Abstract
To characterize the metabolic role of peroxisomes in yeast cells under physiological conditions, we performed a comprehensive meta-analysis of published microarray data. Previous studies of yeast peroxisomes have mainly been focused on the function of peroxisomes under extreme conditions, such as growth on oleate or methanol as the sole carbon source, and may therefore not be representative of the normal physiological role of yeast peroxisomes. Surprisingly, our analysis of the microarray data reveals that the only pathway responding to peroxisome deficiency in mid-log phase is lysine biosynthesis, whereas classical peroxisomal pathways such as beta-oxidation are unaffected. We show that the upregulation of lysine biosynthesis genes in peroxisome-deficient yeasts shares many characteristics with the physiological response to lysine starvation. We provide data that suggest that this is the result of a “pathological” stimulation of the Lys14p transcriptional activator by the pathway intermediate aminoadipate semialdehyde. Mistargeting of the peroxisomal lysine pathway to the cytosol increases the active concentration of aminoadipate semialdehyde, which is no longer contained in the peroxisome and can now activate Lys14p at much lower levels than in wild-type yeasts. This is the first well-documented example of pathway misregulation in response to peroxisome deficiency and will be useful in understanding the phenotypic details of human peroxisome-deficient patients (Zellweger syndrome).
Peroxisomes are single-membrane-bounded organelles present in most eukaryotic cells, with the exception of Plasmodium, Giardia, Trichomonas, Entamoeba, and related species. Peroxisomes typically contain the enzymes of fatty acid beta-oxidation and catalase, which converts the hydrogen peroxide formed by fatty acyl-coenzyme A (CoA) oxidase to water and oxygen. Several other oxidases, e.g., monoamine oxidase, urate oxidase, hydroxyacid oxidase, methanol oxidase, and acetyl-spermidine oxidase, are also present in peroxisomes of some species and benefit from the same detoxification mechanism. In addition to these oxidative reactions, peroxisomes may contain many other biosynthetic pathways. Enzymes of these supplemental pathways are usually absent or localized in nonperoxisomal compartments in most eukaryotes, and their peroxisomal localization is a specialization of relatively few species. Examples of facultative peroxisomal pathways are glycolysis and purine salvage (in the peroxisomes of kinetoplastids, such as Trypanosoma and Leishmania [23]), isoprenoid biosynthesis (in vertebrates [1]), ether phospholipid biosynthesis (in kinetoplastids and animals [15, 33]), and the glyoxylate cycle, which channels the products of beta-oxidation into gluconeogenesis in plants, yeasts, and Caenorhabditis elegans (7).
Peroxisome-deficient mutants of most organisms are viable (with the possible exception of kinetoplastids), but a lack of functional peroxisomes can lead to severe pathologies in multicellular organisms (3, 5, 8, 16, 20, 24). In all organisms examined, the most severely affected metabolic pathway is the beta-oxidation of fatty acids, but in human patients bile acid production and cholesterol biosynthesis also seem to be impaired. Affected human patients show a characteristic complex of symptoms, including metabolic deficiencies, hepatomegaly, facial malformations, and severe neurological disturbances (Zellweger syndrome [24]).
Peroxisome biogenesis and physiology has been examined in detail in several yeast species (Pichia, Yarrowia, Hansenula, and Saccharomyces spp. [27, 28, 31]). Most of these studies were, however, focused on the function of peroxisomes under extreme conditions, such as growth on oleate or methanol as the sole carbon source, and may therefore not be representative of the normal physiological role of yeast peroxisomes. In order to characterize the function of peroxisomes in Saccharomyces cerevisiae, we performed a meta-analysis of two large-scale microarray experiments (13, 18). Our analysis demonstrates that the only pathway affected by peroxisome deficiency in mid-log phase on complete medium is lysine biosynthesis. Furthermore, growth rate determinations show that the upregulation of lysine biosynthesis after peroxisome disruption is not due to a severe deficiency of the enzymes but is most likely the result of the mislocalized production of the transcriptional modulator aminoadipate semialdehyde.
MATERIALS AND METHODS
Microarray analysis.
The microarray analysis is based on two large published datasets (Gasch et al. [13] and Hughes et al. [18]). The data were obtained from these authors and then analyzed by using customized Perl scripts to identify coregulated genes (these programs are available from R. Breitling upon request).
Data set 1: PEX12 disruption.
PEX12 disruption was described previously (18). Mutant (pex12::kanR/− MATa/b his3-1/− leu2-0/− met15-0/+ ura3-0/− lys2-0/+) and wild-type (BY4743, MATa/b his3-1/his3-1 leu2-0/leu2-0 ura3-0/ura3-0 +/met15-0 +/lys2-0) cells were grown in synthetic complete (SC) medium with 2% glucose at 30°C overnight to mid-log phase. They were diluted to 0.4 to 1 million cells/ml and grown an additional 5 to 7 h to reach 4 to 10 million cells/ml).
Data set 2: Stress response.
The stress response was evaluated as described previously (13). Wild-type (DBY7286: ura3-52 GAL2) cells or variants thereof were grown in rich medium (yeast extract-peptone-dextrose) at 30°C to early log phase (optical density = 0.2 to 0.4); a time zero reference sample was then collected. The cells then were grown for some time under one of several stress conditions (e.g., heat shock, temperature shift, osmolarity, hydrogen peroxide, dithiothreitol, nitrogen depletion, and stationary phase). During this time data were collected over the course of 2 to 3 h in most cases. A total of 142 samples was analyzed.
Two-dimensional visualization.
