Abstract
Malaria parasite antigens encoded by multigene families are important factors in virulence and in disease pathology. In Plasmodium falciparum, the virulence factor PfEMP-1 is encoded by the var multigene family and is exposed at the infected erythrocyte surface. PfEMP-1 is clonally variant, allowing the parasite to evade host immunity. The recently identified P. falciparum stevor multigene family and its products also have the potential to be involved in similar important aspects of host-parasite interactions. Here, we show tightly regulated stage-specific transcription of stevor occurring over just a few hours of the asexual parasite life cycle. Only a subset of stevor genes are transcribed in parasite populations maintained in cultures and in single micromanipulated parasites. Antibodies against STEVOR recognize proteins of the expected size (∼37 kDa) and localize STEVOR in Maurer's clefts, unique membranous structures located in the cytoplasm of infected erythrocytes. The fact that the timing of stevor expression and the location of STEVOR are clearly distinct from those of other parasite variant antigens suggests that this gene family may have a novel role in P. falciparum biology.
To facilitate intraerythrocytic growth and survival, the asexual stage of malaria parasites makes extensive modifications to the parasitized host red blood cell (pRBC) (1). These modifications include the presentation of immunogenic parasite proteins at the surface of the pRBC, exposing the parasite to the effects of host immunity (14, 25). Several of these surface proteins, for example, PfEMP-1 in Plasmodium falciparum and SICA antigen in P. knowelsi, induce antibody responses in the infected host which correlate with protection (11, 12). These molecules, encoded by multigene families, have also been shown to be clonally variant, allowing the parasite to evade host immunity. PfEMP-1, encoded by the var multigene family, facilitates the binding of pRBCs to a variety of host receptors on the endothelium, leading to the obstruction of blood vessels and contributing to the pathology and disease severity seen with P. falciparum. PfEMP-1 is thus considered a major virulence factor in P. falciparum infections in humans.
Research on malaria multigene families has so far focused on var and rif in P. falciparum and py235 in P. yoelii. However, the Malaria Genome Project has now revealed additional families, including stevor in P. falciparum. Although relatively little is known about stevor, the possibility that its expression product, STEVOR, is involved in virulence, pathology, immunity, and immune evasion warrants more detailed investigation. First reported as 7h8, stevor was initially identified as an expressed sequence detected by a monoclonal antibody (29). There are about 30 to 40 copies of stevor per haploid genome, clustered within 50 kb of the telomeres on all chromosomes (13). Like rif, stevor has short first and longer (∼1-kb) second exons. The protein (STEVOR) has a predicted size of 30 to 40 kDa (13) and structurally may be similar to the rif-encoded RIFINs. Structure predictions indicate a membrane-bound cell surface protein containing a highly conserved N-terminal sequence (exon 1) encoding a putative signal peptide and three putative transmembrane domains flanking the highly polymorphic region predicted for exon 2. However, although certain stevor and rif sequence motifs are similar in size and structure, detailed analysis reveals stevor to represent a distinct, more conserved, lower-copy-number gene family (13). Preliminary work has indicated that stevor is transcribed in both asexual and sexual stages of P. falciparum (13, 40).
In this study, we show that in infected erythrocytes, the parasite transcribes some, but not all, stevor genes during the mid-trophozoite stage of parasite development. Furthermore, analysis of micromanipulated single trophozoites indicates that although individual parasites contain multiple stevor transcripts, again only a subset of genes is transcribed. We demonstrate that, in late-stage trophozoites and schizonts, STEVOR is located in Maurer's clefts (MC), unique membranous structures located just beneath the RBC membrane. Finally, we show that STEVOR is also expressed in gametocytes. The differential and stage-specific timing of the transcription and expression of stevor, together with the unique location of STEVOR in asexual parasites, suggests that the stevor family may have a novel role in P. falciparum biology, different from that of either the var or the rif family.
MATERIALS AND METHODS
Parasites and cell lines.
Except where stated otherwise, P. falciparum 3D7 was used in all experiments and maintained in vitro as previously described (41). P. falciparum expressing a truncated form of PfEMP-3 was kindly donated by A. Cowman (Melbourne, Australia) and maintained as previously described (43). Parasite cultures were synchronized by sorbitol lysis and fractionation on a Percoll gradient (6, 7). Schizonts collected during synchronization were used to make thin blood smears for indirect immunofluorescence assays (IFAs) and for protein extraction (see below). For the micromanipulation of single pRBCs, trophozoites were isolated over sorbitol-Percoll gradients (24). Uninfected RBC ghosts and schizont ghosts were obtained by hypotonic lysis as previously described (16).
Micromanipulation of single-cell parasites.
Trophozoites were collected as described above, pelleted cells (500 × g for 10 min at room temperature) were washed twice in Krebs buffered saline, and single parasites were micromanipulated (34).
PCR.
DNA was extracted from asynchronous parasites at 5 to 10% parasitemia as previously described (31). The sequences of internal primers RepF1, RepF2, and RepR, designed around the polymorphic region of stevor, have been published elsewhere (13). External primers smkf1 [GA(C/T) (C/G)CA GAA CTC AA(A/G) GAA AT(A/T) ATT G] and smkr1 [GCA G(A/C)A CCA AAG (C/T) (T/A)G (C/T)AA TAC C] were designed to represent a conserved region of the gene based on six full-length stevor sequences (accession no. AF065198 to AF065201 from the National Center for Biotechnology Information database [http://www.ncbi.nlm.nih.gov/]).
The PCR mixture used with primers smkf1 and smkr1 contained 1 μM each primer, 5 μl of PCR buffer (100 mM Tris-HCl, 500 mM KCl), 2 mM MgCl2 (Roche), 1 mM each deoxynucleoside triphosphate (Amersham Pharmacia Biotech), and 5 U of AmpliTaq polymerase (Roche) in a volume of 50 μl. The PCR program was 1 cycle of 94°C for 3 min; 35 cycles of 94°C for 1 min, 62.5°C for 1 min, and 72°C for 1 min 30 s; and finally one 10-min cycle at 72°C. A 2-μl aliquot from the first reaction was directly transferred to the nested PCR mixture, which contained 0.5 μM each RepF1 and RepF2, 1 μM RepR, 2.5 μl of PCR buffer, 3 mM MgCl2, 1 mM each deoxynucleoside triphosphate, and 2.5 U of AmpliTaq polymerase. The PCR program was as follows: 1 cycle of 94°C for 3 min; 40 cycles of 93°C for 30 s, 55°C for 50 s, and 70°C for 30 s; and finally one 10-min cycle at 72°C.
RT-PCR and single-cell RT-PCR.
RNA was isolated from asynchronous parasite cultures (5 to 10% parasitemia) by using TRIzol (Life Technologies) and was stored in formamide at −70°C as described previously (26). Reverse transcriptase (RT) PCR with random primers was performed as described previously (33).
Southern and Northern blot analyses.
