPK | FUS3 | spk1 | Supercontig 234 | e-109 |
| MAPK | KSS1 | spk1 | Supercontig 234 | e-107 |
| Transcription factor | STE12 | * | Supercontig 59 | 3e-58 |
| MAPK | FAR1 | * | Supercontig 39 | 3e-04 |
| Farnesytransferase α | RAM2 | cpp1 | Supercontig 25 | 2e-21 |
| Farnesytransferase β | RAM1 | cwp1 | Supercontig 180 | 5e-35 |
| ‘AAX' endoprotease | RCE1 (AFC1) | SPAC1687.02 | Supercontig 8 | 1e-13 |
| Farnesyl cysteine carboxymethyltransferase | STE14 | mam4 | Supercontig 329 | 1e-49 |
| Aminopeptidase1 | STE24 | SPAC3h1.05 | Supercontig 180 | 1e-80 |
| Aminopeptidase2 | AXL1 | irp1 | Supercontig 118 | 5e-36 |
| Aminopeptidase2 | STE23 | irp1 | Supercontig 108 | 0.0 |
| ATP binding cassette (ABC) transporter | STE6 | mam1 | Supercontig 61 | e-122 |
Amino acid sequences of the S. cerevisiae proteins were compared with N. crassa nucleotide databases (using tBLASTN) available through NCBI and/or WICGR (Whitehead Institute Center for Genome Research). E (expected) values of N. crassa are with respect to the corresponding S. cerevisiae proteins. An E value cutoff of 10−3 (e-3) was used except for the mating type and mating factor genes. Asterisks indicate that no homologs were identified in S. pombe.
The mfa-1 gene has multiple roles in sexual development.
In heterothallic fungi, pheromones play an important role in mating by facilitating recognition between strains of opposite mating type and by launching the complex pheromone response MAPK pathway. Recent studies have suggested that fungal pheromones are required for postfertilization events as well, such as internuclear recognition for karyogamy and/or meiosis (9, 22, 50). Even though it is homothallic, S. macrospora contains the mfa-1 homolog ppg2 and expresses it in perithecia at high levels which peak around the time ascospores are maturing and being ejected (50). Also, mfa-1 (or its homolog) is transcribed in sexual tissues of the homothallic N. africana, which does not contain the mat a sequence (Fig. 5C).
In order to analyze the function(s) of the pheromone precursor gene of N. crassa, apparent null mutants (YAAX, in which the essential cysteine residue was changed to tyrosine) and hypomorphs (CAAX, with an intact ORF but multiple mutations in the 3′ UTR) were isolated. The developmentally regulated expression of mfa-1 and the pleiotropic nature of mfa-1 mutants suggest that mfa-1 has multiple functions in the development of N. crassa, expanding the range of known pheromone functions. While normal levels of mfa-1 mRNA were detected in a CAAX a strain, no expression was detected in a YAAX a strain (not shown).
YAAX mutant strains of mating type a, used as males, were unable to attract and to mate with mat A trichogynes, which suggests that the cysteine residue is absolutely essential for prenylation and therefore for the pheromone function of mfa-1, as shown for other CAAX pheromones of fungi. S. cerevisiae a-factor lacking the farnesyl group shows 1,000-fold reduced pheromone activity (12). In U. maydis, unfarnesylated analogs of Uhmfa1 and Uhmfa2 are inactive, while farnesylated analogs of modified amino acid sequence do induce mating with cells of opposite mating type (35).
