Abstract
The trypanosome variant surface glycoprotein (VSG) is first expressed during differentiation to the infective, metacyclic population in tsetse fly salivary glands. Unlike the VSG genes expressed by bloodstream form trypanosomes, metacyclic VSGs (MVSGs) have their own promoters. The scarcity of metacyclic cells has meant that only indirect approaches have been used to study these promoters, and not even their identities have been agreed on. Here, we isolated trypanosomes by dissection from salivary glands and used an approach involving 5′ rapid amplification of cDNA ends to identify the transcription start site of three MVSGs. This shows that the authentic start site is that proposed for the MVAT series of MVSGs (K. S. Kim and J. E. Donelson, J. Biol. Chem. 272:24637-24645, 1997). In the more readily accessible procyclic trypanosome stage, where MVSGs are normally silent, we used reporter gene assays and linker scanning analysis to confirm that the 67 bp upstream of the start site is a promoter. This is confirmed further by accurate initiation in a homologous in vitro transcription system. We show also that MVSG promoters become derepressed when tested outwith their endogenous, subtelomeric loci. The MVSG promoters are only loosely conserved with bloodstream VSG promoters, and our detailed analysis of the 1.63 MVSG promoter reveals that its activity depends on the start site itself and sequences 26 to 49 bp and 56 to 60 bp upstream. These are longer than those necessary for the bloodstream promoter.
The protozoan parasite Trypanosoma brucei is transmitted between different mammalian hosts by its tsetse fly (Glossina morsitans) vector. Within mammals, T. brucei replicates extracellularly, and its cell surface is enshrouded by approximately 107 copies of a single protein species, the variant surface glycoprotein (VSG). Periodic switching from the expression of one VSG gene to another—antigenic variation—allows the parasite population to avoid eradication by the host immune system (5). The VSG coat is not expressed by the trypanosome stages that proliferate in the tsetse fly. However, the coat is probably a necessary preadaptation for infection of the mammalian host, and it is synthesized during differentiation of infective metacyclic trypanosomes within tsetse fly salivary glands (35). At this point in the life cycle, the expressed VSGs are regulated differently from those used in the bloodstream.
In the mammalian host, VSGs are transcribed from specialized transcription units known as bloodstream expression sites (BESs). Estimates suggest that up to 30 BESs exist in each trypanosome (27, 39), although only one is transcribed completely at any time. BESs occur within subtelomeric loci organized such that the VSG is the final coding sequence on the chromosome, upstream of the telomere tract repeats. Several expression site-associated genes, or ESAGs, are transcribed together with the VSG, and each transcription unit is under the control of a single promoter, usually located 45 to 60 kb upstream of the VSG gene (reviewed in reference 5). Transcription is resistant to α-amanitin, suggesting the use of RNA polymerase I or a modified RNA polymerase II (23, 24). In the procyclic trypanosomes in the tsetse fly midgut, VSGs are not expressed, and transcription of BESs is down-regulated. Although many or all of the BES promoters remain active at a low level, transcription does not proceed throughout the length of the expression site (32, 39). Metacyclic VSG gene (MVSG) expression is unusual in two respects that have relevance to a basic understanding of gene expression within the early-diverging kinetoplastid lineage. First, MVSG expression sites (MESs) are the only clear cases of monocistronic transcription units for protein-coding genes in a trypanosomatid (1, 11). Second, the stage-specific regulation of MVSG expression is an example of absolute regulation at the level of transcription initiation (11, 29), which is unusual in a family of organisms that utilizes posttranscriptional control processes to regulate the expression of housekeeping genes (38).
Up to 27 MVSG coats are expressed by the metacyclic trypanosome population (36). During the differentiation from the epimastigote stage to the metacyclic stage, each cell makes a decision to initiate, at random, the transcription of just one of the MVSG genes that are available (35). This gene is then expressed throughout metacyclic development and for several days following infection of a mammalian host. Thereafter, in a process that is not understood, the MES is silenced and the transcription of VSGs from BESs occurs. MVSG gene promoters are also transcriptionally silent during the procyclic stage and under normal circumstances can be reactivated only by differentiation to the metacyclic form in the salivary glands of the tsetse fly. The random nature of MVSG activation during metacyclic development ensures heterogeneity with respect to the VSG coats expressed by the parasite population and, as argued previously, is likely to enhance the probability of reestablishing infection in a previously infected host (3, 4). Like BESs, MESs are telomeric, with no coding sequences between the MVSG gene and the telomeric tract, and their transcription is resistant to α-amanitin (5).
Although data on the repression of MVSG expression in bloodstream and procyclic-form trypanosomes and on the stochastic nature of differential activation in the metacyclic form are available (11, 29, 35), we do not know how individual promoters function to recruit RNA polymerase, nor do we know anything about the molecular processes that mediate stage-specific regulation. The obvious difficulties in working with a nondividing cell type that cannot be obtained through existing in vitro culture techniques and the need to transmit trypanosomes through tsetse flies have meant that only indirect approaches have been possible. After repeated serial passaging of trypanosomes through laboratory mice Donelson and coworkers were able to identify low numbers of bloodstream form cells that had activated MVSGs de novo. The transcription of two of the MVSGs was monocistronic (1, 20), and one population had transcriptionally activated a MES without detectable genomic rearrangements (1). Transcription start sites for these MVAT4 and MVAT7 genes were mapped to elements referred to as putative MVAT promoters that varied between 67 and 73 bp in length and that were able to drive reporter gene expression from plasmids in transient-transfection experiments with procyclic culture form parasites (1, 20). A similar element was isolated in a promoter trap conducted in procyclic trypanosomes (25). In a different approach, using 5- to 7-day-old trypanosome populations that were expanded in mice from a single metacyclic trypanosome and therefore that were still transcribing a MES within the natural context of the life cycle, Graham et al. (11, 13) reported transcription start sites for the 1.22 and 1.61 MVSG genes. Although partly similar to each other, these sites differed from those for the MVAT4 and MVAT7 genes. Nevertheless, an upstream segment of the 1.22 MVSG element resembled the MVAT promoter-like element and was able to drive reporter gene expression in bloodstream form trypanosomes but was inactive when tested in procyclic cells (11, 13-15). The conclusions from the papers by Graham et al. therefore indicated a promoter structure that was an alternative to that of the putative MVAT promoters studied by Donelson and coworkers and a complex pattern of transcriptional regulation that involved chromosomal context and the presence or absence of trans-acting factors in different life cycle stages (14, 15). In the present work, we resolve the discrepancies between the previously published work from our and other laboratories through direct examination of transcription of MVSGs in trypanosomes isolated from the salivary glands of infected tsetse flies. We also demonstrate for the first time accurate in vitro initiation of transcription of these putative core promoters by using procyclic extracts, validating a detailed mutational analysis of the core promoter in a life cycle stage that is more amenable to experimental study.
MATERIALS AND METHODS
Maintenance of trypanosomes.
T. brucei EATRO 795 parasites that maintain tsetse fly transmissibility (12) were used for all experiments except the transient transfection of bloodstream form trypanosomes, which used an ILTat 1.2 subclone of EATRO 795. Procyclic trypanosomes were obtained by differentiation from bloodstream trypanosomes by standard methods (10) and were maintained in SDM-79 medium supplemented with 10% (vol/vol) heat-inactivated fetal calf serum as described previously (9). Transmission of bloodstream form cells through tsetse flies was also carried out as described previously (12).
Plasmid constructs.