Pulsed-field gel electrophoresis (PFGE) of intact parasite chromosomal DNA was carried out by lysing parasites in agarose plugs (9). Whole chromosomes were resolved by using a custom-built contour-clamped homogeneous electric field apparatus (42) and gels containing 1% chromosome-grade agarose (Gibco BRL) in 0.025 M Tris- 0.025 M boric acid-0.625 M EDTA. Gel run pulse times and conditions were as follows: a 90- to 300-s ramp at 95 V for 36 h followed by a 300- to 720-s ramp at 85 V for 36 h, with a 3-s pause between each pulse. Gels were then transferred by alkali blotting to charged nylon membranes (Hybond N+; Amersham) according to the manufacturer's instructions. Electrophoresis of parasite RNA was carried out with 1% agarose gels (26). RNA was transferred to Hybond N+ as described elsewhere (26). DNA and RNA were fixed on the membranes by cross-linking with a Stratalinker (Stratagene).
PCR or RT-PCR products were labeled for hybridization reactions by using a Prime-it II DNA labeling system (Stratagene). Purified PCR or RT-PCR products (25 ng) were labeled in a 50-μl reaction mixture by using 1.1 MBq of [α-32P]dATP according to the manufacturer's instructions. Filters were prehybridized for 1 h in hybridization buffer (Sigma). They were hybridized overnight in the same buffer with the addition of the probe and washed with buffers containing 0.1% (vol/vol) sodium dodecyl sulfate (SDS) (Sigma) and between 0.5× and 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) at 65°C to room temperature depending on the stringency required.
Sequencing and analysis.
PCR and RT-PCR products were cloned and sequenced as previously described (33). DNA sequence data were analyzed by using ABI SeqED and DNASTAR EditSeq. Sequences were compared against database sequences (http://www.ncbi.nlm.nih.gov/Malaria/plasmodiumbl.html) by using a tblastn search.
Antiserum and monoclonal antibody preparation.
Synthetic peptide 1 (CNPHYHNDPELKEII, amino acids 57 to 70), designated MKSt1196, and peptide 2 (CIWLYRRRKNSWKHECKKHLC, amino acids 281 to 300), designated MKSt1341, were designed on the basis of conserved regions of STEVOR sequences obtained from the National Center for Biotechnology Information database (13). A cysteine residue was added at the N termini of the peptide sequences in order to conjugate them to Imject Maleimide Activated Mariculture KLH (Pierce), a procedure which was performed according to the manufacturer's instructions. The solution was then transferred to Slide-A-Lyzer dialysis cassettes (Pierce) and dialyzed twice with 5 liters of phosphate-buffered saline (PBS). BALB/c mice were immunized intraperitoneally with 50 μg of the peptide-KLH conjugate in complete Freund's adjuvant followed by three boosts of the conjugate in incomplete Freund's adjuvant at 14-day intervals. Serum was collected 7 to 10 days after the second and third boosts. With the same protocol, rabbits were immunized subcutaneously with 200 μg of the peptide-KLH conjugate.
Monoclonal antibodies against peptide 1 were made as described by Shulman et al. (37).
Protein analysis by Western blot and immunoprecipitation assays.
Schizonts (approximately 100 μl) were solubilized in Triton X-100 and SDS buffer in the presence of protease inhibitors (7). Proteins were resolved by electrophoresis on NuPAGE 4 to 12% bis-Tris gels (Invitrogen) in MES sample buffer (Invitrogen) under reducing conditions and according to the manufacturer's instructions. Broad-range prestained protein standards (New England Biolabs) were also loaded as markers. Proteins were electrophoretically transferred to nitrocellulose extra blotting membranes (Sartorius AG) by using a NuPAGE electrophoresis system (Invitrogen). Specific proteins were detected by using mouse polyclonal sera and mouse monoclonal sera, and binding of these antibodies was followed by treatment with horseradish peroxidase-linked secondary antibodies (Bio-Rad) and enhanced chemiluminescence (Pierce).
Asynchronous parasite cultures were metabolically radiolabeled for 4 h with [35S]methionine-cysteine (100 μCi of Promix [Amersham]/ml in methionine-cysteine-free RPMI-Albumax medium). Labeled proteins were immunoprecipitated (5) and then separated on NuPAGE 4 to 12% bis-Tris precast gels (Invitrogen).
IFAs.
IFAs were carried out with acetone-fixed thin blood smears enriched for schizonts (20). The primary antibodies used were 2F10 (anti-merozoite surface protein 1 [MSP-1] mouse monoclonal antibody immunoglobulin G [IgG]; 1:50) (7), 61.2 (anti-rhoptry mouse monoclonal antibody IgG; 1:100) (A. A. Holder), B28 (anti-PfSBP1 mouse polyclonal sera; 1:100) (8), DG662 (anti-PfEMP3 mouse polyclonal sera; 1:100) (19), anti-peptide 1 (MKSt1196) rabbit preimmune and anti-MKSt1196 rabbit polyclonal sera (1:100), anti-MKst1196 mouse polyclonal sera (1:100), anti-MKSt1196 monoclonal antibody 27E11 (neat), and normal and anti-MKSt1341 mouse polyclonal sera (1:20). The secondary antibodies used were goat anti-rabbit IgG-fluorescein isothiocyanate (FITC) (1:200) (Sigma), goat anti-mouse IgG-FITC (1:50) (Sigma), goat anti-mouse IgG-Texas red (1:100) (4), and goat anti-rabbit IgG-tetramethyl rhodamine isothiocyanate (1:50) (Sigma). For double staining, primary and secondary antibodies were added sequentially. The slides were viewed at a magnification of ×63 with a fluorescence microscope or at a magnification of ×100 with an Olympus Delta Vision imaging system.
ELISA.
The reactivity of anti-STEVOR serum to the corresponding peptide was measured by an enzyme-linked immunosorbent assay (ELISA). Polysorb ELISA plates (Nunc, Naperville, Ill.) were coated overnight with 50 μl of peptide (at 4 μg/ml in PBS), and the ELISA was carried out as previously described (28). Briefly, 12 twofold serial dilutions (starting at 1:10) were prepared by using 50 μl each of rabbit or mouse immune serum and preimmune serum (as a negative control) in diluent buffer (PBS containing 1% bovine serum albumin), and these were added to assigned wells. To detect bound STEVOR peptide-specific antibodies, we used 50 μl of goat anti-rabbit alkaline phosphatase-conjugated IgG (1:10,000 dilution in diluent buffer) or goat anti-mouse alkaline phosphatase-conjugated IgG (1:2,000 dilution in diluent buffer) (Southern Biotechnology Associates, Birmingham, Ala.).
RESULTS
Transcription of stevor in a population of parasites.
