Abstract
Three type 2C Ser/Thr phosphatases (PTCs) are negative regulators of the yeast Saccharomyces cerevisiae high-osmolarity glycerol mitogen-activated protein kinase (MAPK) pathway. Ptc2 and Ptc3 are 75% identical to each other and differ from Ptc1 in having a noncatalytic domain. Previously, we showed that Ptc1 inactivates the pathway by dephosphorylating the Hog1 MAPK; Ptc1 maintains low basal Hog1 activity and dephosphorylates Hog1 during adaptation. Here, we examined the function of Ptc2 and Ptc3. First, deletion of PTC2 and/or PTC3 together with PTP2, encoding the protein tyrosine phosphatase that inactivates Hog1, produced a strong growth defect at 37°C that was dependent on HOG1, providing further evidence that PTC2 and PTC3 are negative regulators. Second, overexpression of PTC2 inhibited Hog1 activation but did not affect Hog1-Tyr phosphorylation, suggesting that Ptc2 inactivates the pathway by dephosphorylating the Hog1 activation loop phosphothreonine (pThr) residue. Indeed, in vitro studies confirmed that Ptc2 was specific for Hog1-pThr. Third, deletion of both PTC2 and PTC3 led to greater Hog1 activation upon osmotic stress than was observed in wild-type strains, although no obvious change in Hog1 inactivation during adaptation was seen. These results indicate that Ptc2 and Ptc3 differ from Ptc1 in that they limit maximal Hog1 activity. The function of the Ptc2 noncatalytic domain was also examined. Deletion of this domain decreased Vmax by 1.6-fold and increased Km by 2-fold. Thus Ptc2 requires an additional amino acid sequence beyond the catalytic domain defined for PTCs for full activity.
The yeast Saccharomyces cerevisiae high-osmolarity glycerol (HOG) mitogen-activated protein kinase (MAPK) pathway regulates the response to osmotic stress (8) and is also activated by heat stress (38). Like other MAPK pathways, the HOG pathway contains a three-kinase module: a MEKK (MAPK/ERK kinase kinase) which phosphorylates a MEK (MAPK/ERK kinase), which in turn phosphorylates a MAPK (8, 25). Activated MEKK phosphorylates MEK at conserved Ser and Thr residues, resulting in MEK activation. Activated MEK phosphorylates the conserved Thr-X-Tyr motif in the activation loop of MAPK, where X is Glu, Pro, or Gly. In the HOG pathway, three MEKKs, Ssk2, Ssk22, and Ste11, activate the MEK Pbs2 and the MAPK Hog1 (8). The HOG pathway contains two upstream branches that converge at Pbs2. One branch is the two-component signaling system containing Sln1, Ypd1, and Ssk1, which negatively regulates the downstream MAPK cascade (15, 28). The second branch contains the membrane-bound Sho1 (13), which positively regulates the MAPK cascade via Cdc42, Ste20, Ste50, and Ste11 (23, 26, 27, 29, 30).
Critical to the proper regulation of MAPK pathways is their inactivation. For example, mutations that block the inactivation of these pathways are detrimental for development and cell growth. In Drosophila melanogaster, inactivation of puckered, which encodes a dual-specificity phosphatase that regulates c-Jun N-terminal kinase, results in defects in dorsal closure during embryogenesis (17, 41). In the S. cerevisiae HOG pathway, deletion of the negative regulator SLN1 is lethal (24) due to hyperactivation of Hog1 (15). Furthermore, deletion of PTP2, encoding a protein tyrosine phosphatase (PTP) that dephosphorylates Hog1, and PTC1, coding for a type 2C Ser/Thr phosphatase (PP2C or PTC in yeast) that dephosphorylates Hog1, produces a severe growth defect due to hyperactivation of Hog1 (12, 14).
Two groups of protein phosphatases inactivate the HOG pathway. The PTPs Ptp2 and Ptp3 inactivate the Hog1 MAPK by dephosphorylating the phosphotyrosine residue in the activation loop (12, 40). Ptp2 plays the major role in inactivating Hog1 in vivo. For example, a strain lacking PTP2 more poorly dephosphorylates Hog1-phosphotyrosine (Hog1-pY) than a strain lacking PTP3. However, Ptp3 is important for Hog1 inactivation, as deletion of both PTP2 and PTP3 results in a severe defect in Hog1-pY dephosphorylation (12, 40). The differences in the activities of Ptp2 and Ptp3 may be explained by their relative abilities to bind Hog1, as well as their differential subcellular localizations. Ptp2 binds Hog1 more effectively than Ptp3 (12), and Ptp2 is nuclear whereas Ptp3 is cytoplasmic (19). Since Hog1 accumulates in the nucleus upon activation (5, 18, 31), the nuclear localization of Ptp2 might contribute to its greater activity. A second group of phosphatases, the PTCs, have been identified as negative regulators of the HOG pathway and of MAPK pathways in vertebrates and in plants (9, 10, 20, 35). These phosphatases differ from other Ser/Thr phosphatases in being monomeric enzymes that require Mg2+ or Mn2+ for catalytic activity (1, 36). Three genes, PTC1, PTC2, and PTC3, encode PTCs which have been linked to the HOG pathway. They are multicopy suppressors of sln1Δ lethality, which is due to hyperactivation of the downstream MAPK cascade (15, 37). We showed that the catalytic activity of each of the PTCs was important for HOG pathway inactivation, as mutation of conserved Asp residues that constitute the metal binding site prevented them from suppressing the sln1Δ lethal phenotype (37). Ptc2 and Ptc3 differ from Ptc1 in that they contain carboxy-terminal noncatalytic domains of ∼170 residues not found in Ptc1. A fourth PTC, Ptc4, most closely related in primary structure to Ptc1, Ptc2, and Ptc3, was not able to suppress sln1Δ lethality (37), suggesting that PTC1, PTC2, and PTC3 are the most important genes for HOG pathway inactivation.
Previously, we showed that Ptc1 inactivates the HOG pathway by acting on Hog1 (37). PTC1 overexpression blocked osmotic-stress-induced Hog1 activation. In contrast, deletion of PTC1 elevated Hog1 basal activity and inhibited Hog1 inactivation during adaptation. Ptc1 performed these functions by specifically dephosphorylating the Hog1-phosphothreonine residue in the activation loop. Ptc1 did not have a strong effect on the Pbs2 MEK in vivo (37), although vertebrate PTCs could inactivate stress-activated MEKKs and MEKs (9, 35).
In this work we examined the function of the Ptc2 and Ptc3 phosphatases. Ptc2 and Ptc3 may have similar functions, as they are 75% identical to each other. We believed that their functions might be different from those of Ptc1, as Ptc2 and Ptc3 contain an additional noncatalytic domain. Ptc2 and Ptc3 were similar to Ptc1 in that they inactivated Hog1. However, unlike deletion of PTC1, deletion of PTC2 and PTC3 did not alter basal Hog1 activity or adaptation but rather resulted in hyperactivation of Hog1 upon osmotic stress. Therefore, Ptc2 and Ptc3 limit maximal Hog1 activation, while Ptc1 regulates Hog1 basal activity and its dephosphorylation during adaptation. In this way, the three PTCs acting on this pathway have complementary roles in regulating Hog1.
MATERIALS AND METHODS
Yeast strains, media, and genetic techniques.