Pheromones in fungi have been considered mating type-specific. Based on Northern analysis of RNA from well-nourished vegetative cells, the expression of mfa-1 appeared to be mating type a specific; that is, it required the presence of the mat a gene (Fig. 5B). Paradoxically, the unsubtracted cDNA library from starved mycelia of mat A wild type contained mfa-1 as the most abundant clone (4.6% to date) (44). In addition, the mfa-1 gene or its homolog was highly expressed in sexual tissues of homothallic N. africana, which contains only the mat A idiomorph (Fig. 5C) (26). This species has no need to attract or be attracted to a mate, and it makes no trichogynes. These results show that mfa-1 is expressed, possibly at a specific time(s) of development, in mat A strains of Neurospora and suggest that mfa-1 has at least one important function in addition to that of promoting fertilization. Furthermore, the severely reduced fertility of mfa-1 mutants and the extremely high accumulation of mfa-1 mRNA in maturing perithecia (Fig. 5B, lanes 5 and 6) suggest that mfa-1 is required even after karyogamy and/or meiosis in the sexual development of N. crassa. Also, it was recently shown that expression of the probable pheromone receptor genes of N. crassa is not mating type specific (51).
Possible conglutinin-like role for MFA1.
One hypothesis postulates an additional role for MFA1 in “conglutination” in perithecial development, or the cementing together of hyphae to stabilize the sclerotial structure of the maturing perithecium. This would require prenylation and could involve a second peptide (longer or shorter). An analogous situation may occur in S. cerevisiae, where a second peptide (a-factor related peptide) is produced from the pro-a-factor (17). In Neurospora, the pheromone-like peptide, provisionally named conglutinin, would interact with the normal pheromone receptor to constitute the “glue” that holds together a perithecium. The conglutinin function would require that mfa-1 and the gene encoding the receptor both be expressed in female supporting tissue of the perithecium, whether that tissue is mat a or mat A. We suggest that when the female tissue is mat A, synthesis of the conglutinin is triggered by the processed pheromone from the mat a male, and that the regulation is positive and autogenous. Thus, adjacent hyphae would stick together, since each would be coated with conglutinin and receptor, with both proteins being anchored in the plasma membrane. Presumably the cell walls of adjacent hyphae would have been resorbed at the surfaces of contact, as has long been known to occur during fusion of vegetatively growing hyphae (11).
The conglutinin hypothesis would account for these observations: (i) the extremely high level of mfa-1 mRNA in normal perithecial tissue, (ii) the highly abnormal perithecial development or absence thereof in homozygous mfa-1 crosses (if fertilization occurred at all, the sclerotial tissue would still be completely deficient in conglutinin, so perithecia would fail to develop), and (iii) the severely deficient sexual development in specific heterozygous crosses. The crosses of mfa-1 mutants as males to wild-type strains as females could be explained as follows. In crosses of mat A as males (CAAX A or YAAX A) to wild-type mat a strains as females, attraction and fertility would be normal, since the mat A pheromone is presumably normal and the wild-type mat a female would produce normal conglutinin. In crosses with a CAAX mat a strain as male, fertilization would be an uncertain matter. If a molecule of product from the hypomorphic gene somehow got the process started, then the mat A, as female, would transcribe its normal mfa-1 gene, produce MFA1, process it to pheromone (or conglutinin), and induce synthesis of more MFA1, with the result that the cross would be fertile. In crosses of YAAX a acting as males to wild-type mat A as females, the male would be unable to attract and fuse with trichogynes. In the rare event that fertilization were nevertheless to occur, the wild-type female would not be triggered to initiate synthesis of the MFA1 conglutinin, with the result that the cross would be completely infertile. Finally, in crosses of CAAX or YAAX mutants acting as females to wild type as males, all female parents would be hypomorphic (CAAX) or null (YAAX) in the capacity to make MFA1, and thus little or no sexual development would be expected.
mfa-1 may be involved in filamentous growth.