The distance between gene expression profiles was calculated from the Pearson correlation coefficient (PCC), with distance calculated as follows: (1 − PCC)/2. For visualization, the resulting n × n matrix describing the pairwise distances between n genes was projected into a plane. This was done by a modified multidimensional scaling, determining the two-dimensional arrangement that minimizes the discrepancy between actual and displayed distances.
Nearest-neighbor analysis.
Because of the nonnormality of the correlation coefficients, a close distance in the two-dimensional representation described above does not necessarily indicate whether two genes are highly correlated or not. This problem is aggravated by the fact that only a selection of genes is included in the pictures. To get a more reliable impression of the significance of the clustering, we determined the nearest neighbors of each gene, i.e., the genes among the complete data set that show the highest correlation coefficient to a given gene. If gene A is among the ten nearest neighbors of gene B in the complete data set, the two genes are connected by a line in the two-dimensional representations. In this way, significant coregulated clusters are easily identified.
Yeast experiments.
Diploid gene-deleted yeast strains in the BY4743 background were obtained from Research Genetics. They were maintained in yeast extract-peptone-dextrose medium plus 200 μg of Geneticin/ml. Growth experiments were performed in SC medium with or without lysine. A yeast strain disrupted in the hydroxymethyl-glutaryl-CoA reductase (HMG1) gene, which is unrelated to lysine biosynthesis and peroxisomal metabolism, served as a reference.
Complete genomes.
Genomic sequences for the white-rot fungus Phanerochaete chrysosporium were obtained from the Department of Energy Joint Genome Institute website (http://www.jgi.doe.gov). All other sequences are available from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
RESULTS AND DISCUSSION
Gene expression changes in peroxisome-deficient yeasts.
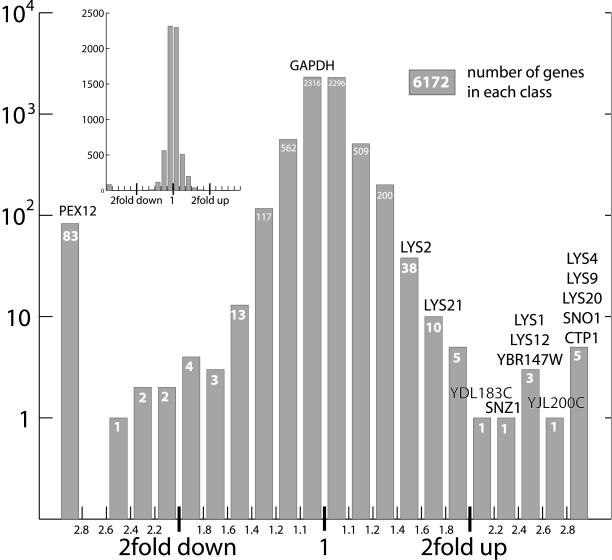
To characterize the function of yeast peroxisomes under physiological conditions, we first analyzed the transcriptional response of a PEX12-disrupted yeast strain (data provided by Hughes et al. [18]). Pex12p is a central member of the peroxisomal import machinery and the disruption of Pex12p leads to a complete loss of matrix protein import and absence of functional peroxisomes (2). Mutations of the human homologue lead to Zellweger syndrome (complementation group 3 [6]). Figure 1 shows that the expression of most of the more than 6,000 yeast genes present on the microarray does not change in response to peroxisome disruption. PEX12 expression is undetectable, as expected. Only a very small fraction of genes are highly upregulated (Table 1): 11 genes show upregulation by more than a factor of 2 (11/6,172 = 0.18%). Of these, five are involved in lysine biosynthesis (LYS9, LYS20, LYS4, LYS1, and LYS12). Of the six other highly upregulated genes, two (SNZ1/SNO1) encode a complex that is involved in pyridoxal phosphate biosynthesis, i.e., production of an essential cofactor of lysine biosynthesis, one (YBR147W) shows similarity to a nitrogen-starvation sensor of Schizosaccharomyces pombe, and two (CTP1 and YJL200C) are supposed to convert citrate and might also interact with homocitrate, the first product of the lysine biosynthesis pathway. The eleventh gene (YDL183C) was previously uncharacterized. Two further lysine biosynthesis genes (LYS21 and LYS2) are upregulated by factors of 1.64 and 1.57, respectively. Downregulation by a factor of 2 or more was observed in only a few genes coding for unrelated proteins (heat shock protein HSP12, inositol-3-phosphate synthase INO1, zinc uptake transporter ZRT1, and the glucose transporter HXT10). Taken together, this expression pattern indicates the activation of a specific lysine starvation response in the peroxisome-deficient yeasts. This is especially surprising since the PEX12-deficient yeasts were grown in SC medium, containing sufficient amounts of lysine. In contrast to the activation of lysine biosynthesis, no significant upregulation of classical peroxisomal pathways was observed (e.g., catalase CTA1 [upregulated by a factor of 1.14], multifunctional beta-oxidation enzyme FOX2 [upregulated by a factor of 1.01], and glyoxylate cycle isocitrate lyase ICL1 [upregulated by a factor of 1.06]). Since in wild-type cells these pathways are predominantly activated in stationary phase (data not shown), a lack of upregulation in PEX12-disrupted yeasts in mid-log phase might be expected.
FIG. 1.
Gene expression changes in response to disruption of PEX12. The histogram shows how many genes are up- or downregulated by a certain factor. The data were extracted from Hughes et al. (18) by using yMGV (21). A total of 4,612 genes are changed by a factor of 1.1 or less. PEX12 expression is undetectable. The names of 11 genes that are upregulated by a factor of 2 or more are indicated above the graph. The inset shows the same data on a linear scale.