The yeast strains used in this work are described in Table 1. The temperature sensitivity of strains lacking PTCs and PTP2 was assessed in the BBY45 background (2). ACB5 (MATaptc2Δ::TRP1 ptp2Δ::HIS3 hog1Δ::kanMX) was produced by crossing IMY126 (MATaptc2Δ::TRP1 ptp2Δ::HIS3) (37) with ACB4 (MATα hog1Δ::kanMX). JMY1 (MATα ptc3Δ::HIS3 ptp2Δ::HIS3 hog1Δ::kanMX) and JMY2 (MATα ptc2Δ::TRP1 ptc3Δ::HIS3 ptp2Δ::HIS3 hog1Δ::kanMX) were produced by transforming IMY127b (37) and IMY128 (37), respectively, with the hog1Δ::kanMX allele. The hog1Δ allele was produced by PCR using oligonucleotides complementary to HOG1 and kanMX contained in the plasmid pRS400 (3). Media to culture yeast were produced essentially as described by Sherman et al. (33). Hog1 kinase assays were performed with the following strains. IMY105 (MATα hog1::hisG) (37), derived from 334 (11), overexpressed PTC2 from pKT-PTC2 (see below) and expressed Hog1 fused to the hemagglutinin (HA) epitope (Hog1-HA) from the pHOG1-ha2 plasmid (37). Hog1 kinase assays of ptcΔ strains were performed in the BBY45 background (2). CYY1 (MATaptc2Δ::TRP1 hog1Δ::TRP1) was produced by mating IMY120b (MATα ptc2Δ::TRP1) (2) to IMY100 (MATahog1Δ::TRP1) (12) carrying pHOG1-ha2. CYY2 (MATaptc3Δ::HIS3 hog1Δ::TRP1) was produced by mating IMY121b (MATα ptc3Δ::HIS3) (37) to IMY100 carrying pHOG1-ha2. Tetrads were dissected to isolate the desired strains. CAY9 (MATaptc2Δ::TRP1 ptc3Δ::HIS3 hog1Δ::TRP1) originated from a cross between IMY100b (MATα hog1Δ::TRP1) (12) and IMY124 (MATaptc2Δ::TRP1 ptc3Δ::HIS3) (37). CAY9 was transformed with pHOG1-ha3 (2μm LEU2), which expressed the same endogenous promoter-driven Hog1-HA as pHOG1-ha2 (2μm HIS3) (37), but from YEplac181 (2μm LEU2) (7).
TABLE 1.
Yeast strains
| Strain | Genotype | Reference or source |
|---|---|---|
| Strain BBY45 and strains derived from it | ||
| BBY45 | MATatrp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 2 |
| IMY21a | MATaptp2Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 24 |
| IMY124 | MATaptc2Δ::TRP1 ptc3Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 37 |
| IMY126 | MATaptc2Δ::TRP1 ptp2Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 37 |
| IMY127b | MATα ptc3Δ::HIS3 ptp2Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 37 |
| IMY128 | MATα ptc2Δ::TRP1 ptc3Δ::HIS3 ptp2Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 37 |
| ACB4 | MATα hog1Δ::kanMX trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| ACB5 | MATaptc2Δ::TRP1 ptp2Δ::HIS3 hog1Δ::kanMX trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| JMY1 | MATα ptc3Δ::HIS3 ptp2Δ::HIS3 hog1Δ::kanMX trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| JMY2 | MATα ptc2Δ::TRP1 ptc3Δ::HIS3 ptp2Δ::HIS3 hog1Δ::kanMX trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY100 | MATahog1Δ::TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 12 |
| IMY120b | MATα ptc2Δ::TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 37 |
| CYY1 | MATaptc2Δ::TRP1 hog1Δ::TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY121b | MATα ptc3Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 37 |
| CYY2 | MATaptc3Δ::HIS3 hog1Δ::TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| CAY9 | MATaptc2Δ::TRP1 ptc3Δ::HIS3 hog1Δ::TRP1 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | This work |
| IMY101 | MATasln1Δ::HIS3 trp1-1 ura3-52 his3-Δ200 leu2-3,112 lys2-801 gal | 12 |
| Strain 334 and a strain derived from it | ||
| 334 | MATα leu2-3,112 gal1 reg1-501 ura3-52 pep4-3 prb1-1122 | 11 |
| IMY105 | MATα hog1::hisG leu2-3,112 gal1 reg1-501 ura3-52 pep4-3 prb1-1122 | 37 |
The ability of Ptc2 mutant proteins lacking portions of the noncatalytic domain to inactivate the HOG pathway was examined by a phenotypic assay that we described previously (12). Deletion of SLN1 is lethal due to hyperactivation of the downstream HOG MAPK cascade (15). Increased expression of protein phosphatases that inactivate the MAPK cascade allows the sln1Δ strain to survive (12, 37). Plasmids carrying PTC2 and truncated PTC2 mutants were transformed into IMY101, a sln1Δ::HIS3 strain containing the plasmid pSLN1-URA3, a low-copy-number, CEN-based plasmid carrying the wild-type SLN1 gene and the URA3 gene (12). Growth of the strain on 5-fluoroorotic acid (5-FOA)-containing media occurs when the pSLN1-URA3 plasmid is lost, indicating that the phosphatase is active.
Plasmids.
The 6xHis-Ptc2 protein was produced in Escherichia coli by using the pRSETA-PTC2 plasmid. A BamHI site was introduced directly upstream of the start codon with the oligonucleotides 5′-GCGGATCCATGGACAAATTCTATCAAAC-3′, containing the restriction site (underlined), and 5′-CGGAATTCTCACAGCGCGACAACCACTATACTC-3′. The resulting fragment, containing the 5′ end of PTC2, was digested with BamHI and ClaI (an endogenous site) and ligated together with the 3′ end of PTC2 contained in an 800-bp ClaI-PstI fragment into pRSETA (Invitrogen) to produce pPRSETA-PTC2.
Ptc2 truncation mutant proteins Ptc2 (1-312), Ptc2 (1-355), and Ptc2 (1-412) were expressed in yeast and in E. coli. PCR was performed with a common 5′ primer containing a BamHI site (underlined), 5′-GCGGGATCCAATGATGGCAAATACGC-3′, and one of three 3′ primers, either 5′-GCAAGCTTTTAGCGGTGGGCCTTGGACTTCATAC-3′, 5′-GCAAGCTTTTAGTGGTCGCGAGTGAATTTGTCC-3′, or 5′-GCAAGCTTTTATCCTGTAGCACCAAGTAGCGCC-3′ (HindIII sites are underlined and stop codons are italicized), which were used to produce Ptc2 (1-312), Ptc2 (1-355), and Ptc2 (1-412), respectively. For expression in yeast, the PCR products were digested with BamHI and HindIII and ligated into the multicopy vector YEplac181 (7) to produce the plasmids pPTC2 (1-312), pPTC2 (1-355), and pPTC2 (1-412). The Ptc2 truncation mutants were also expressed in E. coli by using the pRSETA vector. The plasmids pPTC2 (1-312), pPTC2 (1-355), and pPTC2 (1-412) were digested with ClaI and HindIII to isolate the 3′ end of the gene and cloned into pRSETA-PTC2 digested with the same enzymes.