In S. cerevisiae, six different MAPK pathways have been identified, which are involved in distinct physiological processes including mating, spore formation, cell wall biosynthesis, and sensing of hyperosmotic environments. Due to the high specificity of the constituent protein kinases in their recognition of substrates, and to regulation at multiple levels, normally no significant functional crossover of kinases occurs in the MAPK pathways. Nevertheless, some components of the pheromone response MAPK cascade for mating are known to be shared by three other MAPK cascades, those controlling filamentous growth, cell wall integrity, and osmolyte synthesis (53). In particular, the fundamental elements of the pheromone response pathway—Ste20p (MAPK kinase kinase kinase), Ste11p (MAPK kinase kinase), Ste7p (MAPK kinase), Kss1p (MAPK), and Ste12 (transcription factor)—are common to the filamentous growth pathway and the response to nutrient deprivation, and cross talk can occur under some experimental conditions. The N. crassa genome contains homologs of all known fundamental elements involved in the yeast pheromone response pathway (Table 1). As in yeast, some of the components may also function in other MAPK cascade pathways in N. crassa, either normally or under special circumstances. These could include the pathway controlling filamentous growth. In the heterobasidiomycete U. maydis, pheromone alone is sufficient to induce filamentous growth. Autocrine stimulation of the pheromone response pathway is required for the maintenance of the filamentous form after the actual cell fusion (61). Downstream components of the MAPK cascade, activated by the MFA1 pheromone (possibly via autocrine stimulation), may also be required for filamentous growth of N. crassa.
All mutants isolated in this study showed great reduction in vegetative growth and protoperithecial formation, both of which involve filamentous differentiation. Although the vegetative growth defect of mfa-1 mutants was largely reversed by arginine, citrulline, or ornithine, mfa-1 is not likely to be directly involved in the arginine or ornithine pathways, since a minute amount of supplement was sufficient to suppress the defect, in sharp contrast to the nutritional needs of typical arginine auxotrophs. However, other metabolites made from ornithine, particularly the hydroxamate siderophores needed for transport of iron, are made in much smaller amounts, and the demands on the supply of ornithine, and hence of arginine, are much lower (16). Many models could explain the stimulation by amino acids of the arginine cycle, and we suggest only one. It is conceivable that MFA1 is needed for the sensing and correction of transient nutritional deficits that arise during ordinary growth. Insufficient iron is a plausible example of a deficit, and MFA1 might have a role in requisitioning a small fraction of the internal pool of ornithine toward the synthesis of siderophores. In the absence both of MFA1 and of an external supply of ornithine or arginine, siderophores and hence iron would become growth-limiting.
The 3′ UTR of mfa-1 is required for the full range of function.
Partial male fertility defects of CAAX mat a mutants suggested that the 3′ untranslated region of mfa-1 may be required for the full range of pheromone activity. The sequences of 3′ untranslated regions have been implicated in control of mRNA localization (5, 6, 60), as well as in control of mRNA stability and of translational efficiency (7, 23, 65). mRNA species that must be located very precisely within the cell often have long 3′ UTRs, and the mRNA itself may contain signals, targeting it to preferred subcellular sites. The 1.2-kb mfa-1 transcript is mostly noncoding (over 80% is 3′ UTR), and it has an unusual predicted secondary structure, containing several long double-stranded regions. When compared to the wild type, the predicted mRNA secondary structures of CAAX and YAAX alleles showed noticeable differences. If MFA1 indeed acts as a conglutinin in perithecium formation, precise mRNA localization could be important. Thus, CAAX mutants might be hypomorphs because much of the MFA1 product is not correctly localized, even if it is made in normal amounts. Long 3′ UTRs have also been identified in the hydrophobic pheromone precursor genes of other filamentous fungi, including MF1-1 of M. grisea (75% of the 0.8-kb transcript [58]), ppg2 of S. macrospora (80% of the 1.0-kb transcript [50]), and mf2-1 of Cryphonectria parasitica (86% of the 0.7-kb transcript [68]), suggesting that mRNA localization might be important in those organisms.
In summary, the mfa-1 gene appears to play complex and essential roles in the vegetative growth and sexual development of N. crassa. It encodes a hydrophobic pheromone, required for mat a, acting as a male, to attract and fuse with a mat A trichogyne. It may also encode a peptide, conglutinin, identical in part or in entirety with the mat a pheromone, that is essential for perithecial maturation in both mating types. Finally, the mfa-1 gene is involved in vegetative and protoperithecial development in both mating types, and its 3′ UTR is required for the full range of mfa-1 function.