TABLE 1.
Genes that are upregulated by a factor of 2 or more in response to a disruption of the PEX12 genea
| Gene | ORFb | Extent of upregulation (factor) | Function | PTS | Lysine-auxotroph? |
|---|---|---|---|---|---|
| LYS9 | YNR050C | 5.06 | Saccharopine dehydrogenase | -SLx5HP- | Yes |
| LYS20 | YDL182W | 3.96 | Homocitrate synthase | Yes | |
| CTP1 | YBR291C | 3.39 | Citrate transporter (mitochondrial) | Partially† | |
| LYS4 | YDR234W | 2.99 | Homoaconitase | -SQL* | Yes |
| SNO1 | YMR095C | 2.83 | Stationary-phase induced gene | No | |
| YJL200C | YJL200C | 2.74 | Aconitase homologue | Partially† | |
| LYS1 | YIR034C | 2.56 | Saccharopine dehydrogenase | -SRL* | Yes |
| LYS12 | YIL094C | 2.47 | Homoisocitrate dehydrogenase | -SRL | Yes |
| YBR147W | YBR147W | 2.45 | STM1-homologue, 7-TM nitrogen starvation sensor | No | |
| SNZ1 | YMR096W | 2.26 | Stationary-phase induced gene | No | |
| YDL183C | YDR183C | 2.07 | Uncharacterized | No | |
| LYS21 | YDL131W | 1.64 | Homocitrate synthase | Yes | |
| LYS2 | YBR115C | 1.57 | Aminoadipate semialdehyde dehydrogenase | -KLx5HL- | Yes |
| LYS5 | YGL154C | 1.17 | Lys2p-phosphopantetheinyl transferase | Yes |
For reference, three other genes involved in lysine biosynthesis are shown at the bottom of the table. Functional properties according to the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/). PTS, predicted peroxisomal targeting signal. Peroxisomal targeting has been shown by GFP fusion (14) and is indicated by an asterisk. The doubling time is increased by ca. 150% in YJL200C-disrupted and 10% in CTP1-disrupted yeast in lysine-deficient medium and is indicated by a dagger.
ORF, open reading frame.
Phylogenetic profile of peroxisomal targeting signals in lysine biosynthesis genes.
The specific upregulation of lysine biosynthesis gene expression in a peroxisome-deficient yeast strain is a strong indication that lysine biosynthesis is at least partly a peroxisomal pathway. Indeed, Geraghty et al. (14) has shown that two enzymes of the pathway (Lys4p and Lys1p) contain C-terminal peroxisomal targeting signals (PTS-1) and are targeted to peroxisomes. We identified an additional putative PTS-1 (-serine-arginine-lysine-COOH) in Lys12p. In addition, possible PTS-2-like sequences are present in the N-terminal sequence of Lys9p and Lys2p (Table 1). To determine whether peroxisomal localization is a general feature of the lysine biosynthetic pathway, we identified the homologues of Saccharomyes-Lys1p, -Lys4p, and -Lys12p in the complete genomic sequences of four other yeast species (Fig. 2.) In none of these species are the C-terminal targeting signals conserved, even though the sequences -IKRL and -IKRI in Lys12p of Aspergillus fumigatus and Phanerochaete chrysosporium might function as degenerate signals (25). This is in marked contrast to the case of classical peroxisomal proteins, such as multifunctional beta-oxidation protein (Fox2p), 2,4-dienoyl-CoA reductase (Sps19p), or catalase (Cta1p), and indicates that the peroxisomal localization of lysine biosynthesis is a rather recent specialization of baker's yeast. Just like the other supplemental peroxisomal pathways described above, lysine biosynthesis does not require an oxidase reaction. One reason for its localization in peroxisomes may be facilitated metabolic channeling; another might be the reduction of cytosolic aminoadipate semialdehyde, a pathway intermediate that is toxic at a high concentration (32). This analysis does not imply that the whole pathway is peroxisomal. Peroxisomal targeting signals are conspicuously absent from Lys5p (which could be piggy-backing into peroxisomes with its dimerization partner Lys2p [12]) and from the amino acid aminotransferases (Aro8p and Aro9p) that are proposed to act in the lysine biosynthesis pathway (19). A cytosolic detour is also observed for the peroxisomal glyoxylate cycle in yeasts and plants, where the central aconitase reaction takes place in the cytosol.
FIG. 2.
Phylogenetic profile of peroxisomal targeting signals in lysine biosynthesis enzymes. The six C-terminal amino acid residues are shown for three lysine biosynthesis enzymes from S. cerevisiae containing a canonical PTS1-sequence (in bold uppercase letters) and their homologues from four completed yeast genomes. The C termini of three classical peroxisomal enzymes are shown for comparison. The phylogenetic relationship between the yeast species is indicated on the right (the tree is based on data in Heckman et al. [17]). Mya, million years ago.
Growth on lysine-deficient medium.
Given the role of peroxisomes in lysine biosynthesis, we predicted that peroxisome-deficient yeasts would show a severe defect of lysine biosynthesis, when grown in the absence of lysine. However, when we grew PEX5-, PEX7-, and PEX12-disrupted yeast strains—which are deficient in PTS-1, PTS-2, and both peroxisomal import pathways, respectively—in lysine-deficient SC medium, we observed no significant changes in doubling times compared to a HMG1-disrupted strain, which should show a wild-type response. Growth was retarded by at most 10% in PEX5- and PEX12-deficient strains and was equal to wild-type growth in the PEX7 mutant. This indicates that even if substrate dilution and/or enzyme instability may affect the mislocalized pathway, lysine biosynthesis is still effectively carried out in the peroxisome-deficient yeasts.