Constructs used in transient transfections were all originally derived from pHD52 (19). The substitution of the luciferase gene with a chloramphenicol acetyltransferase (CAT) reporter gene and the subsequent removal of the BES promoter to create p-HD52CAT or replacement of the BES promoter with a sequence from the 1.22 MVSG locus to create p122sHD52CAT have been described previously (11). To create p1.61nfHD52CAT and p1.61nrHD52CAT, plasmid PMT1.61-X6 (13) was digested with NaeI and a 2.7-kb insert containing the transcription start site was cloned into pHD52CAT that had been digested with Acc65I and SmaI and then blunt ended. p1.61nfHD52CAT contains the NaeI insert with a start site and promoter cloned in the forward orientation with respect to the CAT gene; p1.61nrHD52CAT contains the promoter orientated in the opposite direction to the CAT open reading frame. A smaller insert from within the 2.7-kb NaeI fragment, also containing the putative metacyclic promoter element, was isolated after digestion of pMT1.61-X6 with EcoRV and HindIII and cloned into EcoRV- and HindIII-digested pBluescript. This insert was then released from pBluescript by digestion with HindIII and SacI, the 5′ overhang was blunted, and the insert was cloned into SacI- and SmaI-digested pHD52CAT to yield p1.61ehHD52CAT. All of the transient-transfection plasmids that contained the 1.63 MVAT-type promoter and mutants thereof were obtained by annealing the appropriate oligonucleotides to obtain double-stranded DNA fragments with SacI and SmaI ends and cloning these DNA molecules into SacI- and SmaI-digested pHD52CAT. The basic constructs for the analysis of promoter activities in the 1.22 MVSG subtelomeric locus and the nontranscribed ribosomal DNA (rDNA) locus (41) were built through a series of cloning steps, the full details of which are available upon request. The reporter gene used to assay promoter activity was a fusion of the green fluorescent protein (GFP) gene to a truncated form of the trypanosome paraflagellar rod protein A gene (PFRA) (6). This translational fusion was chosen because, in our experience, the expression of native GFP in trypanosomes is less consistent. The selectable marker subcassette, which was designed to operate independently of the test subcassette, consisted of a procyclin promoter and a phleomycin resistance gene flanked by procyclin-processing regions. The reporter subcassette consisted of the test promoter fragment and the GFP gene-PFRAΔ reporter gene flanked at its 5′ end by a β/α tubulin intergenic region and at its 3′ end by an intergenic region from the calmodulin locus. The test promoters analyzed were the rDNA promoter, the 221 BES promoter, the 1.61 MVAT-type promoter, and the 1.2-kb EcoRV-HindIII insert from pMT1.61-X6. p1.63-trm, p1.61-trm, and 1.61 M-trm, used to test the ability of MVAT-type promoters to transcribe DNA templates in vitro, were all derived from pRib-trm (23). Again, appropriate oligonucleotides were annealed together, and KpnI/SmaI DNA inserts were cloned into KpnI- and SmaI-digested pRib-trm. All constructs were prepared by Qiagen Maxi-prep or through CsCl density gradients prior to use.
Recombinant DNA procedures.
Trypanosome genomic DNA was extracted as described previously (7). The ILTat 1.63 MVSG expression site was identified by using a partial cDNA sequence for the 1.63 VSG to screen an ILTat 1.2 genomic library constructed (by Nils Burman) in bacteriophage lambdaGEM12 (Promega). Library screening and Southern blotting were carried out according to standard protocols (33). DNA probes were labeled with [α-32P]dCTP by random priming using the Prime-It II kit (Stratagene), and all blots were washed to high stringency (0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% [wt/vol] sodium dodecyl sulfate; 65°C). A partial sequence of the ILTat 1.64 MES was isolated by PCR using a 1.64 cDNA-specific primer (5′GTAACAGTCAGTGCAGCGAATC3′) and a degenerate primer (5′AATIYRIKYAAIRCRRGGTTC3′). The 50-μl reaction mixture consisted of Herculase polymerase (Stratagene), 125 ng of a cDNA-specific primer and 1 μmol of degenerate primer, and 0.2 mM deoxynucleoside triphosphates. The reaction involved 30 cycles of 94°C for 1 min, 48°C for 1 min, and 72°C for 5 min, with a final extension of 72°C for 12 min. From this reaction a DNA fragment containing the complete sequence of the 1.64 MVSG promoter region was then isolated by a TOPO-Walker (Invitrogen) approach following the manufacturer's guidelines and using the locus-specific primer 5′CGTGTGTAAACCCTGC3′, in conjunction with SspI-digested DNA and Taq polymerase (Abgene). Finally, to obtain an accurate sequence for the promoter region, DNA fragments were amplified from genomic DNA by high-fidelity PCR using Herculase in conjunction with the primer pair 5′CTAATCGCTGGATACCGCCATC3′ and 5′CTACTCGTAGCATTGC3′. The annealing temperature was 60°C. DNA sequences were determined by ABI-PRISM automated sequencing with custom-designed primers.
5′ RACE analysis of MVSG transcription start sites during metacyclic development.
Salivary glands were collected from tsetse flies approximately 35 days after infection with bloodstream form trypanosomes. Glands were massaged under the microscope to release a maximum of 2 × 105 parasites into a small volume of HMI-9 medium (16), and the RNA was extracted with 1 ml of TRIzol (Gibco Life Technologies). After division into aliquots (approximately 50,000 cell equivalents), the RNA was used in rapid amplification of cDNA ends (RACE) reactions, including reverse transcriptase (RT)-negative controls, to map 5′ cDNA ends; the procedures strictly adhered to the manufacturer's protocols (Gibco Life Technologies). Following purification and oligo(dC) tailing of gene-specific cDNA, two rounds of PCR, in which the second reaction was nested with respect to the first, were necessary for DNA bands to be seen by ethidium bromide staining. PCR products were cloned into TOPO vectors (Invitrogen) and sequenced as described above. The 3′ end positions of the primers used to map the transcription start sites in the 1.63 and 1.61 loci, relative to MVSG promoters, are shown in Table 1; the primer sequences are available upon request. All primers were tested for their integrity by use in PCRs with upstream primers to amplify specific DNA targets from genomic DNA templates.
TABLE 1.
5′ RACE analysis of transcription start sites in salivary gland trypanosomes
| MVSG locus | Expt | Primer cocktaila | PCR product size (bp) | No. of PCR clones analyzed | 5′ ends mappedb |
|---|---|---|---|---|---|
| 1.63 | 1 | +291, +207, +114 | ∼140 | 12 | 5 at +1 or −1, 2 at +2, 5 trans-spliced at +53 |
| −64, −86, −101 | None | ||||
| 2 | +291, +207, +114 | ∼140 | 10 | 8 at +49, 1 at +48, 1 at +50 | |
| −64, −86, −101 | None | ||||
| +291, +207, +42 | None | ||||
| +291, +207, −13 | None | ||||
| 3 | +291, +207, +114 | ∼140 | 10 | 5 at +2, 2 at +3, 1 at +6, 2 at +10 | |
| −64, −86, −101 | ∼700 | 3 | ingi | ||
| 1.61 | 1 | +1141, +958, +855 | ∼250 | 7 | 7 trans-spliced at +641 |
| +1141, +958, +637 | ∼90 | 9 | 6 at +560, 2 at +581, 1 at +615 | ||
| 2 | +1141, +958, +855 | ∼250 | 6 | 6 trans-spliced at +640 | |
| +1141, +958, +637 | ∼90 | 17 | 1 at +561, 1 at +562, 15 at +565 | ||
| 3 | +534, +408, +395 | ∼420 | 8 | 8 at +1 | |
| +191, +105, +78 | ∼100 | 8 | 7 at +1, 1 at +2 | ||
| −41, −92, −101 | None |
The position of the 3′ end of each locus-specific oligonucleotide, relative to the in vitro transcription start site, is indicated.