Both CAAX and YAAX mfa-1 mutants showed delayed germination and reduced vegetative growth on Vogel's minimal medium. At 25 or 30°C, linear growth rates were three to four times lower than that of the wild type; growth was slightly improved at 37°C. When supplemented with 0.5% yeast extract, the mutants showed growth rates similar to that of wild type. Individual components of yeast extract were tested, and it was determined that supplementation with ornithine, citrulline, or arginine suppressed the mfa-1 growth defect, while addition of lysine reduced the growth rate. A minute amount (∼30 μM) of ornithine, citrulline, or arginine was sufficient to suppress the mfa-1 growth defect, while much higher levels (e.g., 5 mM) are required to supplement typical arginine auxotrophs, such as arg-1 (49). The arginine analogs N-omega-nitro-l-arginine methyl ester and l-canavanine did not differentially affect the growth of wild-type and mfa-1 strains (not shown).
(ii) Sexual development and the mating response.
Fertility of the mfa-1 mutants was examined. Female fertility was delayed and drastically reduced to approximately the same degree in CAAX and YAAX mfa-1 mutants crossed to wild type, irrespective of their mating type. However, male fertility varied greatly, depending both on the presence of the critical CAAX motif and the mating type of the mutants. For CAAX a, results were almost normal if the mutant was spotted directly on the female; no trichogyne attraction was observed if spotted at a distance. For YAAX a, males were sterile and rarely fertilized; if the males did fertilize, sexual development aborted very early. For CAAX A or YAAX A, male fertility was normal, with trichogyne attraction like that of the wild type.
Under the standard low-nitrogen conditions, protoperithecial development was delayed and the number of protoperithecia was reduced approximately 1,000-fold in all mfa-1 mutants. In crosses of mutants acting as females to wild types acting as males, the fertilized protoperithecia developed to perithecia after a delay of a day or two. The perithecia were smaller than those of wild-type crosses, yet a small number of viable ascospores was produced. Germination of the ascospores from crosses of mfa-1 acting as female by wild types acting as male was no higher than 30%, while the corresponding number from homozygous wild-type crosses was 95 to 99%.
In crosses of mfa-1 mutants of mating type A acting as male to wild-type or fluffy strains acting as female, the mutants (both CAAX A and YAAX A) attracted distant trichogynes (∼5 mm away), perithecia were formed without a delay along the border of the fl a strain, and these produced fertile ascospores (Fig. 7).
Acknowledgments
We thank David Perkins and Peter Stadler for personal communications.
This research was supported by NSF grant MCB-9874488 to M.A.N. and NIH grants GM47374 to M.A.N. and GM08995 to R.L.M.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderegg, R. J., R. Betz, S. A. Carr, J. W. Crabb, and W. Duntze. 1988. Structure of Saccharomyces cerevisiae mating hormone a-factor: identification of S-farnesyl cysteine as a structural component. J. Biol. Chem. 263:18236-18240. [PubMed] [Google Scholar]
- 4.Anderson, C. M., D. A. Willits, P. J. Kosted, E. J. Ford, A. D. Martinez-Espinoza, and J. E. Sherwood. 1999. Molecular analysis of the pheromone and pheromone receptor genes of Ustilago hordei. Gene 240:89-97. [DOI] [PubMed] [Google Scholar]
- 5.Bashirullah, A., R. L. Cooperstock, and H. D. Lipshitz. 1998. RNA localization in development. Annu. Rev. Biochem. 67:335-394. [DOI] [PubMed] [Google Scholar]
- 6.Beach, D. L., E. D. Salmon, and K. Bloom. 1999. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 9:569-578. [DOI] [PubMed] [Google Scholar]
- 7.Beelman, C. A., and R. Parker. 1995. Degradation of messenger RNA in eukaryotes. Cell 81:179-183. [DOI] [PubMed] [Google Scholar]
- 8.Bistis, G. N. 1983. Evidence for diffusible, mating-type-specific trichogyne attractants in Neurospora crassa. Exp. Mycol. 7:292-295. [Google Scholar]
- 9.Bölker, M., and R. Kahmann. 1993. Sexual pheromones and mating responses in fungi. Plant Cell 5:1461-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun, E. L., E. K. Fuge, P. A. Padilla, and M. Werner-Washburne. 1996. A stationary-phase gene in Saccharomyces cerevisiae is a member of a novel, highly conserved gene family. J. Bacteriol. 178:6865-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buller, A. H. R. 1933. Researches on fungi, vol. 5. Longman, London, United Kingdom.