We also show that a yeast strain carrying a targeted disruption of the YJL200C gene shows severely retarded growth on lysine-deficient medium (doubling time ca. 200 min, compared to 80 min on lysine-containing medium). Yjl200cp is predicted to be a citrate aconitase isozyme but might also be a homocitrate aconitase acting in concert with Lys4p. A slight, but consistent, lysine dependence is also seen in a CTP1-deficient yeast strain (doubling time increased ca. 10% in the absence of lysine). Ctp1p is a mitochondrial citrate transporter and might be involved in the transport of homocitrate produced by nuclear homocitrate synthase (Lys20p/Lys21p) into the peroxisome for further conversion by homocitrate aconitase (Lys4p). These data again confirm the lysine specificity of the transcriptional response seen in the peroxisome-disrupted yeast.
Regulation of peroxisome biogenesis and peroxisomal pathways in wild-type yeast.
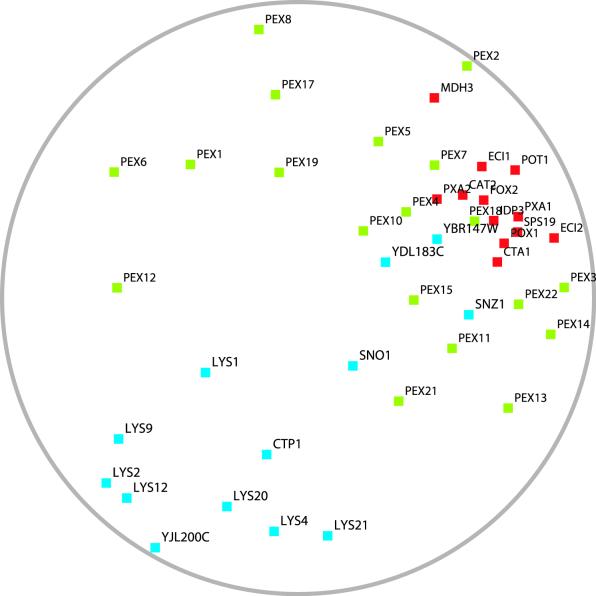
To compare the transcriptional response observed in the PEX12-disrupted yeast to the wild-type situation, we analyzed the most comprehensive microarray data set examining wild-type gene expression under a variety of physiological stress conditions (13). Figure 3 shows in a condensed way the similarity of the expression patterns of peroxin genes under these conditions compared to one another, as well as compared to genes of beta-oxidation or lysine biosynthesis. It can be seen that peroxin genes do not form a tight cluster, but their expression profiles are only weakly correlated, i.e., scattered over a large part of the plane in a two-dimensional visualization. This may be due to the fact that most peroxins are stable proteins and are needed only at relatively low levels, so that regulatory responses may be spurious. This is corroborated by the fact that the most typical expression pattern of all peroxin genes is shown by PEX18, which has the highest average correlation with other peroxins (average correlation coefficient = 0.456, compared to an average of 0.275 for other peroxins). Pex18p is an unstable protein that is constitutively degraded (26), so its expression pattern should most accurately reflect the demands of peroxisome biogenesis.
FIG. 3.
Two-dimensional visualization of similarities of expression profiles in wild-type cells under environmental stress. Genes are arranged in the plane in such a way that genes with highly correlating expression patterns are shown closest together. The diameter of the circle corresponds to the maximal distance of (anticorrelated) genes. The data were extracted from Gasch et al. (13). Red, beta-oxidation genes; blue, lysine biosynthesis genes; green, peroxisome biogenesis genes.
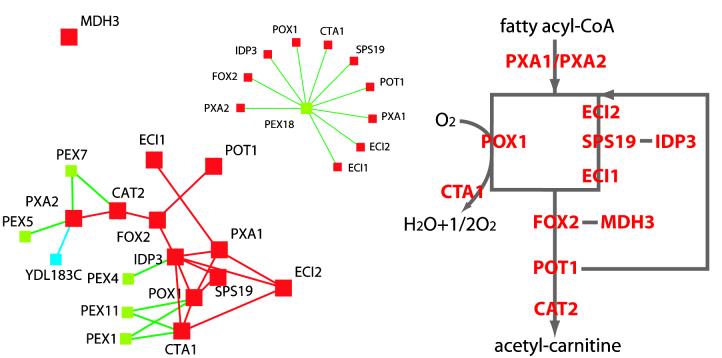
Figure 4 shows a more detailed analysis of the coregulation of peroxisome biogenesis and beta-oxidation. The distances and arrangement of the genes are the same as in Fig. 3, i.e., genes that have highly correlated expression patterns are shown close to one another, but only beta-oxidation genes and closely coregulated genes are shown. Genes that are among each other's 10 nearest neighbors in the complete data set are connected by lines. It can be seen that beta-oxidation genes have highly coregulated expression patterns, i.e., they are connected by a dense network of mutual next-neighbor relationships. The topography of this network has no relationship to the topography of the biosynthetic pathway, e.g., genes for the beta-oxidation of unsaturated fatty acids (ECI2, SPS19, ECI1, and IDP3) are not preferentially coregulated. This indicates that the genes of beta-oxidation form a single regulatory module. They also show a close relationship to peroxisome biogenesis: 100% of the beta-oxidation genes include at least one peroxin gene among their 10 nearest neighbors, and 10 of 11 genes have an expression pattern that is closely similar to that of PEX18, which (as discussed above) is the most typical peroxin with regard to its expression pattern.