Glutathione S-transferase (GST) or green fluorescent protein (GFP) was fused to Ptc2 and Ptc3, and the fusion proteins were expressed in yeast from the following plasmids. The pKT-PTC2 plasmid was used to overexpress GST-Ptc2. PTC2 was isolated from pRSETA-PTC2 and cloned into p(EG)KT (21). Ptc2 and Ptc3 localization was examined by fusing GFP to their carboxy-terminal ends. PCR was used to replace the PTC2 and PTC3 stop codons with NotI sites, and a NotI fragment containing the GFP coding sequence (a gift from J. Hirsch, Columbia University) was inserted. The Ptc2-GFP fusion protein was expressed from its endogenous promoter in pRS426 (2μm URA3) and pRS316 (CEN URA3). Ptc3-GFP was expressed from its endogenous promoter in pRS424 (2μm TRP1) and in pRS314 (CEN TRP1) (34). Both the Ptc2-GFP and Ptc3-GFP fusion proteins were functional, as they suppressed sln1Δ lethality due to HOG pathway hyperactivation, as judged by the 5-FOA test described above (12). For the sln1Δ suppression assay, Ptc2-GFP was subcloned into the plasmid pRS424 (2μm TRP1) and Ptc3-GFP was expressed from the same pRS424 plasmid described above.
Expression and purification of Ptc2 and Ptc2 truncation mutant proteins from E. coli.
6xHis-Ptc2 and 6xHis-Ptc2 truncation mutants were expressed in E. coli and purified by metal affinity chromatography; the methods used were similar to those described by Warmka et al. (37). The plasmids pRSETA-Ptc2, pRSETA-Ptc2 (1-312), pRSETA-Ptc2 (1-355), and pRSETA-Ptc2 (1-412) were transformed into E. coli strain BL21(DE3)pLysS (Novagen), and expression was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 23°C. The 6xHis-Ptc2 proteins were purified with Co2+-immobilized metal affinity resin (TALON; Clontech) and eluted with elution buffer I (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 75 mM imidazole). Fractions containing 6xHis-Ptc2 proteins were identified by immunoblotting with an anti-His6 antibody (Babco). The isolated 6xHis-Ptc2 proteins were greater than 99% pure as judged by Coomassie staining and were quantified by using the bicinchoninic acid protein assay (Pierce).
Ptc2 inactivation of Hog1 kinase in vitro.
Ptc2 inactivation of Hog1 was performed essentially as described previously (37). 6xHis-Ptc2 purified from E. coli was incubated with osmotic-stress-activated GST-Hog1 isolated from yeast strain IMY105 carrying pKT-HOG1 (37). GST-Hog1 activity was assessed by examining its ability to phosphorylate myelin basic protein (MBP; Sigma). Buffers A and C, used to isolate GST-Hog1 (37), were modified so that each contained 5 mM MnCl2 instead of 5 mM MgCl2. Phosphatase assay buffer (37) was modified to contain 20 mM MnCl2 instead of 5 mM MgCl2. GST-Hog1 bound to glutathione-Sepharose (Pharmacia) was aliquoted into fractions containing ∼0.5 μg of GST-Hog1 and dephosphorylated with 0 to 1.2 μg of 6xHis-Ptc2 for 30 min at 37°C. GST-Hog1 was incubated with 1 μM MBP and 0.1 mM [γ-32P]ATP (8,000 cpm/pmol) for 30 min at 30°C to examine the remaining kinase activity (37). Incorporation of 32P into MBP was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and PhosphorImager analysis (Molecular Dynamics).
Phosphoamino acid analysis.
32P-labeled Hog1 was untreated or was incubated with 6xHis-Ptc2, and phosphoamino acid analysis was performed as described previously (37). Briefly, GST-Hog1, isolated from yeast strain IMY105, carrying pKT-HOG1 (37), was phosphorylated in vitro with hyperactive MEK mutant Pbs2EE and [γ-32P]ATP. Approximately 2.5 μg of 32P-GST-Hog1 bound to glutathione-Sepharose was incubated with 1.5 μg of 6xHis-Ptc2 for 30 min at 37°C. GST-Hog1 was isolated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride. GST-Hog1 was excised from the membrane and subjected to acid hydrolysis, and phosphoamino acid analysis was performed.
Ptc2 inactivation of Hog1 in vivo.
Hog1 kinase activity was assayed according to the methods described by Warmka et al. (37). Strains overexpressing PTC2 or deleted for PTCs and expressing Hog1-HA as described above were grown in selective media to an A600 of 1 and were untreated or exposed to 0.4 M NaCl for various times. Cells were disrupted in buffer A with glass beads, and Hog1-HA was immunoprecipitated with an anti-HA antibody (HA.11; Covance) and protein G-agarose (Santa Cruz Biotechnology) (37). Hog1 kinase activity was assayed by using 1 μM MBP (Sigma) and 0.1 mM [γ-32P]ATP (8,000 cpm/pmol) and by incubating cells for 30 min at 30°C
Immunoblotting.
Endogenous Hog1 Tyr phosphorylation in the yeast strain 334 (11), overexpressing PTC2 from the plasmid pKT-PTC2 described above or carrying empty vector pEG(KT), was examined (21). Cells in exponential growth phase were untreated or exposed to 0.4 M NaCl for various times, and samples were prepared for immunoblotting as described previously (12). Protein in cell lysates was quantified by the bicinchoninic acid protein assay (Pierce), and Hog1 Tyr phosphorylation was monitored by immunoblotting with an antiphosphotyrosine antibody (PY99; Santa Cruz Biotechnology). GST-Ptc2 and GST were detected with an anti-GST antibody (Pharmacia), and Hog1 was visualized with an anti-Hog1 antibody (yC-20; Santa Cruz Biotechnology).
Assay for phosphatase activity using pNPP.
The activity of 6xHis-Ptc2 and 6xHis-Ptc2 truncation proteins was assessed by using p-nitrophenyl phosphate (pNPP) as the substrate. Seven-picomole samples of 6xHis-Ptc2, 6xHis-Ptc2 (1-312), 6xHis-Ptc2 (1-355), and 6xHis-Ptc2 (1-412) contained in 100 μl of elution buffer I were incubated at 30°C with 100 μl of phosphatase buffer (100 mM imidazole [pH 7.5], 10 mM dithiothreitol, 10 mM MnCl2) containing 400 μM to 200 mM pNPP. Hydrolysis of pNPP was monitored by recording changes in absorbance at 405 nm over time and was linear from 0 to 100 min of incubation. The change in A405 versus time was plotted by using Kaleidagraph software, and best-fit-line equations were generated to determine velocity.
Fluorescence microscopy.
The subcellular localization of Ptc2-GFP and Ptc3-GFP in the ptc2Δ strain IMY120b (37) and the ptc3Δ strain IMY121b (37), respectively, was examined. Cultures in exponential growth phase were untreated or exposed to 0.4 M NaCl at 30°C for various times before examining PTC-GFP localization. DAPI (4′,6′-diamidino-2-phenylindole) was used to visualize the nucleus. Images were captured with a Zeiss Axioplan fluorescence microscope with a Sensys digital charge-coupled device camera system and processed by using IP-LAB Spectrum software.
RESULTS
Strains lacking PTC2, PTC3, and PTP2 have a temperature-sensitive defect due to HOG1.