Relative to the in vitro transcription start site.
Transfections.
For transient transfections, 2.5 × 107 procyclic trypanosomes in 0.5 ml of Zimmerman postfusion (ZM) medium (132 mM NaCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, 0.5 mM Mg acetate, 0.09 mM Ca acetate, pH 7.0) were pulsed twice with 5 μg of supercoiled plasmid by using a Bio-Rad Gene Pulser II set at 1.5 kV and 25 μF capacitance. Cells were allowed to recover in 10 ml of medium for 18 h before harvesting and processing for the determination of CAT activity as described previously (11). CAT activity was determined on the linear scale by using as the substrate [14C]chloramphenicol at a specific activity of 52 to 57 mCi/mmol (NEN; catalog no. NEN408). Butyrylated products were extracted into mixed xylenes and processed for liquid scintillation counting. Bloodstream form cells were recovered from Wistar rats as a buffy coat from whole blood and resuspended in HMI-9 medium. Cells (5 × 107) in 0.5 ml of ZM buffer containing 1% (wt/vol) glucose were pulsed once with 5 μg of DNA under the conditions described above, recovered in 10 ml of HMI-9 medium for 18 h at 37°C in an atmosphere containing 5% CO2, and then harvested and processed for the determination of CAT activity as described above. For stable transfections, 2.5 × 107 procyclic cells in 0.5 ml of ZM buffer were pulsed twice with ∼3 μg of linearized DNA under the conditions described above and then recovered overnight in SDM-79 medium and selected with phleomycin (5 μg ml−1) by following methods described previously (40).
Fluorescence-activated cell sorter analysis.
Parasites were resuspended in phosphate-buffered saline at a density of between 106 and 107 cells · ml−1. Then, 106 events were analyzed on a FACSCalibur (Becton Dickinson, Oxford, United Kingdom) flow cytometer. The laser beam (15 mW) was tuned to 488 nm, and the FL1 photomultiplier tube (equipped with a 530/30-nm band-pass filter) was used to record GFP fluorescence (emission maximum, 507 nm). Data analysis was carried out with CellQuest software (Becton Dickinson).
In vitro transcription and RNA analysis.
In vitro transcription, RNA preparation, and detection of specific transcripts by primer extension analysis were conducted essentially as described previously (23). Briefly, 40-μl transcription reaction mixtures were incubated for 60 min at 28°C and contained 8 μl of cell extract (23), 20 mM potassium l-glutamate, 20 mM KCl, 3 mM MgCl2, 20 mM HEPES-KOH, pH 7.7, 0.5 mM ATP, 0.5 mM GTP, 0.5 mM CTP, 0.5 mM UTP, 20 mM creatine phosphate, 0.48 mg of creatine kinase ml−1, 2.5% polyethylene glycol, 0.2 mM EDTA, 0.5 mM EGTA, 4.25 mM dithiothreitol, 10 μg of leupeptin ml−1, and template DNA. MES promoter constructs and the control template pSLins19 were added to a final concentration of 7.5 μg · ml−1 each. RNA was prepared by the single-step guanidinium thiocyanate method, and in vitro-synthesized transcripts were specifically detected by primer extension with 32P-end-labeled oligonucleotides and RT Superscript II (Invitrogen) according to the manufacturer's protocol. Transcription signals were separated on 6% polyacrylamide-50% urea gels and visualized by autoradiography.
Nucleotide sequence accession numbers.
GenBank accession numbers of sequences obtained during this study are AJ486954 and AJ486955.
RESULTS
The 1.63 and 1.64 MVSG expression sites.
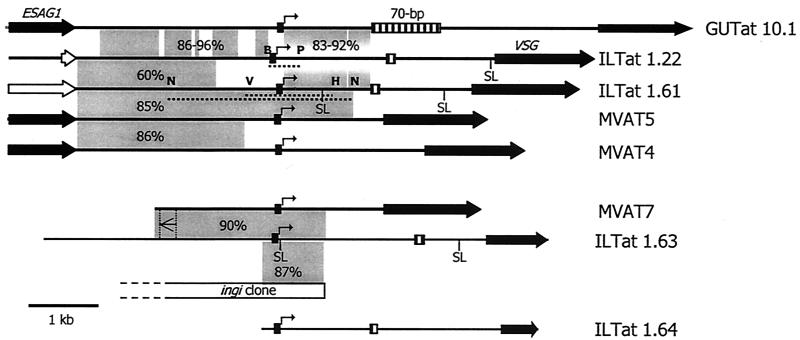
To extend our knowledge of the structure and function of MVSG expression sites and their promoters, we have now analyzed the telomeric 1.63 and 1.64 loci. Using a cDNA probe, we isolated from a genomic library an insert that included 892 bp of the N-terminal coding region for 1.63 MVSG and 16.8 kb of upstream sequence. Sequence analysis indicated that the 1.63 locus shares a number of characteristics with previously characterized MVSG loci, and in particular with the MVAT7 locus studied by Donelson and coworkers (Fig. 1) (20, 34). From bp −854 to −1054 upstream of the ATG initiation codon there is homology to the 70-bp repeats that are either not present or are present in 1 or 2 copies in some MESs and that are present as large repeat arrays in BESs (5). From bp −2335 to −4885 there is a partial copy of the ingi retrotransposon, and, within this region (bp −2983 to −3055), there is also a sequence similar to those of the minimal MVAT promoters. The presence of the MVAT-like promoter element within the ingi flank sequence constitutes an arrangement identical to the arrangement of the putative metacyclic promoter within the MVAT7 locus (20), and indeed, in sequence comparisons with MVAT promoters, this 1.63 MVAT promoter-like sequence is most similar to that of the MVAT7 promoter (92% identity).
FIG. 1.
Structure of the ILTat 1.63 and ILTat 1.64 MVSG loci. Eight characterized MVSG loci are shown. Solid arrows, genes; open arrows, pseudogenes; hatched box, 70-bp repeat that lies upstream of most VSGs; bent arrows, promoters or untested putative promoters resembling them. The relatedness between regions of different loci is shown by grey boxes that depict percent identities between the sequences aligned. GUTat 10.1 is compared with ILTat 1.61 downstream of the promoter. An inverted segment of homology between MVAT7 and ILTat 1.63 is shown. SL, position of a trans-splicing site detected in the present work; dotted lines, test promoter fragments. Restriction sites: B, BamHI; H, HindIII; N, NaeI; P, PstI; V, EcoRV. The loci not described in this paper are in the following references: GUTat 10.1, 21; ILTat 1.22, 3 (accession no. AJ012198); ILTat 1.61, 13 (accession no. AJ012199); MVAT5, 26; MVAT4, 1; MVAT7, 20.
Using PCR in combination with an oligonucleotide specific for the ILTat 1.64 cDNA and a degenerate PCR primer designed to anneal within the most highly conserved region of the consensus MVSG promoter, we were able to amplify an ∼3.2-kb fragment, which was cloned and sequenced. To ascertain an accurate sequence of the putative 1.64 MVSG promoter, we used a TOPO-walking approach to amplify the DNA fragment and locus-specific primers in conjunction with a high-fidelity PCR system to amplify and sequence a putative 1.64 MVSG promoter. Although not yet experimentally tested, this putative promoter contains the functional sequence elements identified in the 1.63 MVSG promoter. The 1.64 MES has only a single, poorly conserved 70-bp repeat element.
MVSG transcription start sites used during metacyclic development.