FIG. 4.
Nearest-neighbor relationships of beta-oxidation genes superimposed on a two-dimensional visualization of expression profile similarities (compare with Fig. 3). The lines between genes A and B indicate that the expression profile of gene A is among the 10 nearest neighbors, i.e., the most highly correlating profiles, of gene B in the complete data set of Gasch et al. (13). For clarity the nearest-neighbor relationship between almost all beta-oxidation genes and PEX18 is shown in an inset. The topology of the beta-oxidation pathway is shown for reference. Red, beta-oxidation genes; blue, lysine biosynthesis genes; green, peroxisome biogenesis genes.
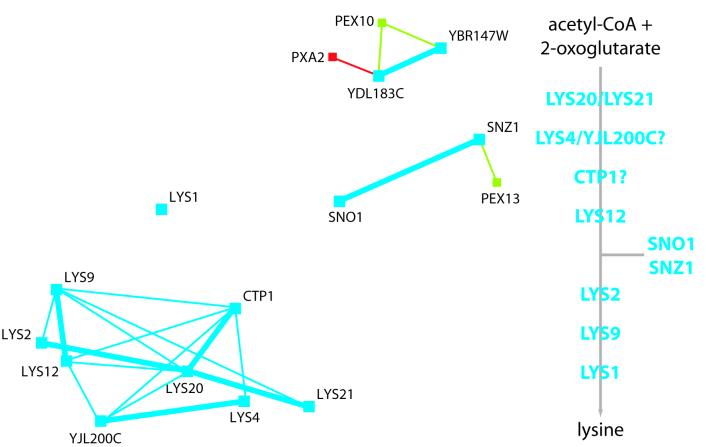
In Fig. 5 the same analysis is shown for the lysine biosynthetic pathway. Again, the genes form a tightly coregulated cluster. This cluster contains all of the lysine biosynthesis genes that are upregulated in the PEX12-disrupted yeasts, including YJL200C and CTP1, which were previously not implied in lysine biosynthesis. On the other hand, it excludes those lysine biosynthesis genes that were not significantly changed in the PEX12-disrupted yeasts (e.g., PEX5 and oxoadipate aminotransferase genes). This indicates that a common mechanism is responsible for the upregulation seen in the peroxisome-deficient yeast and in wild-type yeast under physiological conditions. In fact, the lysine biosynthetic cluster is even tighter than that of beta-oxidation, as can be seen from the fact that many genes are each other's nearest neighbors in the complete data set, i.e., no other nonlysine biosynthesis gene shows a more similar expression pattern (indicated by bold lines in Fig. 5). The four genes that are upregulated in PEX12-deficient yeasts but are not directly involved in lysine biosynthesis form two coregulated pairs (SNZ1/SNO1 and YDL183C/YBR147W), thus confirming that these genes are also specifically upregulated in response to peroxisome deficiency because, if their upregulation were nonspecific, one would not expect concerted upregulation of both genes of a coregulated pair. Somewhat surprisingly, the expression patterns of all genes that are upregulated in the PEX12-disrupted yeast strain do not show any special affinity to peroxisome biogenesis.
FIG. 5.
Nearest-neighbor relationships of lysine biosynthesis genes superimposed on a two-dimensional visualization of expression profile similarities (compare Fig. 3). For details, see legend to Fig. 4 and text. Red, beta-oxidation genes; blue, lysine biosynthesis genes; green, peroxisome biogenesis genes.
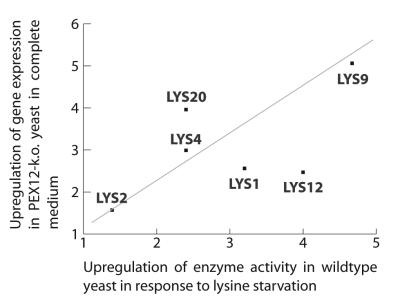
The similarity between the upregulation observed in the peroxisome-deficient yeast strain and the wild-type yeast is also confirmed by the comparison to the response of yeast cells to lysine starvation (Fig. 6). Both the extent of upregulation (two- to fivefold) and the pattern of upregulation (highest for LYS9 and lowest for LYS2) are strikingly similar to the changes of enzyme activity seen in wild-type yeast that are transferred from lysine-containing to lysine-deficient medium (29). Because Lys2p produces the toxic intermediate aminoadipate semialdehyde, which is converted further by Lys9p, this regulatory pattern guarantees that the concentration of the toxic compound is kept at the lowest possible levels even if the pathway is fully activated.
FIG. 6.
PEX12 disruption and lysine starvation. The upregulation of enzyme activity in response to lysine starvation (29) is shown in comparison to the upregulation of gene expression in response to a disruption of PEX12 (18).
Regulation of lysine biosynthesis in other gene-deleted yeast strains.
To further characterize the regulatory mechanisms that may be responsible for the upregulation of lysine biosynthesis genes in the PEX12-deficient yeasts, we searched the data provided by Hughes et al. (18) for other gene-deleted yeast strains that show a similar response. Indeed, several yeast strains disrupted in single genes show a concerted upregulation of all or most of the lysine biosynthesis genes (Table 2). However, in each of these cases, hundreds of other genes show an equally high upregulation. Furthermore, no common denominator is detectable among the disrupted genes that cause this upregulation, indicating that it represents a relatively unspecific response to disturbances in cell cycle (SWI4 and ASE1) or cellular metabolism (ERG3, ERG28, and RPL27A). On the other hand, two yeast strains disrupted in GCN4 and CKB2, respectively, show a concerted downregulation of lysine biosynthesis genes. Both of these genes are involved in the “general control” of amino acid biosynthesis, the pathway that normally upregulates gene expression in response to amino acid starvation (11, 22, 30). These findings confirm and extend earlier observations made by Urrestarazu et al. (29) that lysine biosynthesis is under general control. These data, however, cannot explain the specific upregulation of lysine biosynthesis in the PEX12-disrupted yeasts.