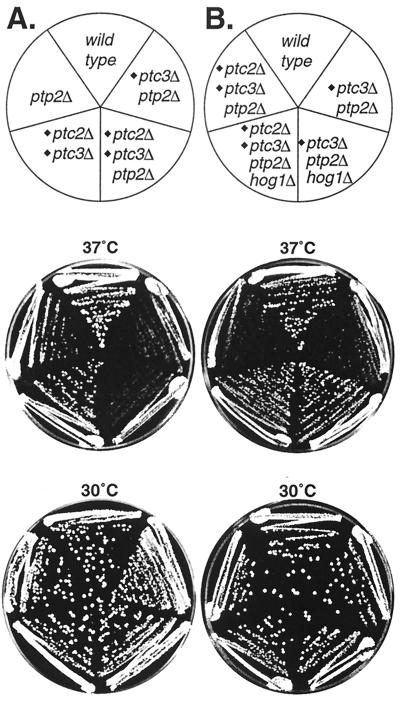
The sln1Δ lethal phenotype, which is due to HOG pathway hyperactivation, is suppressed by PTC2 and PTC3 expression from multicopy plasmids (15, 37). Here we tested whether the phenotype of PTC deletion mutants would also show a connection to the HOG pathway. Previously, we reported that strains lacking PTC2 and/or PTC3 and PTP2, encoding the PTP that inactivates Hog1, exhibited temperature-sensitive defects (27). Briefly, the ptc3Δ ptp2Δ and ptc2Δ ptp2Δ (data not shown) double mutants grew more poorly than the wild type at 37°C but grew normally at 30°C (Fig. 1A). Furthermore, the ptc2Δ ptc3Δ ptp2Δ triple mutant grew slightly worse than the ptc2Δ ptp2Δ and ptc3Δ ptp2Δ double mutants at 37°C but showed no obvious defect at 30°C (Fig. 1A). We tested whether these temperature-sensitive defects could be due to HOG1. This seemed possible, since we recently found that the HOG pathway is activated by heat stress and that a strain lacking the PTPs that inactivate Hog1 is lethal at 37°C due to Hog1 hyperactivation (38). To test this possibility, HOG1 was deleted in the phosphatase-null backgrounds. The ptc2Δ ptc3Δ ptp2Δ hog1Δ strain grew significantly better than the ptc2Δ ptc3Δ ptp2Δ strain at 37°C (Fig. 1B). In addition, ptc3Δ ptp2Δ hog1Δ (Fig. 1B) and ptc2Δ ptp2Δ hog1Δ (data not shown) strains each showed improved growth at 37°C, relative to ptc3Δ ptp2Δ (Fig. 1A) and ptc2Δ ptp2Δ strains. Therefore, strains lacking PTCs and PTP2 exhibit temperature sensitivity which is dependent on HOG1.
FIG. 1.
The temperature sensitivity of strains lacking PTCs and PTP2 is due to HOG1. (A) Strains lacking PTCs and PTP2 exhibit growth defects at 37°C. The growth of ptp2Δ (IMY21a), ptc2Δ ptc3Δ (IMY124), ptc3Δ ptp2Δ (IMY127b), ptc2Δ ptc3Δ ptp2Δ (IMY128), and wild-type (BBY45) strains on rich medium (yeast extract-peptone-dextrose [YPD]) at 37 and 30°C was compared. (B) The growth defects of the ptc3Δ ptp2Δ and ptc2Δ ptc3Δ ptp2Δ strains at 37°C were suppressed by deleting HOG1. The strains lacking HOG1 were JMY1 (ptc3Δ ptp2Δ hog1Δ and JMY2 (ptc2Δ ptc3Δ ptp2Δ hog1Δ) and were compared on YPD at 37 and 30°C.
Ptc2 inactivates Hog1 in vitro by dephosphorylating phosphothreonine.
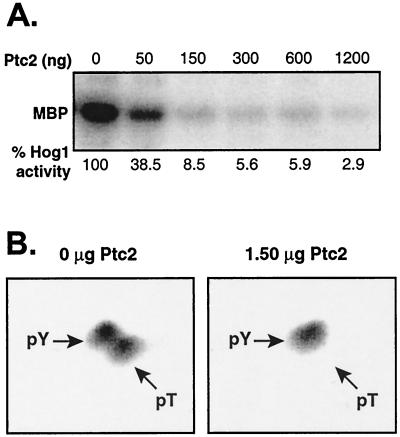
Previously, we showed that Ptc1 inactivates the HOG pathway by dephosphorylating Hog1. Since Ptc2 belongs to the same class of PTC phosphatases, we asked whether Ptc2 could also inactivate Hog1. We first tested whether Ptc2 could inactivate Hog1 in vitro. Activated Hog1 was isolated from osmotic stressed yeast and treated with Ptc2, which was expressed in E. coli and purified by affinity chromatography. Hog1 kinase activity was examined by using MBP and [γ-32P]ATP as substrates. Ptc2 readily inactivated Hog1 (Fig. 2A), similar to Ptc1 (37).
FIG. 2.
Ptc2 inactivates Hog1 in vitro. (A) Activated GST-Hog1 was isolated from osmotic stressed yeast and was untreated or treated with Ptc2. Approximately 0.5 μg of GST-Hog1 bound to resin was incubated with 0 to 1.2 μg of Ptc2 for 30 min at 30°C. The resin was washed extensively to remove Ptc2, and GST-Hog1 was incubated with MBP and [γ-32P]ATP to assess Hog1 kinase activity. Radiolabel incorporated into MBP was examined by PhosphorImager analysis. (B) Ptc2 inactivates Hog1 by dephosphorylating the phosphothreonine residue (pT) in the activation loop. GST-Hog1 was phosphorylated in vitro, and the sample was untreated (left) or incubated with Ptc2 (right) prior to phosphoamino acid analysis. The radiolabeled amino acids were detected with the PhosphorImager. Arrows, positions of phosphoamino acid standards as revealed by ninhydrin staining.
To examine the mechanism by which Ptc2 inactivates Hog1, we asked whether Ptc2 dephosphorylated the Hog1 activation loop Thr or Tyr residue. Since phosphorylation at both sites is required for full activity, Ptc2 could inactivate Hog1 by dephosphorylating either or both residues. Phosphorylated Hog1 was prepared in vitro by using the hyperactive MEK Pbs2EE (37) and [γ-32P]ATP. Prior to treatment with Ptc2, Hog1 was phosphorylated nearly equally at Thr and Tyr residues (Fig. 2B), which are in the activation loop (37). Following treatment with Ptc2, the [32P]phosphothreonine signal was eliminated while the [32P]phosphotyrosine residue was unchanged (Fig. 2B). Thus, Ptc2 inactivates Hog1 by specifically dephosphorylating the activation loop phosphothreonine residue.
Ptc2 inactivates Hog1 in vivo.