- 12.Caldwell, G. A., S.-H. Wang, C.-B. Xue, Y. Jiang, H.-F. Lu, F. Naider, and J. M. Becker. 1994. Molecular determinants of bioactivity of the Saccharomyces cerevisiae lipopeptide mating pheromone. J. Biol. Chem. 269:19817-19825. [PubMed] [Google Scholar]
- 13.Cambareri, E. B., B. C. Jensen, E. Schabtach, and E. U. Selker. 1989. Repeat-induced G-C to A-T mutations in Neurospora. Science 244:1571-1575. [DOI] [PubMed] [Google Scholar]
- 14.Casselton, L. A., and N. S. Olesnicky. 1998. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol. Mol. Biol. Rev. 62:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakraborty, B. N., and M. Kapoor. 1990. Transformation of filamentous fungi by electroporation. Nucleic Acids Res. 18:6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charlang, G., and N. P. Williams. 1983. Siderophore transport mutants (sit) in Neurospora crassa. Neurospora Newsl. 30:6-7. [Google Scholar]
- 17.Chen, P., J. D. Choi, R. Wang, R. J. Cotter, and S. Michaelis. 1997. A novel a-factor-related peptide of Saccharomyces cerevisiae that exits the cell by a Ste6p-independent mechanism. Mol. Biol. Cell 8:1273-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chikashige, Y., D. Q. Ding, Y. Imai, M. Yamamoto, T. Haraguchi, and Y. Hiraoka. 1997. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J. 16:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davey, J. 1992. Mating pheromones of the fission yeast Schizosaccharomyces pombe: purification and structural characterization of M-factor and isolation and analysis of two genes encoding the pheromone. EMBO J. 11:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davey, J., K. Davis, M. Hughs, G. Ladds, and D. Powner. 1998. The processing of yeast pheromone. Semin. Cell Dev. Biol. 9:19-30. [DOI] [PubMed] [Google Scholar]
- 21.Davis, R. H., and F. J. DeSerres. 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17:79-143. [Google Scholar]
- 22.Debuchy, R. 1999. Internuclear recognition: a possible connection between euascomycetes and homobasidiomycetes. Fungal Genet. Biol. 27:218-223. [DOI] [PubMed] [Google Scholar]
- 23.Decker, C. J., and R. Parker. 1994. Mechanisms of messenger RNA degradation in eukaryotes. Trends Biochem. Sci. 19:336-340. [DOI] [PubMed] [Google Scholar]
- 24.Dolan, P. L., D. O. Natvig, and M. A. Nelson. 2000. Neurospora proteome 2000. Fungal Genet. Newsl. 47:7-24. [Google Scholar]
- 25.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Annu. Rev. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 26.Glass, N. L., and M. L. Smith. 1994. Structure and function of a mating-type gene from the homothallic species Neurospora africana. Mol. Gen. Genet. 244:401-409. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths, A. J. F., and A. M. DeLange. 1978. Mutations of the a mating-type gene in Neurospora crassa. Genetics 88:239-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartmann, H. A., R. Kahmann, and M. Bölker. 1996. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 15:1632-1641. [PMC free article] [PubMed] [Google Scholar]
- 29.Hofacker, I. L., M. Fekete, C. Flamm, M. A. Huynen, S. Rauscher, P. E. Stolorz, and P. F. Stadler. 1998. Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acids Res. 26:3825-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofacker, I. L., and P. F. Stadler. 1999. Automatic detection of conserved base pairing patterns in RNA virus genomes. Comput. Chem. 23:401-414. [DOI] [PubMed] [Google Scholar]