TABLE 2.
Concerted expression changes of lysine biosynthesis genes in gene-deleted yeasts
| Gene | Expression change of gene (n)a:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Upregulated
|
Downregulated
|
||||||||||
| PEX12 (11) | SW14 (295) | ERG3 (139) | YMR031W-A (136) | ERG28 (161) | YHL029C (118) | ASE1 (101) | RPL27A (106) | GAS1 (236) | GCN4 (844) | CKB2 (141) | |
| LYS9 | 5.04 | 2.13 | 1.66 | 2.23 | 1.66 | 1.92 | 1.62 | 1.99 | 1.29 | 1.43 | 1.67 |
| LYS20 | 3.96 | 3.08 | 3.30 | 2.88 | 3.48 | 3.33 | 3.07 | 3.08 | 1.37 | 2.17 | 2.70 |
| CTP1 | 3.39 | 1.54 | 1.15 | 1.33 | 0.97 | 1.28 | 1.28 | 2.91 | 1.22 | 1.04 | 1.23 |
| LYS4 | 2.99 | 2.85 | 1.47 | 1.75 | 1.27 | 2.36 | 1.95 | 2.65 | 1.33 | 1.19 | 1.69 |
| YJL200C | 2.74 | 2.73 | 2.06 | 2.27 | 1.86 | 2.22 | 1.88 | 2.19 | 1.80 | 1.59 | 1.85 |
| LYS1 | 2.56 | 6.97 | 4.93 | 4.69 | 5.13 | 4.20 | 4.14 | 2.77 | 2.49 | 3.85 | 4.17 |
| LYS12 | 2.47 | 2.28 | 2.14 | 2.13 | 1.70 | 1.87 | 1.35 | 1.36 | 1.42 | 1.37 | 1.54 |
| LYS21 | 1.64 | 2.90 | 2.33 | 1.94 | 1.87 | 1.57 | 2.12 | 2.42 | 1.93 | 1.54 | 1.89 |
| LYS2 | 1.57 | 2.51 | 2.70 | 1.92 | 0.31 | 1.71 | 2.56 | 1.47 | 1.49 | 1.52 | 1.92 |
| LYS5 | 1.17 | 1.61 | 1.50 | 1.51 | 1.92 | 1.39 | 1.02 | 1.25 | 1.54 | 1.17 | 1.32 |
Promoter analysis of lysine biosynthesis genes.
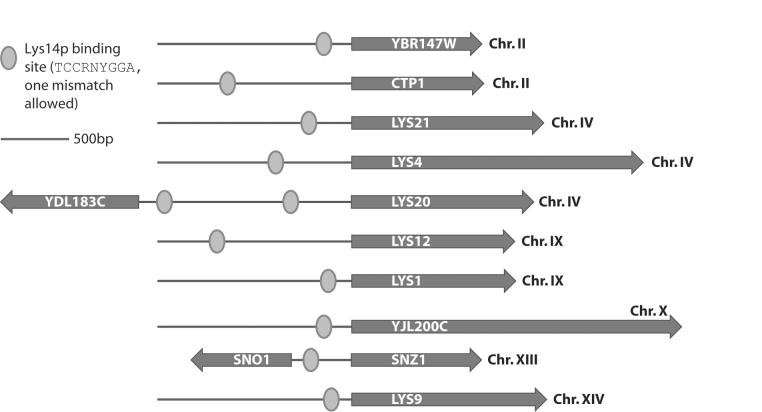
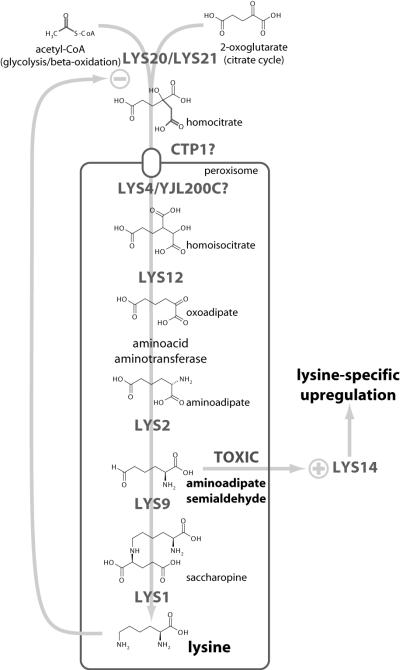
Analysis of the promoter regions of all genes that are upregulated in the PEX12-disrupted yeasts revealed the presence of predicted Lys14p binding sites (TCCRNYGGA, allowing one mismatch [4]) in all promoters, within 1,000 bp of the start codon (Fig. 7). A Lys14p binding site is conspicuously absent in the promoters of LYS5, LYS2, and LYS14 itself, which are not, or only weakly, upregulated in the peroxisome-deficient yeasts, and in the case of LYS5 and LYS14 show no coregulation with other lysine biosynthesis gene in wild-type yeasts. Lys14p is a transcriptional activator specific for the lysine biosynthesis pathway (9). Its regulation is unusual in that it is activated by the pathway intermediate aminoadipate semialdehyde, and apparent repression of Lys14p by lysine is caused indirectly by inhibition of the committed step catalyzed by the two homocitrate synthase enzymes (Lys20p and Lys21p), which are localized in the nucleus (10). This obviously suggests a mechanism for the upregulation of lysine biosynthesis genes in the presence of lysine: mistargeting of the pathway enzymes to the cytosol increases the active concentration of aminoadipate semialdehyde, which is no longer contained in the peroxisome and can activate Lys14p at much lower levels than in wild-type yeasts. Even a slight activation of Lys14p can then trigger a positive feedback loop that fully stimulates the pathway (summarized in Fig. 8). This mechanism does not require that all of the enzymes of the pathway are peroxisomal, and Lys12p (14) and the aminotransferases may well be cytosolic. Indeed, especially in the induced state, an exclusively peroxisomal localization is very unlikely for all of the enzymes, since peroxisomal proliferation is not correlated with stimulation of the pathway, which would make detection by classical techniques of the important role of peroxisomes in the regulation of lysine metabolism very difficult.