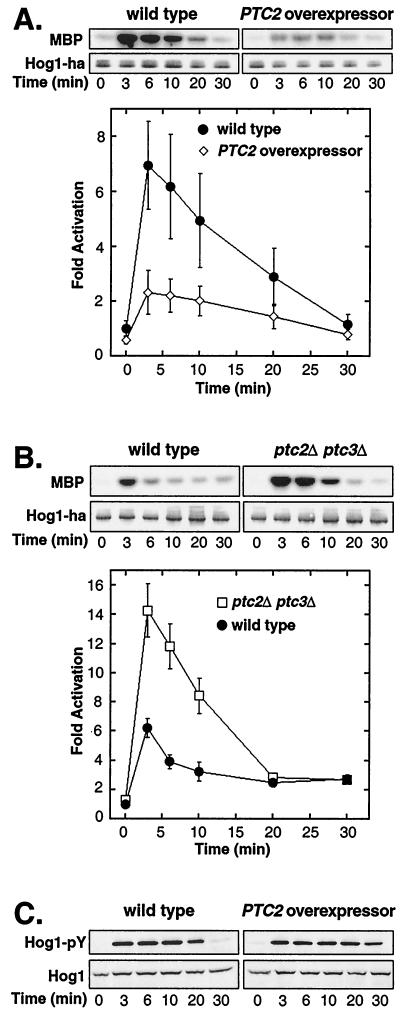
Since Ptc2 inactivated Hog1 in vitro, we tested whether it could also inactivate Hog1 in vivo. Hog1 activity was examined in untreated and osmotic stressed cells exposed to 0.4 M NaCl in a strain overexpressing PTC2 and in a control strain carrying empty vector. Overexpression of PTC2 inhibited maximal activity approximately threefold (Fig. 3A). In the PTC2 overexpressor, Hog1 activity increased fourfold upon osmotic stress, whereas in the wild type it increased sevenfold (Fig. 3A). The level of Hog1 protein was the same in the two strains, suggesting that the decrease in activity in the overexpressor was due to a change in phosphorylation.
FIG.3.
Ptc2 inactivates Hog1 in vivo. (A) Overexpression of PTC2 inhibits osmotic-stress-induced Hog1 activation. Hog1 kinase activity in PTC2 overexpressor IMY105, carrying pKT-PTC2 and pHOG1-ha2, and in the control strain, carrying empty vector pKT and pHOG1-ha2, was examined. Before (time zero) and after exposure to osmotic stress (0.4 M NaCl) for various times, Hog1-HA was immunoprecipitated and incubated with MBP and [γ-32P]ATP. Radiolabel incorporated into MBP was examined with the PhosphorImager. The graph shows the means of three independent experiments ± standard errors of the means (SEM). (B) Hog1 is hyperactivated in a strain lacking PTC2 and PTC3. Hog1 kinase activity was monitored prior to and following osmotic stress in a hog1Δ strain (IMY100) and in a ptc2Δ ptc3Δ hog1Δ strain (CAY9), each carrying a Hog1-HA-expressing plasmid. Hog1 kinase activity was monitored as described for panel A. The graph shows the means of six independent experiments ± SEM. (C) Ptc2 does not inactivate Pbs2 in vivo. Hog1-pY in the 334 strain carrying a plasmid overexpressing PTC2 or an empty vector was examined. The level of Hog1-pY prior to and following osmotic stress was monitored by immunoblotting with an anti-pY antibody. Total Hog1 protein was examined by blotting with an anti-Hog1 antibody.
Since overexpression of PTC2 inhibited Hog1 activation, we tested whether its deletion might elevate Hog1 activity. However, the ptc2Δ strain showed a level of Hog1 activity similar to that for the wild type (data not shown). Since Ptc2 is 75% identical to Ptc3, the effect of deleting PTC2 might be obscured by PTC3. Therefore, we also examined Hog1 activation in ptc3Δ and ptc2Δ ptc3Δ strains. Deletion of PTC3 alone had little effect on Hog1 activation (data not shown). However, deletion of both PTC2 and PTC3 had a significant effect. In the ptc2Δ ptc3Δ strain, Hog1 activity increased 11-fold upon osmotic stress, whereas it increased 6-fold in the wild type (Fig. 3B). The elevated Hog1 activity in the ptc2Δ ptc3Δ strain was likely due to phosphorylation, as the level of Hog1 protein in the mutant was comparable to that in the wild type (Fig. 3B). These results indicate that Ptc2 and Ptc3 have redundant functions in this pathway and that they set a maximum level to which Hog1 can be activated.
The results above suggested that Ptc2 acted on Hog1 in vivo; however, they did not exclude the possibility that Ptc2 could act upstream of Hog1. For example, since the MEK Pbs2 requires phosphorylation on Ser and Thr residues for activity (13), its dephosphorylation would also affect Hog1 activity. If Ptc2 inactivates Pbs2, then overexpressing PTC2 should inhibit Hog1 phosphorylation. We monitored Hog1 Tyr phosphorylation (Hog1-pY), since its Thr phosphorylation would be affected by Ptc2, as shown above (Fig. 2B). Overexpression of PTC2 did not strongly affect the level of Hog1-pY induced by osmotic stress (Fig. 3C), suggesting that Ptc2 has little effect on Pbs2 or activators upstream of Pbs2.
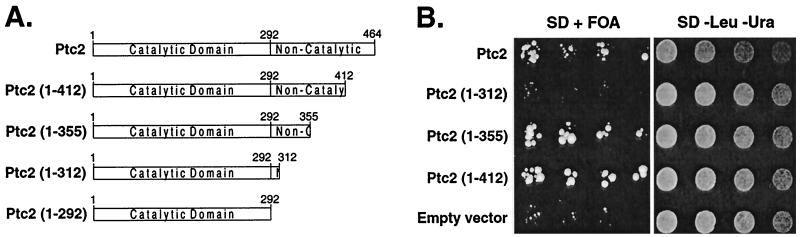
A portion of the Ptc2 noncatalytic domain is needed for activity.
Ptc2 and Ptc3 differ from Ptc1 in that they each have a carboxy-terminal noncatalytic domain of ∼170 residues. To examine the function of this domain, a series of deletion mutants was produced (Fig. 4A) and their ability to suppress the sln1Δ lethal phenotype due to Hog1 hyperactivation was assessed. The truncation mutant proteins Ptc2 (1-312) (Fig. 4B) and Ptc2 (1-292) (data not shown) were unable to suppress sln1Δ lethality. Since Ptc2 (1-292) was very poorly expressed in yeast (data not shown), it was not studied further. Ptc2 (1-355) and Ptc2 (1-412) suppressed sln1Δ lethality similarly to full-length Ptc2 (Fig. 4B). In some cases, they appeared to suppress better than the wild type (data not shown).
FIG. 4.
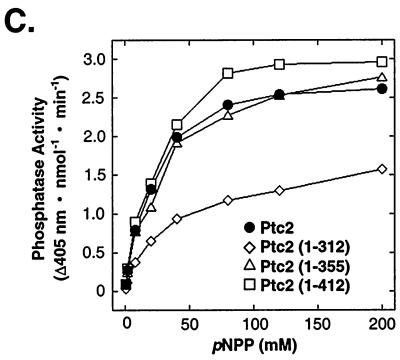
Analysis of Ptc2 noncatalytic domain function. (A) Schematic representation of Ptc2 and Ptc2 truncation mutant proteins. Full-length Ptc2 is 464 residues with an ∼292-residue catalytic domain and an ∼172-residue noncatalytic domain. The catalytic domain was assigned by comparison to S. cerevisiae Ptc1 (32) and human PP2Cα (16). (B) The truncation mutant Ptc2 (1-312) cannot inactivate the HOG pathway. The ability of the Ptc2 truncation proteins to suppress sln1Δ lethality was assayed as described in Materials and Methods. The sln1Δ strain carrying pSLN1-URA3 was transformed with a plasmid expressing full-length Ptc2, the plasmids expressing the truncation mutant proteins Ptc2 (1-312), Ptc2 (1-355), and Ptc2 (1-412), or an empty vector. The transformants were grown in 8 ml of synthetic medium lacking uracil and leucine to an A600 of 1. The cells were concentrated by centrifugation and resuspended in 0.5 ml of water. Twofold serial dilutions were spotted onto synthetic media containing 5-FOA (SD + FOA) or a control lacking FOA and grown at 30°C. Wild-type and mutant Ptc2 proteins were expressed at similar levels as judged by examination of corresponding HA-tagged proteins (not shown). (C) A portion of the noncatalytic domain of Ptc2 is required for full phosphatase activity. Ptc2 and Ptc2 truncation mutants Ptc2 (1-312), Ptc2 (1-355), and Ptc2 (1-412) were purified from E. coli as His6 fusion proteins and incubated with pNPP at the indicated concentrations. Rates were determined by measuring the production of p-nitrophenol at 405 nm and are expressed as the change in absorbance at 405 nm per nanomole of phosphatase per minute.