To study MVSG transcription initiation sites directly and to help resolve conflicting data from two indirect approaches to the identification of MVSG promoters (1, 11), we isolated sufficient amounts of RNA from trypanosomes dissected from tsetse fly salivary glands for transcription start site analysis by 5′ RACE. Although a new generation of 5′ RACE kits that exploit the 5′ cap of transcripts can overcome the problem of premature termination of reverse transcription, the absence of caps from trypanosome primary transcripts forced us to use instead a walking strategy in which priming moves progressively upstream until the point beyond which no products are detectable. Thus, in our first experiments, using gene-specific primers that were complementary to the genomic sequence upstream of the 1.63 MVSG, we repeatedly mapped 5′ cDNA ends to the MVAT promoter-like sequence present in this locus (Table 1). Moreover, in two of the three experiments, the transcription start site was mapped at, or very near to, the CA+1 motif, which characterizes the transcription start sites of BES promoters and the MVAT4 and MVAT7 promoters in bloodstream cells expressing MVSG genes as monocistronic transcripts (1, 20). Using gene-specific primers in 5′ RACE upstream of this transcription start site, we detected no RT-PCR products in two experiments; the transcripts detected in the third transmission (Table 1) could have been derived from the transcription of ingi elements found elsewhere in the genome, as that is what lies immediately upstream of the 1.63 MVSG promoter-like element. As expected for loci that are subjected to transcriptional silencing in the procyclic stage of the life cycle (11, 29), we detected, from procyclic cDNA, no RT-PCR products from the 1.63 MES using either the locus-specific primers or other primer combinations (data not shown). This 1.63 MVSG transcription start site is different from that of the putative 1.22 MVSG and hypothetical 1.61 MVSG promoters identified in a different approach (11, 13). We therefore applied our 5′ RACE walking strategy to identify the transcription start sites of the 1.22 MVSG and 1.61 MVSG in salivary gland trypanosome populations. This indicated that transcription of both genes was initiated from CA+1 motifs present in sequences having identity with those of MVAT promoters lying upstream of the putative elements we reported previously (Table 1 and data not shown). To resolve this further, we walked by 5′ RACE upstream of the 1.61 MVSG gene in 5-day-old bloodstream parasites initiated from metacyclic trypanosomes, which is the indirect approach we used previously (13), and this gave the same CA+1 initiation site for the 1.61 MVSG gene (data not shown). Taken as a whole, our analysis of three MESs indicated that monocistronic transcription of VSG genes in metacyclic populations utilizes MVAT-type promoters but provided no evidence for transcription initiation from common downstream elements.
In our 5′ RACE analyses, we found trans-splice acceptor sites located 51 nucleotides (nt) and 641 nt downstream of the transcription start sites for the 1.63 and 1.61 MVSG genes, respectively (Table 1; Fig. 1). To investigate whether these spliced leader sites were used to form mature MVSG mRNA species with unusually long 5′ untranslated regions (UTR) of 2,936 nt in the 1.63 MVSG mRNA and 2,138 nt in the 1.61 MVSG mRNA, we applied RT-PCRs using metacyclic RNA templates in conjunction with VSG- and spliced-leader-RNA-specific oligonucleotides and determined the lengths of specific products by gel electrophoresis. The PCR products obtained from these reactions indicated that the 5′ UTRs of the 1.61 and 1.63 MVSG genes are only about 400 nt long (data not shown); the observed splicing events at 51 nt and 641 nt from the transcription start do not yield the mature mRNA species.
Locus-associated control prevents MES transcription in procyclic stage trypanosomes.
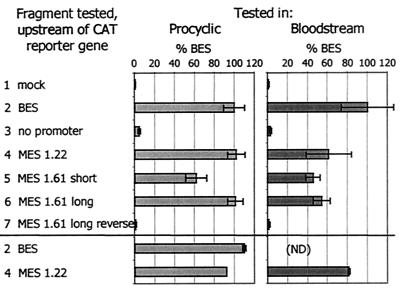
Given our identification of a consensus MVSG transcription initiation site, we have reinvestigated what controls promoter activity. Our previous data suggested that stage-specific silencing of MVSGs in bloodstream form cells is dependent on chromosomal context but also that procyclic cells appeared to be unable to activate promoters, even when they are on plasmids or in the rDNA locus (11, 13, 14). To find out whether MVSG promoters are able to direct transcription on episomal plasmids, we transiently transfected bloodstream form and procyclic-form trypanosomes with gene constructs in which previously examined promoter fragments (Fig. 1) were cloned upstream of a CAT reporter gene; DNA fragments included the published 1.22 promoter short fragment (427 bp) (11, 14, 15) a 2.7-kb sequence from the 1.61 locus encompassing an MVAT element (13), a shorter fragment from the 1.61 locus also containing the 1.61 MVAT element, and appropriate control inserts, namely, one without the promoter and one with the promoter in reverse orientation. The CAT activity measured in extracts of transfected trypanosomes indicated clearly that, contrary to data published previously by Graham et al. (11, 13-15), there were no significant differences between the two life cycle stages in their abilities to use sequences encompassing MVAT promoters to drive gene expression (Fig. 2).
FIG. 2.
Sequences encompassing MVSG transcription start sites drive reporter gene expression from plasmids in different life cycle stages. Each fragment was tested for the ability to drive expression of the CAT reporter gene. Data are means ± standard deviations of at least three experiments, expressed against the BES promoter activity (100%). The MES fragments tested are shown in Fig. 1; the “short” and “long” fragments tested were, respectively, 1.61eh and 1.61n. Constructs containing the BES or 1.22 MES promoter upstream of the CAT gene were retested at the end of each transfection series to determine any effect of the time trypanosomes were maintained in transfection buffer during the experiment; there was no such effect. ND, not determined.
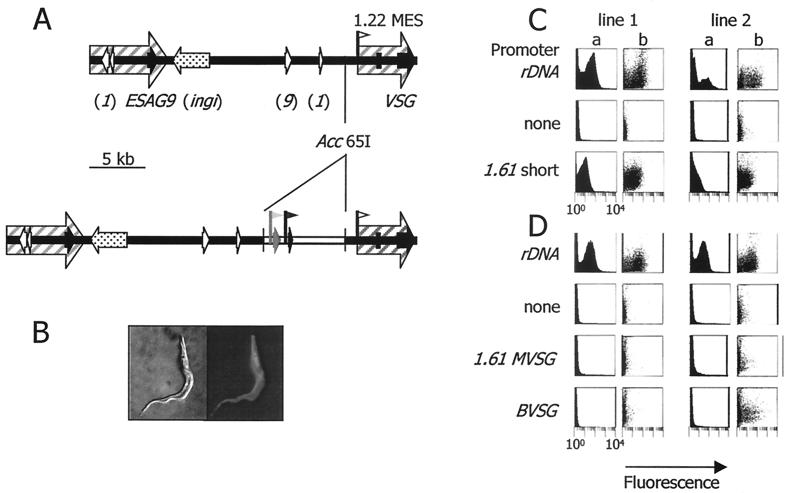
We next asked two questions: whether the transcriptionally silent region upstream of the 1.22 MVSG transcription start site could provide a suitable environment for gene expression from different trypanosome promoters and whether a consensus MVSG promoter would drive gene expression from the nontranscribed spacer in the rDNA array. For this we used stable transfection and a GFP gene fusion as the reporter, which would enable detection to the single-cell level, such as in the tsetse fly. For the 1.22 locus, the reporter cassette was inserted into an Acc65I site, 1,173 bp upstream of the endogenous 1.22 MVSG promoter; taking into account the 2.9-kb pBluescript spacer and other sequences in the cassette, this meant that the test promoter was located about 7 kb upstream of the endogenous promoter. The 1.2-kb EcoRI-HindIII fragment encompassing the 1.61 MVSG promoter was able to drive gene expression from the nontranscribed spacer in the ribosomal array, albeit at a 5-fold-lower level than the ribosomal promoter integrated at the same location (Fig. 3). Assuming that the degree of green fluorescence within the culture correlates with promoter activity, the activity of the 1.61 MVSG promoter fragment was approximately 20% of the activity seen with a ribosomal promoter integrated at this location. This value falls within the range of activities described for a BES promoter from the rDNA array (from 5% [18] to >100% of the activity of a ribosomal promoter [8]). Our results therefore indicate that, within the 300 bp upstream or the 900 bp downstream of the MVSG transcription initiation site, there is no negative regulatory element causing promoter silencing from chromatin at this stage of the life cycle. On the other hand, only the ribosomal promoter was able to direct transcription within the 1.22 telomere. Both the BES promoter and the 1.61 MVSG promoter were inactive at this gene locus, although both were active when assayed away from it, showing construct integrity. The data are in agreement with the hypothesis that a locus-specific control can mediate transcriptional silencing of MVSG telomeres (15). However, while this control also represses the activity of a BES promoter, it is unable to regulate the activity of a ribosomal promoter integrated into transcriptionally silent chromatin in the 1.22 telomere.