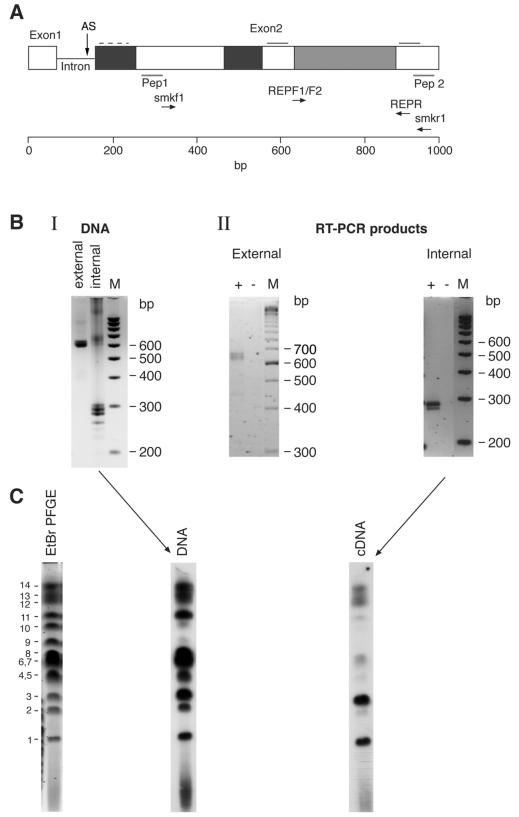
The transcription of stevor in asexual blood stages of P. falciparum was analyzed by using the primers described in Fig. 1A. The primer pairs span the region of highest variability between different stevor genes. Thus, to assess their ability to recognize multiple genes, PCR was performed by using genomic P. falciparum 3D7 DNA. Multiple bands of the expected sizes (∼650 and ∼300 bp for the external and internal primers, respectively) were detected on agarose gels (Fig. 1B, I). RT-PCR with RNA from asynchronous 3D7 parasites (Fig. 1B, II) also gave fragments of sizes similar to those obtained by PCR. Sequencing of a large number of clones obtained from both PCR and RT-PCR confirmed that only stevor-related sequences were amplified (data not shown).
FIG. 1.
(A) Schematic representation of stevor (1,000 bp). The external (smkf1 and smkr1) and internal (RepF1, RepF2, and RepR) primers used in this study are indicated. Dark grey shading, moderately polymorphic. Light grey shading, highly polymorphic. Broken line, weakly predicted transmembrane domain. Solid line (above sequence and at right), strongly predicted transmembrane domain. AS, position of alternative splice site. Positions of peptides (Pep) 1 and 2 are also shown. (B) PCR and RT-PCR analyses of stevor. (I) PCR products of 3D7 genomic DNA amplified with smkf1 and smkr1 and with RepF1, RepF2, and RepR. (II) RT-PCR products obtained from RNA of asynchronous parasites with smkf1 and smkr1 and with RepF1, RepF2, and RepR. The products were separated on Metaphor 3% agarose gels. Lanes +, RT added. Lanes −, no RT added. Lanes M, 100-bp ladder (Bio-Rad). (C) Southern blot analysis of P. falciparum 3D7 chromosomes probed with PCR or RT-PCR products. The left panel shows PFGE of 3D7 chromosomes stained with ethidium bromide (EtBr) and photographed before the DNA from the gels was transferred to nylon membranes and probed. Numbers at left refer to P. falciparum chromosomes. Probing was performed with the PCR product obtained from DNA (middle panel) and with the RT-PCR product obtained from RNA (cDNA) (right panel) of 3D7 parasites.
To determine whether there were any differences in the amplification products obtained by PCR or RT-PCR, 3D7 chromosomes were separated by using PFGE and probed with radioactively labeled products obtained from either PCR or RT-PCR. The genomic PCR product appeared to hybridize with similar intensities to all of the chromosomes except chromosomes 9 and 10 (Fig. 1C). In contrast, the RT-PCR product hybridized strongly only to a subset of the chromosomes, with other chromosomes being recognized weakly or not at all, suggesting that not all stevor genes are transcribed at the same level. To eliminate the possibility that hybridization to multiple chromosomes was due to cross-hybridization, the PCR products were cloned, and individual clones then were used to reprobe PFGE filters. Each cloned probe hybridized only to a single chromosomal band (data not shown). The hybridization results indicating that only a subset of stevor genes are transcribed are consistent with the sequencing data obtained from the products of four independent RT-PCRs and three different PCRs. A total of 21 different sequences were identified in the 53 PCR clones sequenced, while only 9 different sequences were obtained from sequencing of 81 RT-PCR clones. Furthermore, over 50% of all the RT-PCR clones sequenced came from a single gene, indicating strong transcriptional restriction to a subset of stevor genes at a given time in a population of parasites.
Timing of stevor transcription during the asexual parasite life cycle.
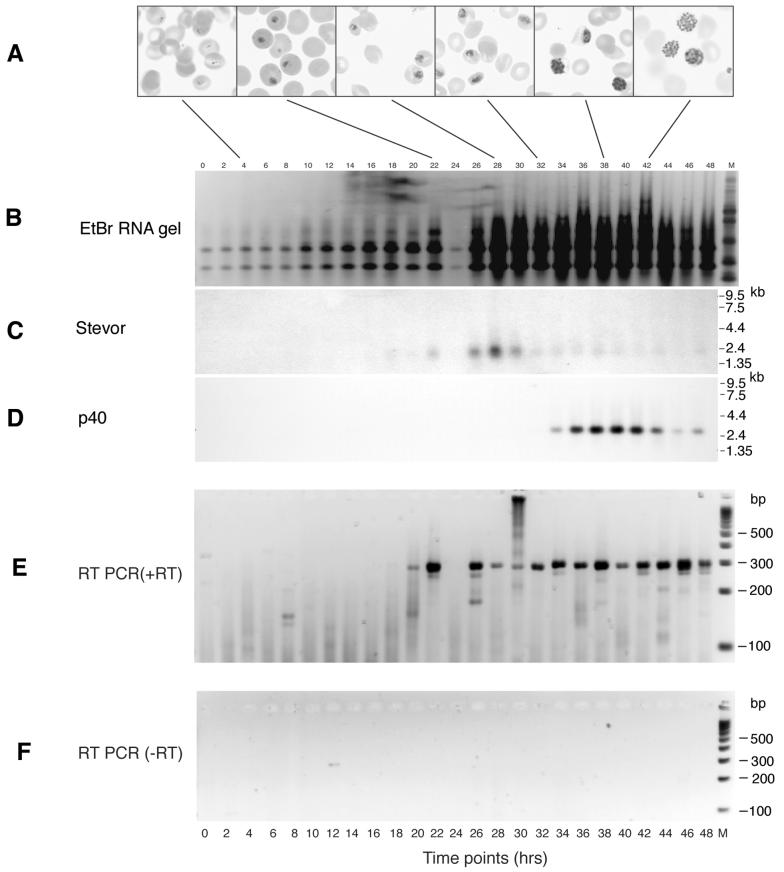
To determine the exact time of stevor transcription during the 48-h erythrocytic cycle, RNA was extracted from samples taken at 2-h intervals from a highly synchronized parasite culture (Fig. 2A shows the corresponding morphological stages of parasite development). After agarose gel electrophoresis, the RNA was transferred to a nylon membrane and probed with radiolabeled stevor nested PCR products obtained from genomic DNA (Fig. 2B). A diffuse band, of approximately 2.4 kb and corresponding to putative multiple stevor genes, was first detected in RNA prepared 22 h after merozoite invasion of RBCs (Fig. 2C). Maximum stevor transcription occurred at 28 h, when the parasites were at the mid-trophozoite stage; after this time, the signal strength was rapidly lost despite the fact that there was an increased amount of total RNA on the filter in the latter half of the life cycle (Fig. 2B). Probing of the same filter with the single-copy gene p40 (msp-7) (32), which is specifically expressed in late trophozoites and schizonts, confirmed the integrity of the RNA samples loaded (Fig. 2D). To confirm that the first appearance of stevor transcripts at 22 h was indeed due to the transcriptional activation of the genes and not due to a lack of sensitivity of the assay, we reanalyzed the time course samples for stevor transcription by using RT-PCR. A stevor-specific RT-PCR product clearly was detected from 20 h after merozoite invasion (Fig. 2E), while no transcripts were detected before. No RT-PCR products were detected in the control (no RT) (Fig. 2F).