To investigate why the Ptc2 truncation mutants showed differences in the phenotypic assay relative to the full-length protein, their phosphatase activity was examined in vitro. Ptc2 and the Ptc2 truncation proteins were expressed as His6 fusions in E. coli and purified by Co2+ affinity chromatography. With pNPP as a substrate, Ptc2 (1-312) had a 1.6-fold lower Vmax and 2-fold-higher Km than full-length Ptc2 (1-464) (Fig. 4C), while the corresponding values for Ptc2 (1-355) and Ptc2 (1-412) were similar to those for the wild type. Therefore, the inability of Ptc2 (1-312) to inactivate the HOG pathway, as judged by the sln1Δ lethality suppression test (Fig. 4B), can be explained by its decreased catalytic activity. Thus, the catalytic domain, defined by similarity to other PTC sequences, is not sufficient for full Ptc2 activity.
Ptc2 and Ptc3 subcellular localization.
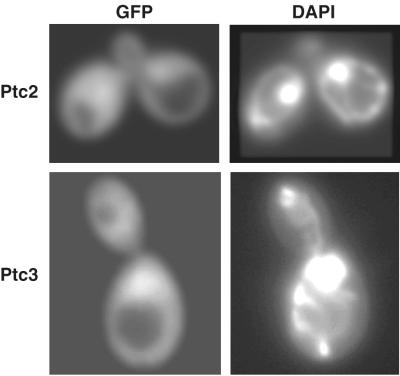
The subcellular localization of MAPK phosphatases is important to consider, as MAPKs including Hog1 accumulate in the nucleus upon activation and redistribute to the cytoplasm during adaptation (5, 18, 31). Therefore, protein phosphatases that regulate maximal Hog1 activity might be expected to be nuclear. The subcellular localization of Ptc2 and Ptc3 was examined by producing functional fusions to GFP. Both Ptc2-GFP and Ptc3-GFP were evenly distributed between the cytoplasm and the nucleus when expressed from low-copy-number or multicopy plasmids in their respective null backgrounds (Fig. 5). Therefore, Ptc2 and Ptc3 could control maximal Hog1 activity. PTC-GFP localization upon osmotic stress was also examined. However, no change in localization was observed with Ptc2-GFP or Ptc3-GFP (data not shown).
FIG. 5.
Ptc2 and Ptc3 are localized to the cytoplasm and nucleus. Ptc2-GFP and Ptc3-GFP were expressed from low-copy-number CEN-based plasmids in ptc2Δ and ptc3Δ strains, respectively. The PTC-GFP fusion proteins were visualized by fluorescence microscopy, and DAPI was used to identify the nucleus.
DISCUSSION
Hog1 MAPK activity is under coordinate regulation by the PTCs Ptc2 and Ptc3 and the PTPs Ptp2 and Ptp3. The PTCs regulate the level of Hog1 activation loop Thr phosphorylation (Fig. 2) (37), while the PTPs regulate the level of Tyr phosphorylation (12, 39). The focus of this study was the Ptc2 and Ptc3 phosphatases, which are 75% identical to each other, being related by a genome duplication event (39), and which show functional similarity in this pathway (37; this work). First, Ptc2 inactivated Hog1 activity in vitro (Fig. 2A) and specifically dephosphorylated the phosphothreonine residue, which is needed for activity (Fig. 2B). Second, Ptc2 inactivated Hog1 in vivo. Its overexpression inhibited Hog1 activation upon osmotic stress (Fig. 3A), and Ptc2 acted together with Ptc3 to inactivate Hog1. Whereas deletion of PTC2 or PTC3 had no effect on Hog1 activity, the ptc2Δ ptc3Δ double mutant showed Hog1 hyperactivation in osmotic stressed cells (Fig. 3B). Taken together, these results indicate that Ptc2 and Ptc3 inactivate the HOG pathway by dephosphorylating Hog1-phosphothreonine.
Our results also showed that Ptc2 had little effect on components of the pathway upstream of Hog1. For example, PTC2 overexpression did not significantly reduce Hog1 Tyr phosphorylation (Fig. 3C), suggesting that it had little effect on Pbs2 or upstream components of the HOG pathway. However, we cannot unequivocally state that Ptc2 has no activity on Pbs2, as we were not able measure Pbs2 activity directly. An unexpected effect of PTC2 overexpression was that Hog1-pY could be detected after 30 min of osmotic stress (Fig. 3C), whereas little Hog1-pY was seen in the wild type. Similar results were obtained with PTC1 overexpression (37). Hog1 Tyr phosphorylation may be prolonged in these PTC overexpressors because they might block the access of Ptp2 and Ptp3 to Hog1. Like S. cerevisiae, the Schizosaccharomyces pombe PTCs that regulate a stress-activated MAPK pathway have little effect on upstream regulators of MAPK (6, 22). The vertebrate PTCs differ from those of the yeasts in that they appear to act on MEKKs, MEKs, and p38, a stress-activated MAPK (9, 10, 35). How these differences in specificity between the yeast and vertebrate PTCs arise is not known, but they are likely to be due to different sequence elements within PTCs that mediate recognition of substrates.
In this work we also investigated the function of the Ptc2 noncatalytic domain. Since there is strong sequence similarity between the C-terminal residues of S. cerevisiae Ptc1 and sequences in Ptc2 and Ptc3 just prior to their noncatalytic domains, D-N-[VM]-[TS]-[VI]-X-[VI]-V-[FA]-L, we predicted that complete deletion of the Ptc2 noncatalytic domain would produce an active protein. However, neither Ptc2 (1-292) nor Ptc3 (1-293) was expressed in yeast (C. Young. and I. Ota, unpublished data). Therefore, other Ptc2 truncation proteins were produced. Ptc2 (1-312) was expressed in yeast similarly to the wild type, but it could not suppress sln1Δ lethality. We showed that this was due to its decreased catalytic activity relative to that of the full-length Ptc2 (Fig. 4C). Thus, unexpectedly, a portion of the Ptc2 noncatalytic domain is required for full catalytic activity.