FIG. 3.
Locus-associated control tested in procyclic cells. (A) The 1.22 MVSG locus before and after integration of the reporter construct used to test for promoter activation. In the upper map the region not enclosed by the hatched arrows represents chromatin that is transcriptionally silent in nuclear run-on assays (3, 11). Speckled arrow, partial copy of the ingi retrotransposon. Other features are marked as described in the legend to Fig. 1. In the lower map, the reporter cassette is depicted as inserted at the Acc65I site, 1,173 bp upstream of the MVSG transcription start site. In the cassette, the selectable drug marker subcassette is shown in black. The reporter subcassette (grey) consists of the test promoter fragment and the GFP-PFRAΔ reporter gene. The same reporter-selectable marker system was used also to target the nontranscribed spacer of the rDNA locus (41). (B) Fluorescent trypanosome expressing the GFP fusion reporter protein. The whole cell fluoresces. (C) Promoters were tested for ability to drive reporter gene expression in the rDNA locus. Data are FACScan fluorescence (x axes; arbitrary units on a logarithmic scale from 1 to 10,000) versus number of events (columns a) or size scatter (columns b) on the y axes. Two independent lines were tested for each construct. (D) Same as for panel C except that promoters were tested at the 1.22 MVSG locus.
In vitro and in vivo dissection of a core 1.63 MVSG promoter activity in procyclic cells.
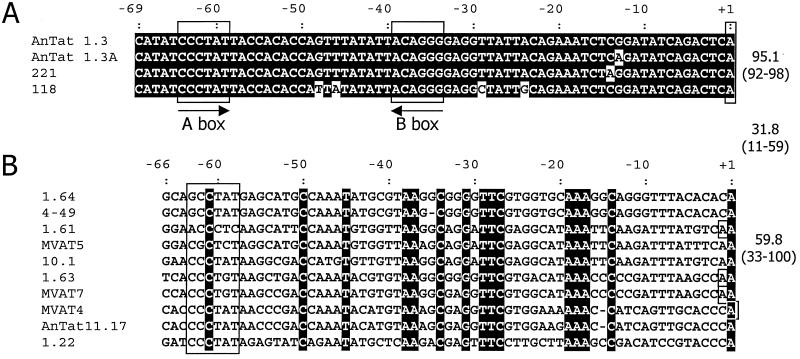
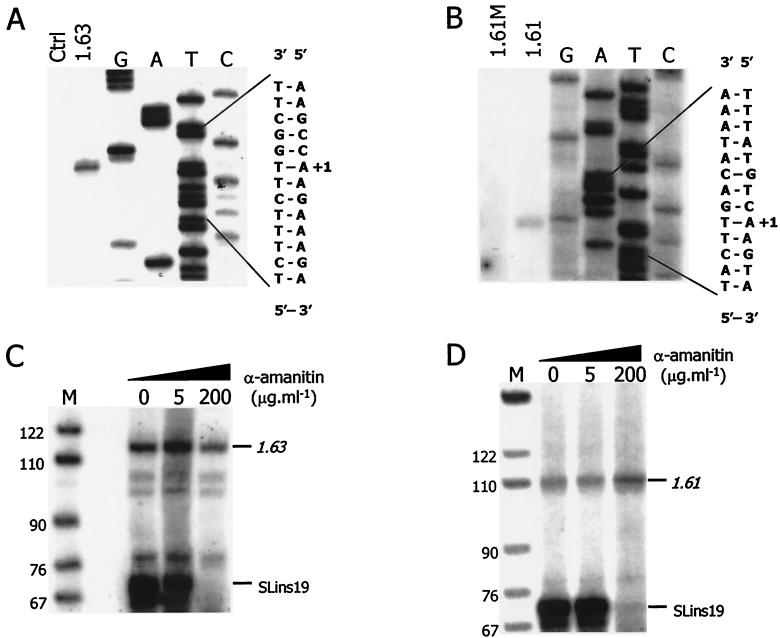
MVSG promoters have limited identity with BES promoters or with one another (Fig. 4). To test whether MVSG promoter sequence conservation correlates with recruitment of the transcription initiation complex, we employed a previously developed in vitro transcription system for T. brucei (23). This system is based on a cytoplasmic extract prepared from procyclic cells, and it was shown to support accurate and α-amanitin-resistant transcription of circular template DNA harboring either the rDNA, the procyclin gene, or a BES promoter as well as correctly initiated and α-amanitin-sensitive transcription of the SL RNA gene. The template constructs contain unrelated tag sequences downstream of the transcription initiation sites, and in vitro-transcribed RNA of these templates is detected specifically by primer extension of total-RNA preparations with a 5′ end-labeled oligonucleotide hybridizing to the tag. Using a transcription-competent extract from an EATRO 795 procyclic culture, we studied the 1.22, 1.61, 1.63, and 1.64 MVSG promoters and detected specific signals in 1.61 and 1.63 MVSG reactions, establishing MVSG promoter transcription in vitro for the first time (Fig. 5). For both 1.61 and 1.63 MVSG promoters, we accurately mapped the in vitro transcription initiation site and found that it coincided with the same A+1 residue that we had mapped in our analysis of tsetse fly salivary gland trypanosomes (Fig. 5A and B). We tested also one mutant form of the 1.61 MVSG promoter, in which eight conserved residues between positions −46 and −26 had been changed (from AATGTGGTTAAGGCAGGATT to ccgGTGGTTccGGCAGGcgg [changed residues are lowercase]). The mutation abolished in vitro transcription, demonstrating that the conserved sequence represents an essential MVSG promoter element (Fig. 5B, lane 1.61M). Furthermore, transcription from both promoters was resistant to α-amanitin (Fig. 5C and D), indicating that, in addition to appropriate trans-acting factors, the correct RNA polymerase was recruited to the promoter outside its life cycle context.
FIG. 4.
ClustalW sequence alignments of bloodstream (A) and metacyclic (B) VSG promoters. Conserved residues within each group are shown by a dark background. The A and B boxes of the BVSG promoters are boxed, as are poorly conserved A box-like regions of the MVSG promoters. The transcription start sites of the BES promoters are also boxed, as are the start sites identified for MVSGs in any life cycle stage. The means and ranges of identity within and between the two promoter groups are shown to the right. Promoter sequences are from the following sources: GUTat 10.1, 21; ILTat 1.22 and 1.61, 13; MVATs, 20; 4-49, 25; BESs and AnTat 11.17, 37 (accession numbers are within these studies).