FIG. 2.
Northern blot and RT-PCR analyses of stevor mRNAs purified from samples collected every 2 h during a 48-h cycle of development of P. falciparum 3D7 asexual blood-stage forms in vitro. The first time point (0 h) corresponds to the time of erythrocyte invasion by merozoites. (A) Images from Giemsa-stained slides of parasites taken at the times indicated (magnification, ×100). (B to D) Total RNA was electrophoresed through agarose, stained with ethidium bromide (EtBr) (B), transferred to nitrocellulose, and hybridized with a radiolabeled DNA probe derived from stevor (C) and a P. falciparum control gene, p40 (msp-7) (D). Nested RT-PCR products were obtained from the different samples by using the external (smkf1 and smkr1) and subsequently the internal (RepF1, RepF2, and RepR) stevor primers. (E) Samples separated on Metaphor 3% agarose gels. (F) Control nested RT-PCR analysis with no RT added. The migration of molecular mass markers is shown.
Multiple stevor transcripts are detected in single micromanipulated trophozoite-stage parasites.
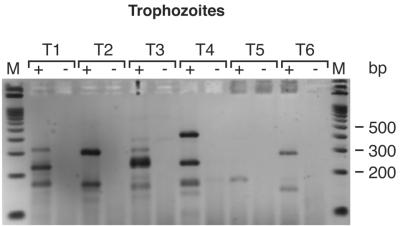
To determine the pattern of stevor transcription in single cells, trophozoites at about 28 h of development were collected individually by micromanipulation and analyzed by RT-PCR. RT-PCR products of the expected size (∼300 bp) were obtained from most of the cells analyzed (Fig. 3), but additional bands were observed in all of the cells (Fig. 3). After cloning and sequencing of the RT-PCR products, a comparison with database sequences confirmed that all of the products were stevor. The ∼300-bp fragments contained the expected full-length stevor sequences, while the smaller bands corresponded to products in which mispriming of one of the nested PCR primers was suspected to have led to smaller products. These smaller products most likely were PCR artifacts due to the mispriming of one of the internal primers, but the finding that two or three full-length products were detected per cell indicates that more than one gene was transcribed.
FIG. 3.
RT-PCR of single micromanipulated trophozoite-infected erythrocytes. The nested PCR products from six individual trophozoites (T1 to T6) were separated on Metaphor 3% agarose gels. Lanes +, RT added. Lanes −, no RT added. Lanes M, 100-bp ladder (Bio-Rad).
STEVOR is expressed in blood-stage parasites.
To determine whether STEVOR is expressed in pRBCs, polyclonal and monoclonal antibodies were raised against STEVOR peptides in rabbits and mice. Multiple stevor sequences obtained from the database were aligned, and two highly conserved regions were identified for peptide synthesis. Peptide 1 comprised a sequence from the conserved amino-terminal region, while peptide 2 was based on the conserved carboxy-terminal domain (Fig. 1A). The specificity of the antisera for the appropriate peptide was tested by an ELISA with normal mouse or preimmune rabbit serum as a negative control. Antisera with the appropriate specificity and titer were used for the subsequent studies (see below).
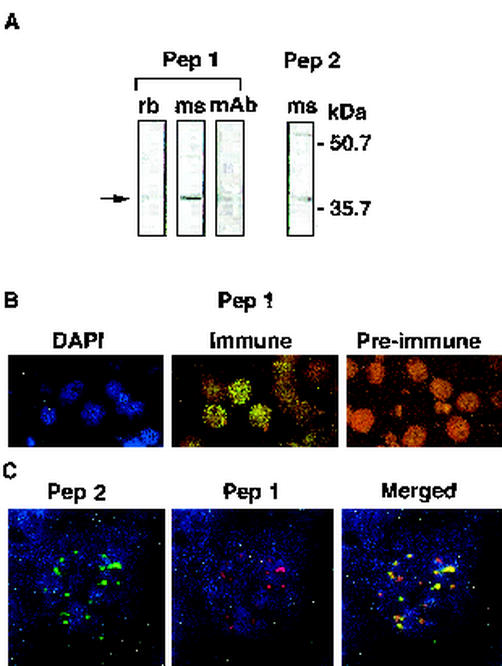
Protein extracts prepared from metabolically labeled asynchronous parasites were precleared for nonspecific binding with preimmune rabbit serum followed by immunoprecipitation with the rabbit serum against peptide 1. A protein of ∼37 kDa was specifically immunoprecipitated (Fig. 4A, lane rb). Furthermore, both mouse polyclonal antibodies and a mouse monoclonal antibody raised against peptide 1 detected a protein of the same size in Western blot analysis (Fig. 4A, lanes ms and mAb). A protein of the same size was also detected by a mouse polyclonal antibody raised against peptide 2 (Fig. 4A). No proteins of the appropriate size were detected by preimmune serum in either Western blotting or immunoprecipitation analysis (data not shown). Accordingly, all the antisera recognized proteins within the predicted size range (30 to 40 kDa) in both immunoprecipitation and Western blotting analyses. stevor genes are thus transcribed and translated into STEVOR in asexual blood-stage parasites.
FIG. 4.