We also noted that in some cases Ptc2 (1-355) and Ptc2 (1-412) suppressed sln1Δ lethality somewhat better than full-length Ptc2 (1-464). For example, the colonies of Ptc2 (1-355)- and Ptc2 (1-412)-bearing strains were often slightly larger than wild-type colonies on solid media containing 5-FOA. However, when these truncated proteins were expressed in E. coli and purified as His6 fusion proteins, no obvious difference in their kinetic properties relative to those of the full-length Ptc2 could be detected (Fig. 4C). Interestingly, when expressed as HA epitope-tagged proteins in yeast, isolated by immunoprecipitation, and assayed with pNPP, Ptc2 (1-355) and Ptc2 (1-412) had slightly greater activity than the full-length Ptc2, although their expression levels were similar (C. Young and I. Ota, unpublished data). These data suggest that Ptc2 activity may be modified in yeast.
The subcellular localizations of Ptc2 and Ptc3 were also examined, and, like those of Ptc1, they were found to be cytoplasmic and nuclear (Fig. 5). Since Ptc2 and Ptc3 are ∼50 kDa, close to the nuclear exclusion limit, ∼40 to 60 kDa (4), and since the Ptc2-GFP and Ptc3-GFP fusions are beyond that limit, nuclear localization signals are likely present in these phosphatases. Thus, the PTCs are unlike the two PTPs that act on Hog1, which show differences in localization; Ptp2 is nuclear and appears excluded from the cytoplasm, whereas Ptp3 is cytoplasmic and is nonnuclear (19). However, like the PTPs, the PTCs showed no change in subcellular localization in response to signal. As far as we are aware, none of the protein phosphatases that inactivate MAPKs in yeast or vertebrates change localization in response to signal. Therefore, the activity of Hog1 and other MAPKs, which translocate from the cytoplasm to the nucleus upon activation (5, 18, 31), is regulated by protein phosphatases whose localization is static.
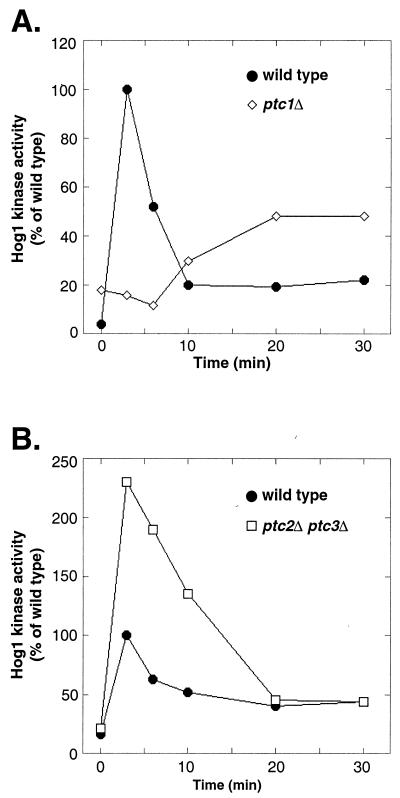
Comparison of the three PTCs in the HOG pathway shows they have complementary functions (summarized in Fig. 6). Previously, we showed that Ptc1 regulates the basal level of Hog1 phosphorylation and is required for dephosphorylation of Hog1 during adaptation (Fig. 6A) (37). Deletion of PTC1 resulted in basal Hog1 activity sixfold greater than that for the wild type. As shown in Fig. 6A, in the absence of stress, Hog1 activity in the ptc1Δ strain was already 20% of the stress-induced activity in the wild type. In addition, the ptc1Δ strain failed to inactivate Hog1 during adaptation (Fig. 6A). The ptc1Δ strain also showed a defect in its ability to activate Hog1, and this may be due to increased levels of negative regulators. For example, PTP2 and PTP3 transcripts are known to be upregulated when Hog1 is activated (12). In contrast, the ptc2Δ ptc3Δ strain did not show a strong change in basal Hog1 activity or in adaptation. Instead, it showed higher Hog1 activation in response to stress (Fig. 3B and 6B).
FIG. 6.
Ptc1 regulation of Hog1 differs from Ptc2 and Ptc3. (A) Deletion of PTC1 results in high basal Hog1 activity and an inability to inactivate Hog1 during adaptation. In addition, Hog1 showed a delay in activation and could not be fully activated in the ptc1Δ strain. The percentages of Hog1 activity relative to its maximal activity in the wild type are shown. Data are taken from Warmka et al. (37). (B) Deletion of PTC2 and PTC3 allowed Hog1 to be activated to a higher maximal activity than that for the wild type. No significant effects on basal activity or inactivation of Hog1 during adaptation were seen. Data are taken from Fig. 3B, and the percentages of Hog1 activity relative to that for the wild type at its maximum are shown.
The phenotypic effect of deleting PTCs was consistent with their roles in this pathway as defined by analysis of Hog1 activity. For example, deletion of PTC1 resulted in a phenotype consistent with regulation of basal Hog1 activity. The ptc1Δ ptp2Δ mutant exhibits a strong growth defect under standard growth conditions at 30°C (14), which is due to HOG1 (12). This result suggests that Hog1 is hyperphosphorylated in the ptc1Δ ptp2Δ mutant in the absence of stress. Indeed, Hog1 activity was increased in the ptc1Δ mutant prior to stress (37), as was the level of Hog1-pY in the ptp2Δ mutant (12, 40). In contrast, deletion of PTC2 and/or PTC3 together with PTP2 produced no obvious defect under standard growth conditions (Fig. 1), consistent with the result that deletion of PTC2 and PTC3 had little effect on basal Hog1 kinase activity (Fig. 3B).
Other phenotypic data supported the idea that Ptc2 and Ptc3 regulate maximal Hog1 activity. We recently showed that Hog1 is activated by heat stress and that deletion of PTP2 and PTP3, which encode PTPs that inactivate Hog1, is lethal at 37°C due to Hog1 hyperactivation (38). In this work we showed that deletion of PTCs together with PTP2 produced a temperature-sensitive defect dependent on HOG1. For example, the ptc3Δ ptp2Δ (Fig. 1A), ptc2Δ ptp2Δ (data not shown), and ptc2Δ ptc3Δ ptp2Δ (Fig. 1A) strains grew poorly at 37°C, and deletion of HOG1 suppressed these defects (Fig. 1B). These results suggest that the growth defect of the ptcΔ ptp2Δ strains at high temperature is due to increased maximal activity of Hog1. An additional phenotype was that the ptc2Δ ptc3Δ and ptc2Δ ptc3Δ ptp2Δ strains showed some enhanced ability to grow on solid media containing 0.9 to 1.4 M NaCl (J. Mapes and I. Ota, unpublished data). This phenotype is also consistent with stress inducing higher Hog1 activity in the phosphatase-null strains. One point that remains unclear is why heat stress is lethal to phosphatase-null strains, while osmotic stress is not. The simplest explanation is that heat stress induces physiological changes different from those induced by osmotic stress, which, when combined with Hog1 hyperactivation, are lethal. For example, as the cell wall integrity MAPK pathway is activated by heat stress and as the PTPs also inactivate this pathway, lethality may be due to misregulation of both this and the HOG pathways.
Ptc1 also differed from Ptc2 and -3 in its effects on adaptation. Previously, we showed that Ptc1 was required for adaptation, as Hog1 is poorly inactivated in the ptc1Δ mutant compared to its inactivation in the wild type (Fig. 6A) (37). For example, the ptc1Δ strain retained high Hog1 activity even after 1 h of osmotic stress (37). However, the ptc2Δ ptc3Δ strain showed no defect in adaptation (Fig. 3B and 6B). How Ptc2 and Ptc3 affect Hog1 maximal activity but not Hog1 inactivation during adaptation is not known. One possibility is that Ptc2 and Ptc3 activity is differentially regulated during Hog1 activation and adaptation. This hypothesis will require further investigation.