FIG. 7.
Gene structure and promoters of genes that are upregulated in PEX12-disrupted yeasts. Predicted Lys14p binding sites in the promoter regions are indicated by circles. Four of the genes form tail-to-tail clusters (YDL183C/LYS20 and SNZ1/SNO1).
FIG. 8.
Summary of the lysine pathway.
Conclusion.
We demonstrated here a surprising misregulation of the lysine biosynthetic pathway in peroxisome-disrupted yeasts. Reanalysis of two large-scale microarray datasets indicates that this upregulation is due to a specific activation of a lysine starvation response pathway. However, disruption of peroxisomal biogenesis does not lead to lysine auxotrophy or serious growth defects in the absence of lysine. In addition, the specific activation of the lysine pathway is observed even in the presence of sufficient amounts of lysine in the medium. Promoter analysis of the genes involved, indicates that the upregulation is mediated by the transcription factor Lys14p. Lys14p can activate expression of the lysine biosynthesis pathway in an all-or-nothing way via a positive feedback loop triggered by the pathway intermediate aminoadipate semialdehyde. The sensitivity of Lys14p toward the semialdehyde might be adjusted to the small amounts leaking out of the peroxisome, so that mistargeting of the pathway to the cytosol can inappropriately activate the starvation response even in the presence of exogenous lysine. To the best of our knowledge, this is the first well-documented example of pathway misregulation in response to peroxisome disruption. This observation is of great relevance for the interpretation of physiological findings in human Zellweger patients, in whom cytosolic accumulation of metabolic intermediates may also interfere with otherwise normal pathways.
The present study also documents the usefulness of public microarray data for the examination of specific cell biological questions. It allowed us to conclusively demonstrate the important role of peroxisomes for lysine biosynthesis and at the same time identified a possible “pathogenic” mechanism for the upregulation of lysine biosynthesis in peroxisome-deficient yeasts. “Wet” experiments were used to confirm the specificity of the observed effects. Although the specificity observed here will most likely remain an exceptional case even for yeast microarrays, the continuous accumulation of new expression datasets holds great potential for further studies.
Acknowledgments
We thank D. Laubner and Zhang Mi for critical reading of the manuscript.
This work was supported in part by National Institutes of Health grants DK58238 and DK58040.
REFERENCES
- 1.Aboushadi, N., W. H. Engfelt, V. G. Paton, and S. K. Krisans. 1991. Role of peroxisomes in isoprenoid biosynthesis. J. Histochem. Cytochem. 47:1127-1132. [DOI] [PubMed] [Google Scholar]
- 2.Albertini, M., W. Girzalsky, M. Veenhuis, and W. H. Kunau. 2001. Pex12p of Saccharomyces cerevisiae is a component of a multi-protein complex essential for peroxisomal matrix protein import. Eur. J. Cell Biol. 80:257-270. [DOI] [PubMed] [Google Scholar]
- 3.Baes, M. 2000. Mouse models for peroxisome biogenesis disorders. Cell. Biochem. Biophys. 32:229-237. [DOI] [PubMed] [Google Scholar]
- 4.Becker, B., A. Feller, M. el Alami, E. Dubois, and A. Pierard. 1998. A nonameric core sequence is required upstream of the LYS genes of Saccharomyces cerevisiae for Lys14p-mediated activation and apparent repression by lysine. Mol. Microbiol. 29:151-163. [DOI] [PubMed] [Google Scholar]
- 5.Burmester, T., M. Mink, M. Pal, Z. Laszloffy, J. Lepesant, and P. Maroy. 2000. Genetic and molecular analysis in the 70CD region of the third chromosome of Drosophila melanogaster. Gene 246:157-167. [DOI] [PubMed] [Google Scholar]
- 6.Chang, C. C., and S. J. Gould. 1998. Phenotype-genotype relationships in complementation group 3 of the peroxisome-biogenesis disorders. Am. J. Hum. Genet. 63:1294-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escher, C. L., and F. Widmer. 1997. Lipid mobilization and gluconeogenesis in plants: do glyoxylate cycle enzyme activities constitute a real cycle? A hypothesis. Biol. Chem. 378:803-813. [PubMed] [Google Scholar]
- 8.Faust, P. L., H. M. Su, A. Moser, and H. W. Moser. 2001. The peroxisome deficient PEX2 Zellweger mouse: pathologic and biochemical correlates of lipid dysfunction. J. Mol. Neurosci. 16:289-297. [DOI] [PubMed] [Google Scholar]
- 9.Feller, A., E. Dubois, F. Ramos, and A. Pierard. 1994. Repression of the genes for lysine biosynthesis in Saccharomyces cerevisiae is caused by limitation of Lys14-dependent transcriptional activation. Mol. Cell. Biol. 14:6411-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feller, A., F. Ramos, A. Pierard, and E. Dubois. 1999. In Saccharomyces cerevisiae, feedback inhibition of homocitrate synthase isoenzymes by lysine modulates the activation of LYS gene expression by Lys14p. Eur. J. Biochem. 261:163-170. [DOI] [PubMed] [Google Scholar]