FIG. 5.
Accurate and α-amanitin-resistant in vitro transcription from MVSG promoters in procyclic cell extract. (A and B) Accurate initiation of transcription. In vitro transcription reactions were carried out with template constructs p1.63-trm (1.63), p1.61 M-trm (1.61M), and p1.61-trm (1.61). In a control reaction, vector DNA was transcribed (Ctrl). Transcription signals were obtained from in vitro-synthesized RNA by primer extension reactions with the 5′ end-labeled oligonucleotide TAG_PE. For comparison, the p1.63-trm (A) and p1.61-trm (B) DNAs were subjected to linear amplification sequencing using the same oligonucleotide. Primer extension products and sequencing ladders were analyzed in parallel on 6% polyacrylamide-50% urea gels. At the right, the double-stranded sequences surrounding the transcription initiation sites are shown; of these, the left strands are those transcribed and correspond to the sequencing ladders. The start residues at position +1 are aligned with the primer extension signal. (C and D) α-Amanitin-resistant transcription. In vitro transcription of 1.63-trm (C) and 1.61-trm (D) was carried out in the presence of α-amanitin at 0, 5, or 200 μg · ml−1, and the corresponding RNAs were detected as described above. In each reaction, the SLins19 template containing the RNA polymerase II-recruiting SL RNA gene promoter was cotranscribed as a control, and its RNA was detected by primer extension of the 5′ end-labeled oligonucleotide Sltag, which is complementary to the SLins19 tag. On the right, the specific signals of 1.63, 1.61, and SLins19 RNAs are indicated, and, on the left, sizes of marker fragments are given.
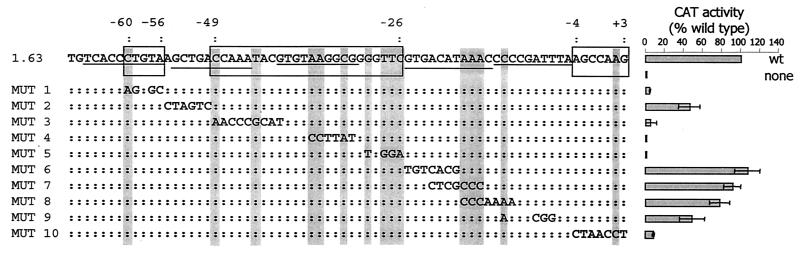
We then carried out a mutational analysis of the 1.63 MVSG promoter in procyclic cells, making base substitutions along its whole length, encompassing both conserved residues and regions that are divergent in sequence between different MVSG promoters. We chose this promoter because it has high homology with the MVAT7 promoter, which has been subjected to preliminary mutational analysis previously (20). The 1.63 MVSG promoter mutants were then assayed for the ability to drive CAT expression from plasmids transfected into cultured procyclic cells. The results shown in Fig. 6 revealed that mutations from positions bp −26 to −49 and bp −56 to −60 with respect to the transcription start site abolished the activity of the promoter. The more downstream of these regions includes nine residues that are conserved in all of the MVSG promoters that have been identified, whereas the region from bp −56 to −60 is not conserved. While there was not an obvious role for the conserved residues at positions −18 to −16 and −13, the bases around the transcription start site were important for efficient CAT expression.
FIG. 6.
Mutational analysis of the 1.63 MVSG promoter. A series of synthetic mutant promoter sequences (MUT 1 to MUT 10) were tested for ability to drive expression of the CAT reporter gene during a transient transfection assay of procyclic cells. Activities relative to that for the wild type (wt) are shown at the right. Positions that yielded low activity when mutated are boxed in the wild-type sequence at the top, and their coordinates relative to the start site are shown. The underlined residues in the 1.63 MVSG sequence show the positions of 10-bp block substitutions that were analyzed in the MVAT4 promoter; all of these substitutions were reported to abolish MVAT4 promoter activity (1). Residues conserved in all known MVSG promoters are shaded. The data are means ± standard deviations for at least three experiments.
DISCUSSION
By a number of approaches, including direct observation of transcription start sites in trypanosomes dissected from tsetse flies, accurate in vitro transcription initiation in a procyclic-stage extract, and mutational analysis in the same stage, we show that there is a consensus promoter for the MVSG genes. Our use of trypanosomes isolated from tsetse fly salivary glands provides the first direct transcriptional analysis of the specialized VSG repertoire that is expressed at this phase of the life cycle. A number of factors had to be taken into consideration when deciding how best to prepare these trypanosomes. Ultrastructural analysis has shown that the VSG coat is first synthesized by the premetacyclic trypanosomes attached to the salivary gland (35). As these divide, whereas metacyclic parasites do not, it is likely that more VSG-specific RNA will be present in the mixed population than in purified metacyclic cells. In any case, the limited availability of salivary gland parasites makes it less feasible to attempt to obtain metacyclic cells by salivary gland probing. It is possible to select the metacyclic stage by trapping epimastigote cells on an ion-exchange matrix or, as the epimastigote cells appear to have an active respiratory chain, by killing them with sodium azide, but we considered that the incubation and washing periods involved in those approaches may lead artifactually to a decrease or cessation of transcription and rapid disappearance of nascent RNA. We were able to isolate, from different fly transmissions, sufficient RNA with which to map transcription start sites by 5′ RACE. To validate the 5′ RACE walking approach, we first applied it to the well-characterized VSG221 transcription site in T. brucei S427 (42) and indeed obtained products specific to the site at which transcription is known to start (data not shown). For all three MVSG genes tested, the salivary gland RNA yielded a number of 5′ RACE products as we successively walked upstream, but none was obtained upstream of the 3′ terminal A+1 residue of the proposed MVAT series of promoters; data for the 1.61 and 1.63 MVSG genes are shown in Table 1. The abundance of products terminating downstream probably reflects limitations in the 5′ RACE technology. These products were of two types. Some presumably represented termination of reverse transcription, possibly due to a persistent secondary structure in the nascent transcript, while others had the spliced leader sequence that is trans-spliced onto trypanosome mature transcripts. We believe that such spliced products represent early splicing events that protect the RNA molecule as transcription proceeds over ∼2.8 kb from the start site to the VSG coding sequence.
Combining the results of all our approaches, including the 5′ RACE analysis of salivary gland trypanosomes, the study of in vitro transcription initiation in procyclic cell extracts, and the study of mutated promoter sequences, we conclude that MVSG transcription begins at the A+1 residue of the element proposed by Donelson and colleagues as the MVSG promoter from their studies of derepression in bloodstream trypanosomes. We believe that this resolves the question of what the authentic promoter is, and it also annuls our previous claim that an initiator-type element acts as the promoter (13, 14). There may be several reasons for the erroneous nature of the previous data, which were obtained from bloodstream trypanosomes still expressing MVSGs that had been activated in tsetse flies, but one obvious difference from the present analysis is that a walking approach was not used, and it is likely that incomplete primer extension products were obtained. Another discrepancy from data presented previously by Graham et al. (11, 15) is that MVSG promoters are capable of acting in the procyclic stage when removed from their endogenous loci. We have tested some of the constructs and a very similar procyclic cell line as studied before, but consistently obtained activity; it is not possible to conclude why the difference exists. The conclusion from the present work is that, when located on a plasmid or in the rDNA locus, the MVSG promoter essentially can be as active as other RNA polymerase I promoters. Again, this is consistent with previous findings in other laboratories (1, 25).