- 31.Imai, Y., and M. Yamamoto. 1994. The fission yeast mating pheromone P-factor: its molecular structure, gene structure, and ability to induce gene expression and G1 arrest in the mating partner. Genes Dev. 8:328-338. [DOI] [PubMed] [Google Scholar]
- 32.Jantzen, H.-M., A. Admon, S. P. Bell, and R. Tjian. 1990. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature 344:830-836. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, T. E. 1979. A Neurospora mutation that arrests perithecial development as either male or female parent. Genetics 92:1107-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinnaird, J. H., M. A. Keighren, J. A. Kinsey, M. Eaton, and J. R. S. Fincham. 1982. Cloning of the am (glutamate dehydrogenase) gene of Neurospora crassa through the use of a synthetic DNA probe. Gene 20:387-396. [DOI] [PubMed] [Google Scholar]
- 35.Kosted, P. J., S. A. Gerhardt, C. M. Anderson, A. Stierle, and J. E. Sherwood. 2000. Structural requirements for activity of the pheromones of Ustilago hordei. Fungal Genet. Biol. 29:107-117. [DOI] [PubMed] [Google Scholar]
- 36.Kothe, E. 1999. Mating types and pheromone recognition in the homobasidiomycete Schizophyllum commune. Fungal Genet. Biol. 27:146-152. [DOI] [PubMed] [Google Scholar]
- 37.Kozak, M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kronstad, J. W., and C. Staben. 1997. Mating type in filamentous fungi. Annu. Rev. Genet. 31:245-276. [DOI] [PubMed] [Google Scholar]
- 39.Kurjan, J. 1993. The pheromone response pathway in Saccharomyces cerevisiae. Annu. Rev. Genet. 27:147-179. [DOI] [PubMed] [Google Scholar]
- 40.Lindegren, C. C., V. Beanfield, and R. Barber. 1939. Increasing the fertility of Neurospora by selective inbreeding. Bot. Gaz. 100:592-599. [Google Scholar]
- 41.Metzenberg, R. L., J. N. Stevens, E. U. Selker, and E. Morzycka-Wroblewska. 1984. A method for finding the genetic map position of cloned DNA fragments. Neurospora Newsl. 31:35-39. [Google Scholar]
- 42.Moore, T. D. E., and J. C. Edman. 1993. The alpha mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol. 13:1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson, M. A., and R. L. Metzenberg. 1992. Sexual development genes of Neurospora crassa. Genetics 132:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson, M. A., S. Kang, E. L. Braun, M. E. Crawford, P. L. Dolan, P. M. Leonard, J. Mitchell, A. M. Armijo, L. Bean, E. Blueyes, T. Cushing, A. Errett, M. Fleharty, M. Gorman, K. Judson, R. Miller, J. Ortega, I. Pavlova, J. Perea, S. Todisco, R. Trujillo, J. Valentine, A. Wells, M. Werner-Washburne, S. Yazzie, and D. O. Natvig. 1997. Expressed sequences from conidial, mycelial and sexual stages of Neurospora crassa. Fungal Genet. Biol. 21:348-363. [DOI] [PubMed] [Google Scholar]
- 45.Nelson, M. A., and D. D. Perkins. 2000. Restriction polymorphism maps of Neurospora crassa: 2000 update. Fungal Genet. Newsl. 47:25-39. [Google Scholar]
- 46.O'Shea, S. F., P. T. Chaure, J. R. Halsall, N. S. Olesnicky, A. Leibbrandt, I. F. Connerton, and L. A. Casselton. 1998. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics 148:1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orbach, M. J. 1994. A cosmid with a HyR marker for fungal library construction and screening. Gene 150:159-162. [DOI] [PubMed] [Google Scholar]
- 48.Perkins, D. D., and E. G. Barry. 1977. The cytogenetics of Neurospora. Adv. Genet. 19:133-285. [DOI] [PubMed] [Google Scholar]
- 49.Perkins, D. D., A. Radford, and M. S. Sachs. 2001. The Neurospora compendium: chromosomal loci. Academic Press, San Diego, Calif.