- 11.Feng, L., H. Yoon, and T. F. Donahue. 1994. Casein kinase II mediates multiple phosphorylation of Saccharomyces cerevisiae eIF-2 alpha (encoded by SUI2), which is required for optimal eIF-2 function in S. cerevisiae. Mol. Cell. Biol. 14:5139-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flynn, C. R., R. T. Mullen, and R. N. Trelease. 1998. Mutational analyses of a type 2 peroxisomal targeting signal that is capable of directing oligomeric protein import into tobacco BY-2 glyoxysomes. Plant J. 16:709-720. [DOI] [PubMed] [Google Scholar]
- 13.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geraghty, M. T., D. Bassett, J. C. Morrell, G. J. Gatto, Jr., J. Bai, B. V. Geisbrecht, P. Hieter, and S. J. Gould. 1999. Detecting patterns of protein distribution and gene expression in silico. Proc. Natl. Acad. Sci. USA 96:2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Hajra, A. K., and A. K. Das. 1996. Lipid biosynthesis in peroxisomes. Ann. N. Y. Acad. Sci. 804:129-141. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, M., K. Nito, K. Toriyama-Kato, M. Kondo, T. Yamaya, and M. Nishimura. 2000. AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 19:5701-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heckman, D. S., D. M. Geiser, B. R. Eidell, R. L. Stauffer, N. L. Kardos, and S. B. Hedges. 2001. Molecular evidence for the early colonization of land by fungi and plants. Science 293:1129-1133. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 19.Iraqui, I., S. Vissers, M. Cartiaux, and A. Urrestarazu. 1998. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 257:238-248. [DOI] [PubMed] [Google Scholar]
- 20.Liu, F., J. D. Thatcher, and H. F. Epstein. 1997. Induction of glyoxylate cycle expression in Caenorhabditis elegans: a fasting response throughout larval development. Biochemistry 36:255-260. [DOI] [PubMed] [Google Scholar]
- 21.Marc, P., F. Devaux, and C. Jacq. 2001. yMGV: a database for visualization and data mining of published genome-wide yeast expression data. Nucleic Acids Res. 29:E63-E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsons, M., T. Furuya, S. Pal, and P. Kessler. 2001. Biogenesis and function of peroxisomes and glycosomes. Mol. Biochem. Parasitol. 115:19-28. [DOI] [PubMed] [Google Scholar]
- 24.Powers, J. M., and H. W. Moser. 1998. Peroxisomal disorders: genotype, phenotype, major neuropathologic lesions, and pathogenesis. Brain Pathol. 8:101-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purdue, P. E., and P. B. Lazarow. 1996. Targeting of human catalase to peroxisomes is dependent upon a novel COOH-terminal peroxisomal targeting sequence. J. Cell Biol. 134:849-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purdue, P. E., and P. B. Lazarow. 2001. Pex18p is constitutively degraded during peroxisome biogenesis. J. Biol. Chem. 276:47684-47689. [DOI] [PubMed] [Google Scholar]
- 27.Sakai, Y., and S. Subramani. 2000. Environmental response of yeast peroxisomes: aspects of organelle assembly and degradation. Cell. Biochem. Biophys. 32:51-61. [DOI] [PubMed] [Google Scholar]
- 28.Titorenko, V. I., J. J. Smith, R. K. Szilard, and R. A. Rachubinski. 2000. Peroxisome biogenesis in the yeast Yarrowia lipolytica. Cell. Biochem. Biophys. 32:21-26. [DOI] [PubMed] [Google Scholar]
- 29.Urrestarazu, L. A., C. W. Borell, and J. K. Bhattacharjee. 1985. General and specific controls of lysine biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 9:341-344. [DOI] [PubMed] [Google Scholar]
- 30.van den Heuvel, J., V. Lang, G. Richter, N. Price, L. Peacock, C. Proud, and J. E. McCarthy. 1995. The highly acidic C-terminal region of the yeast initiation factor subunit 2 alpha (eIF-2α) contains casein kinase phosphorylation sites and is essential for maintaining normal regulation of GCN4. Biochim. Biophys. Acta 1261:337-348. [DOI] [PubMed] [Google Scholar]
- 31.Veenhuis, M., F. A. Salomons, and I. J. Van Der Klei. 2000. Peroxisome biogenesis and degradation in yeast: a structure/function analysis. Microsc. Res. Technol. 51:584-600. [DOI] [PubMed] [Google Scholar]
- 32.Zaret, K. S., and F. Sherman. 1985. alpha-Aminoadipate as a primary nitrogen source for Saccharomyces cerevisiae mutants. J. Bacteriol. 162:579-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zomer, A. W., P. A. Michels, and F. R. Opperdoes. 1999. Molecular characterisation of Trypanosoma brucei alkyl-dihydroxyacetone-phosphate synthase. Mol. Biochem. Parasitol. 104:55-66. [DOI] [PubMed] [Google Scholar]