Compared with other trypanosome RNA polymerase I promoters, the MVSG promoters are as short as the BES promoters, requiring only up to 67 bp for full activity, at least when tested in the readily available procyclic and bloodstream life cycle stages. BES promoters, although highly conserved at the sequence level, require only two elements, boxes 1 and 2, for transcriptional activity (30, 37). Both boxes are required to form a stable DNA-protein complex in electrophoretic mobility shift assays or to compete with a specific in vitro transcription signal (22, 31), which suggests that they cooperate in the recruitment of trans-acting factors. MVSG promoters appear to have a more complex structure, with elements important for activity located along the length of the promoter. In a previously published linker scan of the MVAT4 promoter, 10-bp substitutions through the promoter from positions −4 to −63 all resulted in a loss of activity (Fig. 6). Although all of these mutations encompassed at least one of the residues that are conserved in all MVSG promoters, in our linker scan analysis of the 1.63 MVSG promoter using shorter substitutions we could show that only the regions −56 to −60 and −26 to −49 and the initiation region were essential for promoter activity. These include some, but not all, of the residues conserved in all MVSG promoters. From our data and the results from the MVAT4 linker scan, we can conclude that MVSG promoters require for their activity the partially conserved element centered on position −37 and a shorter, less conserved region just further upstream. These perhaps can operate within a series of sequence contexts, depending on the promoter in question. Vanhamme and colleagues (37), by testing for putative box 2 activity in the possible 11.17 MVSG promoter, also showed that the region corresponding to −36 to −26 of the 1.63 MVSG promoter is essential. In terms of promoter activation, we believe that similarities to box 2 or box 1 might not be relevant. Indeed, the greater number of sequences now available reveals that neither box (minimally CCCT and AGGG, respectively, on the sense strand) is conserved in the MVSG promoters (Fig. 4), suggesting that, while common proteins may be recruited, the BES and MVSG promoters also bind their own specific factors within the life cycle context. Assays in the procyclic stage of the life cycle suggest that different MVSG promoters direct varied levels of transcription, but the relevance of this to the metacyclic stage is uncertain. In our in vitro transcription experiments, we saw a good signal from the 1.63 MVSG promoter, a weaker signal from the 1.61 MVSG promoter, but no signals from the MVSG promoters for the 1.22 and 1.64 MVSG genes. This does not correlate, however, with previous observations that, in trypanosomes of the EATRO 795 genotype, 1.22 MVSG (also known as GUTat 7.1) and 1.61 MVSG (GUTat 7.15) are the most commonly activated MVSGs in metacyclic populations, with 1.63 MVSG (GUTat 7.2) and 1.64 MVSG (GUTat 7.13) appearing at lower frequencies (2). Either the differential-activation levels in the fly are not a consequence of relative promoter affinity for the initiation complex or the in vitro transcription system, using extract from the wrong life cycle stage, does not wholly reflect what occurs in the metacyclic stage. Although the 1.61 MVSG element acts as a core promoter in the assays we used, it is not possible yet to rule out upstream enhancing elements that function in the metacyclic stage.
Defining the molecular mechanisms behind the random activation of MES activation is going to be difficult, but we can perhaps predict some of the general mechanisms that regulate MES silencing in bloodstream and procyclic trypanosomes. Results from our reporter gene experiments indicate that there is no negative cis-acting element within 300 bp upstream or 900 bp downstream of the 1.61 MVSG promoter that represses the activity of the promoter within a chromosomal position in the procyclic stage of the life cycle. A BES promoter integrated upstream of a subtelomeric MES is inactive in the procyclic form, just as it is when integrated upstream of a VSG gene in a BES (17, 18). A ribosomal promoter integrated into the same region upstream of the 1.22 MES is not transcriptionally silent, just as it is not repressed when integrated upstream of a VSG in a BES (17, 18). The similarity in the activities of these promoters at two distinct subtelomeric locations suggests that the regulation on chromatin that silences BESs during replicative stages of the life cycle (18, 28) is a global mechanism that extends to all the telomeres and upstream loci on the megabase and intermediate-size chromosomes in T. brucei. This silencing mechanism may fail to regulate the ribosomal promoter because, in contrast to BES and MVSG promoters, it contains also an upstream control element (UCE). It is conceivable that the UCE may recruit chromatin-remodeling activities, thereby facilitating accessibility of the core promoter to RNA polymerase I transcription machinery in a subtelomeric locus that otherwise does not permit transcription. The finding that the rDNA promoter UCE is not important for in vitro transcription in a cell-free system which may not support the formation of a chromatin template is in accordance with this hypothesis (22).
Acknowledgments
We thank Hubert Denise for his help with the FACScan analysis.
A.G. is supported by the German Research Foundation (DFG). J.D.B. is a Wellcome Trust Principal Research Fellow, and the Wellcome Trust funded this work.
REFERENCES
- 1.Alarcon, C. M., H. J. Son, T. Hall, and J. E. Donelson. 1994. A monocistronic transcript for a trypanosome variant surface glycoprotein. Mol. Cell. Biol. 14:5579-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry, J. D., J. S. Crowe, and K. Vickerman. 1983. Instability of the Trypanosoma brucei rhodesiense metacyclic variable antigen repertoire. Nature 306:699-701. [DOI] [PubMed] [Google Scholar]
- 3.Barry, J. D., S. V. Graham, M. Fotheringham, V. S. Graham, K. Kobryn, and B. Wymer. 1998. VSG gene control and infectivity strategy of metacyclic stage Trypanosoma brucei. Mol. Biochem. Parasitol. 91:93-105. [DOI] [PubMed] [Google Scholar]
- 4.Barry, J. D., S. V. Graham, K. R. Matthews, P. G. Shiels, and O. A. Shonekan. 1990. Stage-specific mechanisms for activation and expression of variant surface glycoprotein genes in Trypanosoma brucei. Biochem. Soc. Trans. 18:708-710. [DOI] [PubMed] [Google Scholar]
- 5.Barry, J. D., and R. McCulloch. 2001. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 49:1-70. [DOI] [PubMed] [Google Scholar]
- 6.Bastin, P., T. H. MacRae, S. B. Francis, K. R. Matthews, and K. Gull. 1999. Flagellar morphogenesis: protein targeting and assembly in the paraflagellar rod of trypanosomes. Mol. Cell. Biol. 19:8191-8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernards, A., L. H. T. van der Ploeg, A. C. C. Frasch, P. Borst, J. C. Boothroyd, S. Coleman, and G. A. M. Cross. 1981. Activation of trypanosome surface glycoprotein genes involves a duplication-transposition leading to an altered 3′ end. Cell 27:497-505. [DOI] [PubMed] [Google Scholar]
- 8.Biebinger, S., S. Rettenmaier, J. Flaspohler, C. Hartmann, J. Penadiaz, L. E. Wirtz, H. R. Hotz, J. D. Barry, and C. Clayton. 1996. The PARP promoter of Trypanosoma brucei is developmentally regulated in a chromosomal context. Nucleic Acids Res. 24:1202-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brun, R., and M. Schonenberger. 1979. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 10.Brun, R., and M. Schonenberger. 1981. Stimulating effect of citrate and cis-aconitate on the transformation of Trypanosoma brucei bloodstream forms to procyclic forms in vitro. Z. Parasitenkd. 66:17-24. [DOI] [PubMed] [Google Scholar]