Expression of STEVOR in 3D7 parasites analyzed by immunoprecipitation, SDS-polyacrylamide gel electrophoresis (PAGE), and IFA. (A) Upon analysis by SDS-PAGE, rabbit (rb) antibodies against peptide 1 precipitated a protein from an SDS-solubilized fraction of metabolically [35S]methionine-cysteine-labeled asynchronous 3D7 parasites. Mouse (ms) antibodies against peptides 1 and 2 and a monoclonal antibody (mAb) against peptide 1 were used to probe an SDS-soluble fraction of a schizont extract analyzed by SDS-PAGE and Western blotting. (B) IFA of fixed 3D7 pRBCs (late trophozoites and schizonts). pRBCs are stained blue with DAPI nucleic acid stain (left panel) and green with immune serum (middle panel) and are not stained with preimmune rabbit polyclonal serum against peptide 1 (right panel). Following incubation with FITC-conjugated goat anti-rabbit IgG, the cells were visualized with a fluorescence microscope. The left and middle panels are the same microscopic field. (C) Colocalization studies of anti-STEVOR antibodies. The images show IFA of fixed 3D7 schizonts stained with anti-peptide 2 antibodies and FITC-conjugated goat anti-mouse IgG (green) (left panel) and with anti-peptide 1 antibodies and tetramethyl rhodamine isothiocyanate-conjugated goat anti-rabbit IgG (red) (middle panel). The right panel shows a merging of the two images. An Olympus Delta Vision imaging system was used (magnification, ×1,000).
STEVOR is located in MC in asexual blood-stage parasites.
To determine the cellular location of STEVOR, we first carried out an IFA with the rabbit and mouse antibodies to peptides 1 and 2. Both sets of antibodies gave identical punctate IFA staining patterns in pRBCs (Fig. 4B, Immune, and 4C, Pep 2 and Pep 1), while the preimmune serum did not react with pRBCs (Fig. 4B, Pre-immune). Staining with all antibodies was observed only in pRBCs, as confirmed with DAPI. Clear differences in the IFA staining patterns were observed when staining obtained with antibodies to peptides 1 and 2 was compared with that obtained with antibodies to MSP-1 (data not shown), indicating different cellular locations. A comparison of the MSP-1 and DAPI staining patterns with those obtained with antibodies to peptides 1 and 2 indicated that only ∼30% of the pRBCs reacted strongly with these antibodies. Colocalization studies with rabbit anti-peptide 1 and mouse anti-peptide 2 antibodies confirmed that they bound to the same subcellular locations within pRBCs (Fig. 4C, Merged). Importantly, the colocalization studies also showed that the majority of the pRBCs stained by anti-peptide 1 antibodies were also stained by anti-peptide 2 antibodies.
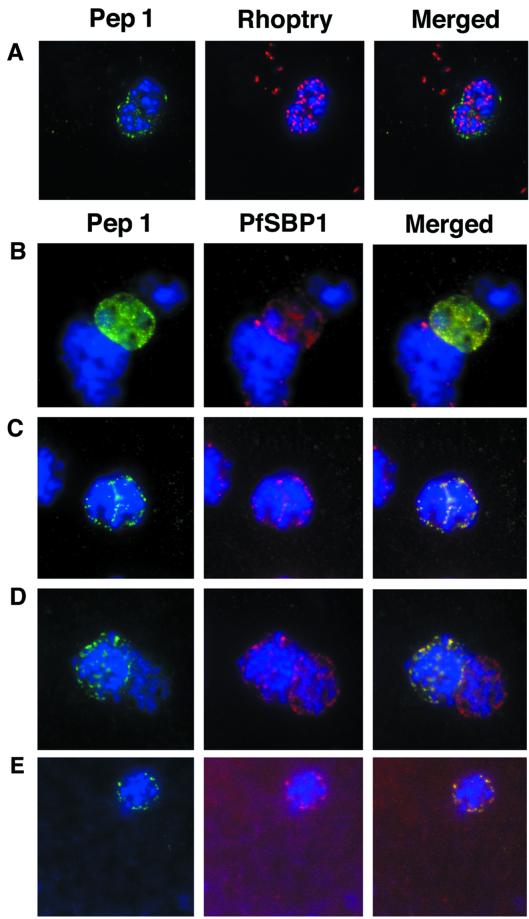
IFA studies with only anti-STEVOR antibodies alone are insufficient to define the cellular location of STEVOR within pRBCs. Therefore, a number of other antibodies against P. falciparum proteins of known locations were used in IFA colocalization experiments. In schizonts, an antibody (61.2) raised against a 52-kDa rhoptry protein gave a punctate staining pattern (Fig. 5A, Rhoptry) similar to that obtained with anti-STEVOR antibodies (Fig. 5A, Pep 1). However, merging of the two images showed that STEVOR was excluded from the rhoptries (Fig. 5A, Merged). Instead, the images suggested that the protein was located external to the rhoptries, possibly within or just underneath the surface of the RBC membrane.
FIG. 5.
Colocalization studies with anti-peptide 1 antibodies and fixed 3D7 pRBCs (late trophozoites and schizonts). Parasite nuclei were stained with DAPI (blue). Double staining was carried out with MAb 61.2 against a 52-kDa rhoptry protein (red) and anti-peptide 1 serum (green) (A) and with mouse serum (B28) against the 77-kDa carboxy terminus of MC protein PfSBP1 (red) and anti-peptide 1 serum (green) (B to E). Schizonts progressd from binucleate to multinucleate in panels B to E, as determined by staining of nuclear material. Anti-peptide 1 serum was detected by using FITC-conjugated goat anti-rabbit IgG. Colocalizing antibody was detected by using Texas red-conjugated goat anti-mouse IgG. Magnification, ×1,000.
In an earlier study, antibodies against an MC-associated protein, P. falciparum skeletal binding protein 1 (PfSBP-1), showed an IFA pattern very similar to the one that we observed with anti-STEVOR antibodies (8). We carried out a direct comparison of the labeling obtained with an anti-PfSBP-1 mouse polyclonal antibody (B28) and that obtained with the rabbit anti-peptide 1 antibody and found that these antibodies gave indistinguishable punctate staining patterns in mature parasites (schizonts) (Fig. 5C to E). Hence, the merged fluorescence pattern conclusively showed that STEVOR colocalized with PfSBP-1 in pRBCs. While the punctate pattern observed with anti-PfSBP-1 antibodies can be observed in relatively young parasites (late rings or early trophozoites) (8), no evidence of reactivity with anti-STEVOR antibodies was seen with these stages (data not shown). STEVOR expression was first seen in late trophozoites or early schizonts as a diffuse fluorescence pattern (Fig. 5B). In these cells, STEVOR seemed to be located within the parasitophorous vacuole as well as the RBC cytosol. As the parasites matured, STEVOR fluorescence was redistributed to give a more punctate pattern identical to that seen for PfSBP-1 (Fig. 5C), while in fully mature schizonts, the punctate pattern appeared to be predominately associated with the periphery of pRBCs (Fig. 5D). These results suggest that the redistribution of STEVOR, possibly exclusively to MC, has occurred by the time that the parasites have reached the segmenting stage (Fig. 5D and E).