- 50.Pöggeler, S. 2000. Two pheromone precursor genes are transcriptionally expressed in the homothallic ascomycete Sordaria macrospora. Curr. Genet. 37:403-411. [DOI] [PubMed] [Google Scholar]
- 51.Pöggeler, S., and U. Kück. 2001. Identification of transcriptionally expressed pheromone receptor genes in filamentous ascomycetes. Gene 280:9-17. [DOI] [PubMed] [Google Scholar]
- 52.Radford, A., and J. H. Parish. 1997. The genome and genes of Neurospora crassa. Fungal Genet. Biol. 21:258-266. [DOI] [PubMed] [Google Scholar]
- 53.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. D. He, H. Y. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saupe, S., L. Stenberg, K. T. Shiu, A. J. F. Griffiths, and N. L. Glass. 1996. The molecular nature of mutations in the mt A-1 gene of the Neurospora crassa A idiomorph and their relation to mating-type function. Mol. Gen. Genet. 250:115-122. [DOI] [PubMed] [Google Scholar]
- 57.Selker, E. U. 1990. Premeiotic instability of repeated sequences in Neurospora crassa. Annu. Rev. Genet. 24:579-613. [DOI] [PubMed] [Google Scholar]
- 58.Shen, W. C., P. Bobrowicz, and D. J. Ebbole. 1999. Isolation of pheromone precursor genes of Magnaporthe grisea. Fungal Genet. Biol. 27:253-263. [DOI] [PubMed] [Google Scholar]
- 59.Shiu, P. K. T., and N. L. Glass. 2000. Cell and nuclear recognition mechanisms mediated by mating type in filamentous ascomycetes. Curr. Opin. Microbiol. 3:183-188. [DOI] [PubMed] [Google Scholar]
- 60.Singer, R. H. 1993. RNA zipcodes for cytoplasmic addresses. Curr. Biol. 3:719-721. [DOI] [PubMed] [Google Scholar]
- 61.Spellig, T., M. Bölker, F. Lottspeich, R. W. Frank, and R. Kahmann. 1994. Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 13:1620-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Staben, C., and C. Yanofsky. 1990. Neurospora crassa a mating-type region. Proc. Natl. Acad. Sci. USA 87:4917-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stevens, J. N., and R. L. Metzenberg. 1982. Preparing Neurospora DNA: some improvements. Neurospora Newsl. 29:27-28. [Google Scholar]
- 64.Sugimoto, A., Y. Iino, T. Maeda, Y. Watanabe, and M. Yamamoto. 1991. Schizosaccharomyces pombe STE11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 5:1990-1999. [DOI] [PubMed] [Google Scholar]
- 65.Tanguay, R. L., and D. R. Gallie. 1996. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol. Cell. Biol. 16:146-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vogel, H. J. 1956. A convenient growth medium for Neurospora (medium N). Microbiol. Genet. Bull. 13:42-43. [Google Scholar]
- 67.Westergaard, M., and H. K. Mitchell. 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34:573-577. [Google Scholar]
- 68.Zhang, L., and R. A. Baasiri. 1998. Viral repression of fungal pheromone precursor gene expression. Mol. Cell. Biol. 18:953-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu, H., M. Nowrousian, D. Kupfer, H. V. Colot, G. Berrocal-Tito, H. S. Lai, D. Bell-Pedersen, B. A. Roe, J. J. Loros, and J. C. Dunlap. 2001. Analysis of expressed sequence tags from two starvation, time-of-day-specific libraries of Neurospora crassa reveals novel clock-controlled genes. Genetics 157:1057-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]