- 11.Graham, S. V., and J. D. Barry. 1995. Transcriptional regulation of metacyclic variant surface glycoprotein gene expression during the life cycle of Trypanosoma brucei. Mol. Cell. Biol. 15:5945-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham, S. V., K. R. Matthews, P. G. Shiels, and J. D. Barry. 1990. Distinct, developmental stage-specific activation mechanisms of trypanosome VSG genes. Parasitology 101:361-367. [DOI] [PubMed] [Google Scholar]
- 13.Graham, S. V., S. Terry, and J. D. Barry. 1999. A structural and transcription pattern for variant surface glycoprotein gene expression sites used in metacyclic stage Trypanosoma brucei. Mol. Biochem. Parasitol. 103:141-154. [DOI] [PubMed] [Google Scholar]
- 14.Graham, S. V., B. Wymer, and J. D. Barry. 1998. A trypanosome metacyclic VSG gene promoter with two functionally distinct, life cycle stage-specific activities. Nucleic Acids Res. 26:1985-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham, S. V., B. Wymer, and J. D. Barry. 1998. Activity of a trypanosome metacyclic variant surface glycoprotein gene promoter is dependent upon life cycle stage and chromosomal context. Mol. Cell. Biol. 18:1137-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirumi, H., and K. Hirumi. 1989. Continuous cultivation of Trypanosoma brucei bloodstream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985-989. [PubMed] [Google Scholar]
- 17.Horn, D., and G. A. M. Cross. 1995. A developmentally regulated position effect at a telomeric locus in Trypanosoma brucei. Cell 83:555-561. [DOI] [PubMed] [Google Scholar]
- 18.Horn, D., and G. A. M. Cross. 1997. Position-dependent and promoter-specific regulation of gene expression in Trypanosoma brucei. EMBO J. 16:7422-7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hug, M., V. B. Carruthers, C. Hartmann, D. S. Sherman, G. A. M. Cross, and C. Clayton. 1993. A possible role for the 3′-untranslated region in developmental regulation in Trypanosoma brucei. Mol. Biochem. Parasitol. 61:87-96. [DOI] [PubMed] [Google Scholar]
- 20.Kim, K. S., and J. E. Donelson. 1997. Co-duplication of a variant surface glycoprotein gene and its promoter to an expression site in African trypanosomes. J. Biol. Chem. 272:24637-24645. [DOI] [PubMed] [Google Scholar]
- 21.LaCount, D. J., N. M. El Sayed, S. Kaul, D. Wanless, C. M. R. Turner, and J. E. Donelson. 2001. Analysis of a donor gene region for a variant surface glycoprotein and its expression site in African trypanosomes. Nucleic Acids Res. 29:2012-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laufer, G., and A. Günzl. 2001. In vitro competition analysis of procyclin gene and variant surface glycoprotein gene expression site transcription in Trypanosoma brucei. Mol. Biochem. Parasitol. 113:55-65. [DOI] [PubMed] [Google Scholar]
- 23.Laufer, G., G. Schaaf, S. Bollgönn, and A. Günzl. 1999. In vitro analysis of α-amanitin-resistant transcription from the rRNA, procyclic acidic repetitive protein, and variant surface glycoprotein gene promoters in Trypanosoma brucei. Mol. Cell. Biol. 19:5466-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, M. G.-S., and L. H. van der Ploeg. 1997. Transcription of protein-coding genes in trypanosomes by RNA polymerase I. Annu. Rev. Microbiol. 51:463-489. [DOI] [PubMed] [Google Scholar]
- 25.McAndrew, M., S. Graham, C. Hartmann, and C. Clayton. 1998. Testing promoter activity in the trypanosome genome: isolation of a metacyclic-type VSG promoter, and unexpected insights into RNA polymerase II transcription. Exp. Parasitol. 90:65-76. [DOI] [PubMed] [Google Scholar]
- 26.Nagoshi, Y. L., C. M. Alarcon, and J. E. Donelson. 1995. The putative promoter for a metacyclic VSG gene in African trypanosomes. Mol. Biochem. Parasitol. 72:33-45. [DOI] [PubMed] [Google Scholar]
- 27.Navarro, M., and G. A. M. Cross. 1996. DNA rearrangements associated with multiple consecutive directed antigenic switches in Trypanosoma brucei. Mol. Cell. Biol. 16:3615-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro, M., G. A. M. Cross, and E. Wirtz. 1999. Trypanosoma brucei variant surface glycoprotein regulation involves coupled activation/inactivation and chromatin remodeling of expression sites. EMBO J. 18:2265-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedram, M., and J. E. Donelson. 1999. The anatomy and transcription of a monocistronic expression site for a metacyclic variant surface glycoprotein gene in Trypanosoma brucei. J. Biol. Chem. 274:16876-16883. [DOI] [PubMed] [Google Scholar]
- 30.Pham, V. P., C. C. Qi, and K. M. Gottesdiener. 1996. A detailed mutational analysis of the VSG gene expression site promoter. Mol. Biochem. Parasitol. 75:241-254. [DOI] [PubMed] [Google Scholar]
- 31.Pham, V. P., P. B. Rothman, and K. M. Gottesdiener. 1997. Binding of trans-acting factors to the double-stranded variant surface glycoprotein (VSG) expression site promoter of Trypanosoma brucei. Mol. Biochem. Parasitol. 89:11-23. [DOI] [PubMed] [Google Scholar]
- 32.Rudenko, G., P. A. Blundell, M. C. Taylor, R. Kieft, and P. Borst. 1994. VSG gene expression site control in insect form Trypanosoma brucei. EMBO J. 13:5470-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Son, H. J., G. A. Cook, T. Hall, and J. E. Donelson. 1989. Expression site associated genes of Trypanosoma brucei rhodesiense. Mol. Biochem. Parasitol. 33:59-66. [DOI] [PubMed] [Google Scholar]
- 35.Tetley, L., C. M. R. Turner, J. D. Barry, J. S. Crowe, and K. Vickerman. 1987. Onset of expression of the variant surface glycoproteins of Trypanosoma brucei in the tsetse fly studied using immunoelectron microscopy. J. Cell Sci. 87:363-372. [DOI] [PubMed] [Google Scholar]
- 36.Turner, C. M. R., J. D. Barry, I. Maudlin, and K. Vickerman. 1988. An estimate of the size of the metacyclic variable antigen repertoire of Trypanosoma brucei rhodesiense. Parasitology 97:269-276. [DOI] [PubMed] [Google Scholar]
- 37.Vanhamme, L., A. Pays, P. Tebabi, S. Alexandre, and E. Pays. 1995. Specific binding of proteins to the noncoding strand of a crucial element of the variant surface glycoprotein, procyclin, and ribosomal promoters of Trypanosoma brucei. Mol. Cell. Biol. 15:5598-5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanhamme, L., and E. Pays. 1995. Control of gene expression in trypanosomes. Microbiol. Rev. 59:223-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanhamme, L., P. Poelvoorde, A. Pays, P. Tebabi, H. V. Xong, and E. Pays. 2000. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol. Microbiol. 36:328-340. [DOI] [PubMed] [Google Scholar]
- 40.Vassella, E., J. van den Abbeele, P. Butikofer, C. K. Renggli, A. Furger, R. Brun, and I. Roditi. 2000. A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev. 14:615-626. [PMC free article] [PubMed] [Google Scholar]
- 41.Wirtz, E., and C. Clayton. 1995. Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science 268:1179-1183. [DOI] [PubMed] [Google Scholar]
- 42.Zomerdijk, J. C. B. M., M. Ouellette, A. L. M. A. ten Asbroek, R. Kieft, A. M. M. Bommer, C. E. Clayton, and P. Borst. 1990. The promoter for a variant surface glycoprotein gene expression site in Trypanosoma brucei. EMBO J. 9:2791-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]