Acknowledgments
We thank Ji Lee for constructing the pKT-PTC2 plasmid and Chris Arkind and Anne Burkholder for constructing the ptc2Δ ptc3Δ ptp2Δ and ptc2Δ ptp2Δ hog1Δ yeast strains, respectively. We thank Chris Mattison and Amanda Reynolds for helpful comments and critical reading of the manuscript.
This work was supported by a grant from the American Cancer Society.
REFERENCES
- 1.Barford, D. 1996. Molecular mechanisms of the protein serine/threonine phosphatases. Trends Biochem. Sci. 21:407-412. [DOI] [PubMed] [Google Scholar]
- 2.Bartel, B., I. Wunning, and A. Varshavsky. 1990. The recognition component of the N-end rule pathway. EMBO J. 9:3179-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 4.Davis, L. I. 1995. The nuclear pore complex. Annu. Rev. Biochem. 64:865-896. [DOI] [PubMed] [Google Scholar]
- 5.Ferrigno, P., F. Posas, D. Koepp, H. Saito, and P. A. Silver. 1998. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 17:5606-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaits, F., K. Shiozaki, and P. Russell. 1997. Protein phosphatase 2C acts independently of stress-activated kinase cascade to regulate the stress response in fission yeast. J. Biol. Chem. 272:17873-17879. [DOI] [PubMed] [Google Scholar]
- 7.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 8.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanada, M., T. Kobayashi, M. Ohnishi, S. Ikeda, H. Wang, K. Katsura, Y. Yanagawa, A. Hiraga, R. Kanamaru, and S. Tamura. 1998. Selective suppression of stress-activated protein kinase pathway by protein phosphatase 2C in mammalian cells. FEBS Lett. 437:172-176. [DOI] [PubMed] [Google Scholar]
- 10.Hanada, M., J. Ninomiya-Tsuji, K. Komaki, M. Ohnishi, K. Katsura, R. Kanamaru, K. Matsumoto, and S. Tamura. 2001. Regulation of the TAK1 signaling pathway by protein phosphatase 2C. J. Biol. Chem. 276:5753-5759. [DOI] [PubMed] [Google Scholar]
- 11.Hovland, P., J. Flick, M. Johnston, and R. A. Sclafani. 1989. Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene 83:57-64. [DOI] [PubMed] [Google Scholar]
- 12.Jacoby, T., H. Flanagan, A. Faykin, A. G. Seto, C. Mattison, and I. Ota. 1997. Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J. Biol. Chem. 272:17749-17755. [DOI] [PubMed] [Google Scholar]
- 13.Maeda, T., M. Takekawa, and H. Saito. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269:554-558. [DOI] [PubMed] [Google Scholar]
- 14.Maeda, T., A. Y. Tsai, and H. Saito. 1993. Mutations in a protein tyrosine phosphatase gene (PTP2) and a protein serine/threonine phosphatase gene (PTC1) cause a synthetic growth defect in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:5408-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeda, T., S. M. Wurgler-Murphy, and H. Saito. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242-245. [DOI] [PubMed] [Google Scholar]
- 16.Mann, D. J., D. G. Campbell, C. H. McGowan, and P. T. Cohen. 1992. Mammalian protein serine/threonine phosphatase 2C: cDNA cloning and comparative analysis of amino acid sequences. Biochim. Biophys. Acta 1130:100-104. [DOI] [PubMed] [Google Scholar]
- 17.Martin-Blanco, E., A. Gampel, J. Ring, K. Virdee, N. Kirov, A. M. Tolkovsky, and A. Martinez-Arias. 1998. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 12:557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattison, C. P., and I. M. Ota. 2000. Two protein tyrosine phosphatases, Ptp2 and Ptp3, modulate the subcellular localization of the Hog1 MAP kinase in yeast. Genes Dev. 14:1229-1235. [PMC free article] [PubMed] [Google Scholar]
- 19.Mattison, C. P., S. S. Spencer, K. A. Kresge, J. Lee, and I. M. Ota. 1999. Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol. Cell. Biol. 19:7651-7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meskiene, I., L. Bogre, W. Glaser, J. Balog, M. Brandstotter, K. Zwerger, G. Ammerer, and H. Hirt. 1998. MP2C, a plant protein phosphatase 2C, functions as a negative regulator of mitogen-activated protein kinase pathways in yeast and plants. Proc. Natl. Acad. Sci. USA 95:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell, D. A., T. K. Marshall, and R. J. Deschenes. 1993. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9:715-722. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen, A. N., and K. Shiozaki. 1999. Heat-shock-induced activation of stress MAP kinase is regulated by threonine- and tyrosine-specific phosphatases. Genes Dev. 13:1653-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ota, I. M., and A. Varshavsky. 1993. A yeast protein similar to bacterial two-component regulators. Science 262:566-569. [DOI] [PubMed] [Google Scholar]
- 25.Pearson, G., F. Robinson, T. B. Gibson, B. E. Xu, M. Karandikar, K. Berman, and M. H. Cobb. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22:153-183. [DOI] [PubMed] [Google Scholar]
- 26.Posas, F., and H. Saito. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702-1705. [DOI] [PubMed] [Google Scholar]
- 27.Posas, F., E. A. Witten, and H. Saito. 1998. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 18:5788-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865-875. [DOI] [PubMed] [Google Scholar]
- 29.Raitt, D. C., F. Posas, and H. Saito. 2000. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19:4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramezani Rad, M., G. Jansen, F. Buhring, and C. P. Hollenberg. 1998. Ste50p is involved in regulating filamentous growth in the yeast Saccharomyces cerevisiae and associates with Ste11p. Mol. Gen. Genet. 259:29-38. [DOI] [PubMed] [Google Scholar]
- 31.Reiser, V., H. Ruis, and G. Ammerer. 1999. Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10:1147-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson, M. K., W. H. van Zyl, E. M. Phizicky, and J. R. Broach. 1994. TPD1 of Saccharomyces cerevisiae encodes a protein phosphatase 2C-like activity implicated in tRNA splicing and cell separation. Mol. Cell. Biol. 14:3634-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takekawa, M., T. Maeda, and H. Saito. 1998. Protein phosphatase 2Calpha inhibits the human stress-responsive p38 and JNK MAPK pathways. EMBO J. 17:4744-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walter, G., and M. Mumby. 1993. Protein serine/threonine phosphatases and cell transformation. Biochim. Biophys. Acta 1155:207-226. [DOI] [PubMed] [Google Scholar]
- 37.Warmka, J., J. Hanneman, J. Lee, D. Amin, and I. Ota. 2001. Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol. Cell. Biol. 21:51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler, A., C. Arkind, C. P. Mattison, A. Burkholder, K. Knoche, and I. Ota. 2002. Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. 1:163-173. [DOI] [PMC free article] [PubMed]
- 39.Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708-713. [DOI] [PubMed] [Google Scholar]
- 40.Wurgler-Murphy, S. M., T. Maeda, E. A. Witten, and H. Saito. 1997. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol. Cell. Biol. 17:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeitlinger, J., and D. Bohmann. 1999. Thorax closure in Drosophila: involvement of Fos and the JNK pathway. Development 126:3947-3956. [DOI] [PubMed] [Google Scholar]