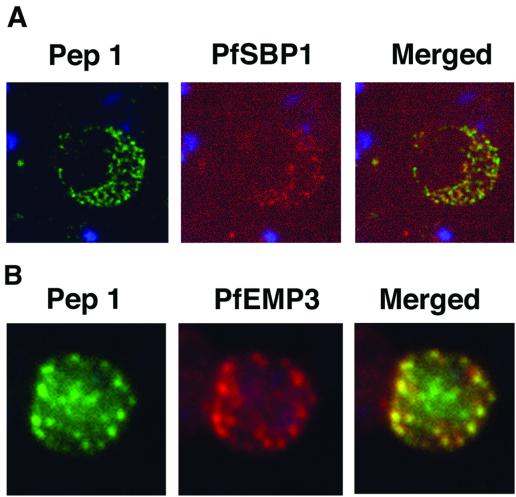
Previous work showed that MC and its associated proteins, including PfSBP-1, remain with the RBC ghost after schizont rupture and merozoite release (8, 22, 30). RBC ghosts were therefore prepared from late-stage schizonts and analyzed by an IFA with both anti-STEVOR and anti-PfSBP-1 antibodies to investigate the colocalization of these two proteins. DAPI staining clearly showed that no parasite nuclei were present in RBC ghosts (Fig. 6A), indicating that no merozoites remained. Nevertheless, the typical STEVOR and PfSBP-1 punctate staining patterns observed in pRBCs were still detected in RBC ghosts (Fig. 6A).
FIG. 6.
Colocalization studies with anti-peptide 1 antibodies and fixed 3D7 pRBC from schizont ghosts and PfEMP3 truncated 3D7 cell lines. (A) Schizont ghosts. (B) PfEMP3 truncated 3D7 cell lines. Parasite nuclei were stained with DAPI (blue). Double staining was carried out with anti-peptide 1 rabbit serum (green) and mouse serum (B28) against the 77-kDa carboxy terminus of MC protein PfSBP1 (red) or mouse serum (DG662) against the carboxy terminus of PfEMP3 (red). Anti-peptide 1 serum was detected by using FITC-conjugated goat anti-rabbit IgG. Colocalizing antibody was detected by using Texas red-conjugated goat anti-mouse IgG. Magnification, ×1,000.
A truncated form of PfEMP-3 obtained by transfection was no longer distributed around the cytoplasmic surface of the erythrocyte membrane but was instead associated with structures believed to be MC (43). IFA experiments with antibodies against PfEMP-3 and STEVOR showed the colocalization of truncated PfEMP-3 and STEVOR (Fig. 6B), further supporting the observation that STEVOR is located in MC.
DISCUSSION
Multigene families coding for variant proteins are a common feature of most, if not all, Plasmodium species. The expression products of various multigene families, var and rif in P. falciparum (3, 17, 27, 38, 39), SICAvar in P. knowlesi (2, 10), vir in P. vivax (15), and py235 in P. yoelii (9, 18, 23, 34), have been linked to one or more parasite functions, including cytoadherence, virulence, antigenic variation, and immune evasion. The possibility exists that STEVOR fulfills one or more similar functions.
Here we have confirmed that stevor is transcribed in asexual parasites in vitro and that this transcription occurs in a tightly regulated fashion. Kyes et al. (26) have shown that both var and rif are transcribed only during the ring and late ring or early trophozoite stages of intraerythrocytic parasite maturation, respectively. We have now shown that stevor transcription and expression are also tightly regulated, occurring after 22 to 28 h of parasite development (mid-trophozoite stage). Thus, var, rif, and stevor appear to be transcribed sequentially in the stated order, and this differential transcription may reflect important functional differences in these gene families and their products during parasite maturation.
We have also shown that stevor is translated and located in MC. Importantly, the timing of expression and unique location of STEVOR indicate a potentially novel role for the stevor multigene family. Only a subset of the total stevor genes are transcribed at any given time in a population of parasites in vitro, and this finding was reflected in individual pRBCs when single trophozoite-infected RBCs were analyzed. There is some evidence that the stevor transcription profile changes over time in a population of parasites (data not shown) and may be similar to what has been observed for var, where switching rates of about 2% have been observed in cultures (36). However, some kind of transcriptional exclusion mechanism seems to be operating, as parasites expressed only defined subsets of stevor at any given time. It is not yet clear from this study whether there are sets of stevor genes that are always expressed together.
Antibodies raised against peptides representative of conserved regions of STEVOR recognized an ∼37-kDa protein in pRBC extracts. The sizes and properties of these proteins were consistent with the expected sizes and membrane binding characteristics of STEVOR (13). In mid-trophozoite to late trophozoite stages, IFAs with anti-STEVOR antibodies showed peripheral punctate, as well as more diffuse, staining associated with the RBC cytosol and/or membrane. The diffuse distribution most likely was indicative of newly expressed STEVOR en route to its final location, since it disappeared during merozoite differentiation to produce a pronounced punctate pattern. The final location of STEVOR, unlike that of PfEMP-1 and the RIFINs, seems not to be the surface of the pRBC but instead MC, membranous structures found in the RBC cytosol at the periphery of the pRBC membrane (1). This finding was confirmed by colocalization studies with PfSBP-1, an MC-associated protein. MC have a potential role in protein export to the RBC membrane (21, 22), and STEVOR also colocalizes with a truncated form of PfEMP-3 whose transport is arrested in these structures (43). Other proteins, such as the knob-associated histidine-rich protein (21, 44), only seem to pass through MC en route to their final destinations. These data contrast with the data shown here, as well as with the results of preliminary biotinylation and iodination experiments (data not shown) suggesting that STEVOR does not appear to be transported beyond the MC to the RBC surface, although the transport of small amounts cannot be totally excluded. Not all of the late-stage trophozoites and schizonts were stained by antibodies to either peptide 1 or 2 in IFAs, and it is not yet clear whether this finding indicates that the antibodies do not recognize all forms of STEVOR or that not all the cells express STEVOR. Since cultured P. falciparum parasites are under no selective pressure from the immune system, it is not surprising that a proportion of the parasites lost their capacity to express STEVOR.
Variant parasite antigens (e.g., PfEMP-1) have been linked to the ability of pRBCs to be sequestered by binding to different host cell ligands. Antigenic variation of PfEMP-1 mediated by var ensures that the parasite is able to evade the variant-specific PfEMP-1 immune response(s) induced in the host. The location and relative lateness of expression of STEVOR are strong indications that it may contribute to parasites some additional as well as different functions compared to PfEMP-1 and RIFINs. There is no evidence to suggest that RIFINs are located in MC, and all evidence to date indicates their surface expression; however, it is possible that RIFINs, like other membrane-associated proteins, are trafficked through MC to the surface of the RBC. Some insight into the role of STEVOR may come from understanding its role within MC. MC have been implicated in regulating protein trafficking to, and in assembly of, the cytoadherence complex at the pRBC surface (21, 44). Components of MC become transiently exposed to the immune system and thus vulnerable to immune attack (22). Interestingly, it has been shown that the accessibility of MC proteins to antibodies occurs in late-stage (schizont) parasites at a time when STEVOR is already present (22). The presence of STEVOR in MC may therefore play a role in shielding essential components of MC when they are exposed to the immune system.
Alternative spliced stevor transcripts have been observed in gametocytes (40), and we have preliminary evidence that they are also expressed in gametocytes (data not shown). Consistent with the alternative splicing pattern, only anti-STEVOR peptide 2 antibodies reacted with gametocyte-expressed protein(s). Whether STEVOR has potentially different functions depending on the stage at which it is expressed needs further investigation. Differential stage-specific expression was recently descibed for the P. yoelii py235 (rhoptry protein) multigene family, which plays a role in the invasion of RBCs by merozoites as well as invasion of hepatocytes by sporozoites (35).
In conclusion, our results show that stevor is fundamentally different from both var and rif in its expression and subcellular location pattern. Moreover, STEVOR is also expressed in gametocytes. The location of STEVOR in MC, believed to be an important site for the sorting and assembly of proteins destined for the surface of infected erythrocytes, indicates a distinct function for the stevor multigene family that requires variability.
Acknowledgments
We thank Muni Grainger and Isabelle Delerieu for providing parasites and Peter Moore for providing PFGE blots. We also thank Catherine Braun-Breton for providing antibody B28 and Ann-Charlotte Grüner for providing antibody DG662. Further thanks are due to Alan Cowman for providing the truncated PfEMP3 parasite line and to R. J. M. Wilson and Georges Snounou for critical reading of the manuscript.
M.K. was supported by a Ph.D. studentship from the Medical Research Council, London, United Kingdom.
REFERENCES
- 1.Aikawa, M. 1988. Morphological changes in erythrocytes induced by malarial parasites. Biol. Cell 64:173-181. [DOI] [PubMed] [Google Scholar]
- 2.al-Khedery, B., J. W. Barnwell, and M. R. Galinski. 1999. Antigenic variation in malaria: a 3′ genomic alteration associated with the expression of a P. knowlesi variant antigen. Mol. Cell 3:131-141. [DOI] [PubMed] [Google Scholar]
- 3.Baruch, D. I., B. L. Pasloske, H. B. Singh, X. Bi, X. C. Ma, M. Feldman, T. F. Taraschi, and R. J. Howard. 1995. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82:77-87. [DOI] [PubMed] [Google Scholar]
- 4.Blackman, M. J., E. D. Dennis, E. M. Hirst, C. H. Kocken, T. J. Scott-Finnigan, and A. W. Thomas. 1996. Plasmodium knowlesi: secondary processing of the malaria merozoite surface protein-1. Exp. Parasitol. 83:229-239. [DOI] [PubMed] [Google Scholar]
- 5.Blackman, M. J., H. Fujioka, W. H. Stafford, M. Sajid, B. Clough, S. L. Fleck, M. Aikawa, M. Grainger, and F. Hackett. 1998. A subtilisin-like protein in secretory organelles of Plasmodium falciparum merozoites. J. Biol. Chem. 273:23398-23409. [DOI] [PubMed] [Google Scholar]
- 6.Blackman, M. J., H. G. Heidrich, S. Donachie, J. S. McBride, and A. A. Holder. 1990. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J. Exp. Med. 172:379-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackman, M. J., T. J. Scott-Finnigan, S. Shai, and A. A. Holder. 1994. Antibodies inhibit the protease-mediated processing of a malaria merozoite surface protein. J. Exp. Med. 180:389-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blisnick, T., M. E. Morales Betoulle, J. C. Barale, P. Uzureau, L. Berry, S. Desroses, H. Fujioka, D. Mattei, and C. Braun Breton. 2000. Pfsbp1, a Maurer's cleft Plasmodium falciparum protein, is associated with the erythrocyte skeleton. Mol. Biochem. Parasitol. 111:107-121. [DOI] [PubMed] [Google Scholar]
- 9.Borre, M. B., C. A. Owen, J. K. Keen, K. A. Sinha, and A. A. Holder. 1995. Multiple genes code for high-molecular-mass rhoptry proteins of Plasmodium yoelii. Mol. Biochem. Parasitol. 70:149-155. [DOI] [PubMed] [Google Scholar]
- 10.Brown, K. N., and I. N. Brown. 1965. Immunity to malaria: antigenic variation in chronic infections of Plasmodium knowlesi. Nature 208:1286-1288. [DOI] [PubMed] [Google Scholar]
- 11.Bull, P. C., B. S. Lowe, M. Kortok, C. S. Molyneux, C. I. Newbold, and K. Marsh. 1998. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat. Med. 4:358-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bull, P. C., and K. Marsh. 2002. The role of antibodies to Plasmodium falciparum-infected-erythrocyte surface antigens in naturally acquired immunity to malaria. Trends Microbiol. 10:55-58. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, Q., N. Cloonan, K. Fischer, J. Thompson, G. Waine, M. Lanzer, and A. Saul. 1998. stevor and rif are Plasmodium falciparum multicopy gene families which potentially encode variant antigens. Mol. Biochem. Parasitol. 97:161-176. [DOI] [PubMed] [Google Scholar]
- 14.Craig, A., and A. Scherf. 2001. Molecules on the surface of the Plasmodium falciparum infected erythrocyte and their role in malaria pathogenesis and immune evasion. Mol. Biochem. Parasitol. 115:129-143. [DOI] [PubMed] [Google Scholar]
- 15.del Portillo, H. A., C. Fernandez-Becerra, S. Bowman, K. Oliver, M. Preuss, C. P. Sanchez, N. K. Schneider, J. M. Villalobos, M. A. Rajandream, D. Harris, L. H. da Silva, B. Barrell, and M. Lanzer. 2001. A superfamily of variant genes encoded in the subtelomeric region of Plasmodium vivax. Nature 410:839-842. [DOI] [PubMed] [Google Scholar]
- 16.Dodge, J. T., C. Mitchell, and D. J. Hanahan. 1963. The preparation and chemical characterisation of haemoglobin-free ghosts of human erythrocytes. Arch. Biochem. Biophys. 100:119-130. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez, V., M. Hommel, Q. Chen, P. Hagblom, and M. Wahlgren. 1999. Small, clonally variant antigens expressed on the surface of the Plasmodium falciparum-infected erythrocyte are encoded by the rif gene family and are the target of human immune responses. J. Exp. Med. 190:1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman, R. R., A. J. Trejdosiewicz, and G. A. Cross. 1980. Protective monoclonal antibodies recognising stage-specific merozoite antigens of a rodent malaria parasite. Nature 284:366-368. [DOI] [PubMed] [Google Scholar]
- 19.Gruner, A. C., K. Brahimi, W. Eling, R. Konings, J. Meis, M. Aikawa, P. Daubersies, C. Guerin-Marchand, S. Mellouk, G. Snounou, and P. Druilhe. 2001. The Plasmodium falciparum knob-associated PfEMP3 antigen is also expressed at pre-erythrocytic stages and induces antibodies which inhibit sporozoite invasion. Mol. Biochem. Parasitol. 112:253-261. [DOI] [PubMed] [Google Scholar]
- 20.Hackett, F., M. Sajid, C. Withers-Martinez, M. Grainger, and M. J. Blackman. 1999. PfSUB-2: a second subtilisin-like protein in Plasmodium falciparum. Mol. Biochem. Parasitol. 103:183-195. [DOI] [PubMed] [Google Scholar]
- 21.Hinterberg, K., A. Scherf, J. Gysin, T. Toyoshima, M. Aikawa, J. C. Mazie, L. P. da Silva, and D. Mattei. 1994. Plasmodium falciparum: the Pf332 antigen is secreted from the parasite by a brefeldin A-dependent pathway and is translocated to the erythrocyte membrane via the Maurer's clefts. Exp. Parasitol. 79:279-291. [DOI] [PubMed] [Google Scholar]
- 22.Hui, G. S., and W. A. Siddiqui. 1988. Characterization of a Plasmodium falciparum polypeptide associated with membrane vesicles in the infected erythrocytes. Mol. Biochem. Parasitol. 29:283-293. [DOI] [PubMed] [Google Scholar]
- 23.Keen, J., A. Holder, J. Playfair, M. Lockyer, and A. Lewis. 1990. Identification of the gene for a Plasmodium yoelii rhoptry protein. Multiple copies in the parasite genome. Mol. Biochem. Parasitol. 42:241-246. [DOI] [PubMed] [Google Scholar]
- 24.Kutner, S., W. V. Breuer, H. Ginsburg, S. B. Aley, and Z. I. Cabantchik. 1985. Characterization of permeation pathways in the plasma membrane of human erythrocytes infected with early stages of Plasmodium falciparum: association with parasite development. J. Cell Physiol. 125:521-527. [DOI] [PubMed] [Google Scholar]
- 25.Kyes, S., P. Horrocks, and C. Newbold. 2001. Antigenic variation at the infected red cell surface in malaria. Annu. Rev. Microbiol. 55:673-707. [DOI] [PubMed] [Google Scholar]
- 26.Kyes, S., R. Pinches, and C. Newbold. 2000. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol. Biochem. Parasitol. 105:311-315. [DOI] [PubMed] [Google Scholar]
- 27.Kyes, S. A., J. A. Rowe, N. Kriek, and C. I. Newbold. 1999. Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 96:9333-9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, C., and J. Langhorne. 2000. Tumor necrosis factor alpha p55 receptor is important for development of memory responses to blood-stage malaria infection. Infect. Immun. 68:5724-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Limpaiboon, T., M. W. Shirley, D. J. Kemp, and A. Saul. 1991. 7H8/6, a multicopy DNA probe for distinguishing isolates of Plasmodium falciparum. Mol. Biochem. Parasitol. 47:197-206. [DOI] [PubMed] [Google Scholar]
- 30.Martinez, S. L., C. A. Clavijo, and E. Winograd. 1998. Identification of peripheral membrane proteins associated with the tubo-vesicular network of Plasmodium falciparum infected erythrocytes. Mol. Biochem. Parasitol. 91:273-280. [DOI] [PubMed] [Google Scholar]
- 31.Owen, C. A., K. A. Sinha, J. K. Keen, S. A. Ogun, and A. A. Holder. 1999. Chromosomal organisation of a gene family encoding rhoptry proteins in Plasmodium yoelii. Mol. Biochem. Parasitol. 99:183-192. [DOI] [PubMed] [Google Scholar]
- 32.Pachebat, J. A., I. T. Ling, M. Grainger, C. Trucco, S. Howell, D. Fernandez-Reyes, R. Gunaratne, and A. A. Holder. 2001. The 22 kDa component of the protein complex on the surface of Plasmodium falciparum merozoites is derived from a larger precursor, merozoite surface protein 7. Mol. Biochem. Parasitol. 117:83-89. [DOI] [PubMed] [Google Scholar]
- 33.Preiser, P. R., and W. Jarra. 1998. Plasmodium yoelii: differences in the transcription of the 235-kDa rhoptry protein multigene family in lethal and nonlethal lines. Exp. Parasitol. 89:50-57. [DOI] [PubMed] [Google Scholar]
- 34.Preiser, P. R., W. Jarra, T. Capiod, and G. Snounou. 1999. A rhoptry-protein-associated mechanism of clonal phenotypic variation in rodent malaria. Nature 398:618-622. [DOI] [PubMed] [Google Scholar]
- 35.Preiser, P. R., S. Khan, F. T. Costa, W. Jarra, E. Belnoue, S. Ogun, A. A. Holder, T. Voza, I. Landau, G. Snounou, and L. Renia. 2002. Stage-specific transcription of distinct repertoires of a multigene family during Plasmodium life cycle. Science 295:342-345. [DOI] [PubMed] [Google Scholar]
- 36.Roberts, D. J., A. G. Craig, A. R. Berendt, R. Pinches, G. Nash, K. Marsh, and C. I. Newbold. 1992. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature 357:689-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shulman, M., C. D. Wilde, and G. Kohler. 1978. A better cell line for making hybridomas secreting specific antibodies. Nature 276:269-270. [DOI] [PubMed] [Google Scholar]
- 38.Smith, J. D., C. E. Chitnis, A. G. Craig, D. J. Roberts, D. E. Hudson-Taylor, D. S. Peterson, R. Pinches, C. I. Newbold, and L. H. Miller. 1995. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82:101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su, X. Z., V. M. Heatwole, S. P. Wertheimer, F. Guinet, J. A. Herrfeldt, D. S. Peterson, J. A. Ravetch, and T. E. Wellems. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82:89-100. [DOI] [PubMed] [Google Scholar]
- 40.Sutherland, C. J. 2001. stevor transcripts from Plasmodium falciparum gametocytes encode truncated polypeptides. Mol. Biochem. Parasitol. 113:331-335. [DOI] [PubMed] [Google Scholar]
- 41.Trager, W. 1977. Cultivation of Plasmodium falciparum. Arch. Pathol. Lab. Med. 101:277-278. [PubMed] [Google Scholar]
- 42.Vollrath, D., and R. W. Davis. 1987. Resolution of DNA molecules greater than 5 megabases by contour-clamped homogeneous electric fields. Nucleic Acids Res. 15:7865-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterkeyn, J. G., M. E. Wickham, K. M. Davern, B. M. Cooke, R. L. Coppel, J. C. Reeder, J. G. Culvenor, R. F. Waller, and A. F. Cowman. 2000. Targeted mutagenesis of Plasmodium falciparum erythrocyte membrane protein 3 (PfEMP3) disrupts cytoadherence of malaria-infected red blood cells. EMBO J. 19:2813-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickham, M. E., M. Rug, S. A. Ralph, N. Klonis, G. I. McFadden, L. Tilley, and A. F. Cowman. 2001. Trafficking and assembly of the cytoadherence complex in Plasmodium falciparum-infected human erythrocytes. EMBO J. 20:5636-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]