Abstract
Both normal development and neoplastic progression involve cellular transitions from one physiological state to another. Whereas much is being discovered about signal transduction networks involved in regulating these transitions, little progress has been made in identifying the higher order genetic determinants that establish and maintain mammary cell identity and dictate cell type-specific responses to mammotropic signals. Homeobox genes are a large superfamily of genes whose members function in establishing and maintaining cell fate and cell identity throughout embryonic development. Recent genetic and expression analyses strongly suggest that homeobox genes may perform similar functions at specific developmental transition points in the mammary gland. These analyses also suggest that homeobox genes may play a contributory or causal role in breast cancer.
Keywords: breast cancer, homeodomain, HOX genes, embryogenesis, organogenesis
Introduction
The mammary gland is a remarkable organ with respect to its development and functional differentiation. It is also remarkable with respect to the consequences on mammalian life should development become abnormal, leading either to lactational failure or, most importantly, mammary cancer.
Unlike most mammalian organs, which develop primarily embryonically with a more or less linear progression toward functional maturity, development of the mammary gland is primarily postpubertal and may be divided into both a linear and a cyclical phase (Fig. 1) (see [1,2,3] for detailed reviews). These phases can be characterized further as a series of highly orchestrated transitions, or switches, in which critical developmental decisions are made concerning cell differentiation, pattern formation and cell function. While mutations have been identified that block or delay most of these transitions (primarily in signal transduction networks), the higher order genetic determinants of cell identity that dictate cell type specific responses are largely unknown.
Figure 1.
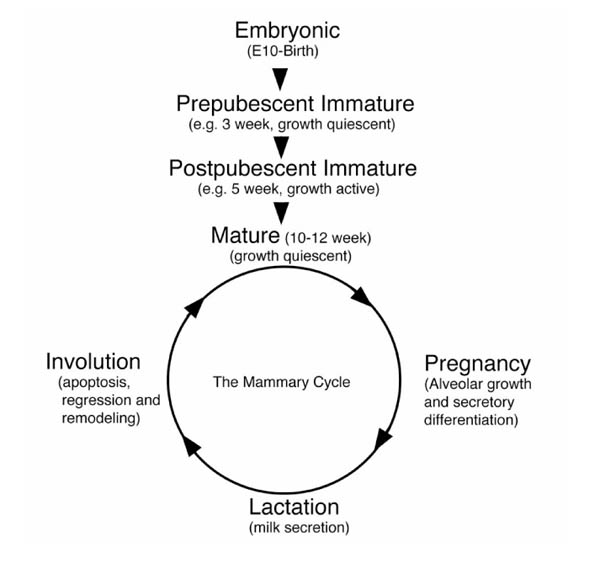
Phases of mammary gland development. Proliferative development in virgin animals is represented by the linear portion of the diagram. Cyclical development is represented by the circular portion of the diagram.
Among other candidates, one superfamily of genes presents itself as capable of regulating developmental decisions during these transitions: the homeobox genes. As a general principle, homeobox genes encode transcription factors that play key roles in the determination and maintenance of cell fate and cell identity [4,5,6,7]. Homeobox genes share a common nucleotide sequence motif (the homeobox) encoding the roughly 61 amino acid homeodomain. The homeodomain, in turn, is a helix-turn-helix DNA binding domain of the functional transcription factor. Evolutionary relationships and family classifications are determined based upon the degree of identity and similarity among homeodomains followed by comparative analyses of amino acid sequences both amino-terminal and carboxyl-terminal to the homeodomain [8,9,10]. These terminal sequences vary considerably from protein to protein and, indeed, may demonstrate no evidence of evolutionary or functional relationship whatsoever.
Homeobox genes are found in animals ranging from hydra to humans (as well as fungi and plants). Over evolutionary time, the number of homeobox genes has increased and their functions have been reengineered to meet the demands of increasingly diverse developmental processes. To date, there are well over 100 homeobox genes identified in the human, with a comparable number of homologs identified in the mouse [10].
In mammals, homeobox genes reign over the specification of the overall body plan and are known to play key roles in a variety of developmental processes including central nervous system and skeletal development, limb and digit specification, and organogenesis. Mutations in homeobox genes can cause dramatic developmental defects including loss of specific structures as well as 'homeotic transformations', in which one body part or segment is converted to the likeness (identity) of another. Some homeobox genes appear to serve cell autonomous functions in differentiation and cell cycle control; others serve noncell autonomous functions such as pattern formation and mediation of reciprocal tissue interactions.
At least five difficulties present themselves when trying to understand the functions of individual members of such a large superfamily of genes. First and foremost, exceptionally little is known about the number and identity of target genes controlled directly or indirectly by any mammalian homeobox gene, although it appears that much of the Drosophila genome is under homeobox gene control [11]. Second, only slightly more is known about upstream genes that control homeobox gene expression and function. Recent efforts in both Drosophila and mice demonstrate that establishment and maintenance of restricted homeobox gene expression is controlled, in part, by combinatorial activity of trithorax and polycomb group genes (which themselves are highly regulated) [12,13,14,15]. Third, homeobox genes do not generally act alone to determine cell identity. Rather, in many cases, it appears to be the combinatorial spatially and temporally regulated pattern of homeobox genes functioning in a given cell (a 'homeobox code') that determines the cell's identity [6,16,17]. In fact, in some cases, important functions of a given homeobox gene can be masked by compensatory function of a related homeobox gene. Fourth, to further complicate the issue, some homeodomain proteins are known to interact physically with other homeodomain proteins (eg PBX and MEIS or PBX and HOX) or other cofactors to control downstream target gene specificity [18,19,20]. Finally, many homeobox gene mutations are (or will be) embryonic or perinatal lethal, making analysis of the development of adult organs particularly challenging. This current state of affairs makes it difficult to develop mechanistic models for the function of a given homeobox gene, especially in the mammary gland where none of these issues have been investigated adequately.
In addition to roles in normal development, altered homeobox gene function is implicated in the development of cancers, particularly leukemias and rhabdomyosarcomas, as well as those of the breast, prostate, kidney, colon, skin and brain. With the exception of a role in breast cancer, this subject has been reviewed extensively [21,22,23,24].
Homeobox genes clearly occupy a prominent position in the developmental regulatory heirarchy, yet homeobox genes have received little attention with respect to mammary gland organogenesis, functional differentiation and cancer. It is the purpose of this review to highlight a few known and suspected roles of homeobox genes in controlling developmental decisions and neoplasia in the mammary gland, and to point out some gaping holes in our understanding that study of homeobox genes may help to fill.
Transitions and Switches in mammary gland development
The linear phase
Differentiation of the embryonic mammary gland
The mammary epithelium is an ectodermal derivative. As such, among the first distinctions that must be made is the differentiation of presumptive mammary epithelium from tissue that would otherwise form skin, hair follicles or other ectodermally derived structures. This differentiation occurs in at least two major stages. The first stage begins about day 10 of gestation (E10) with the establishment of the mammary streaks, two lines of epidermally derived thickened epithelium that run anterior --> posterior, symmetrically displaced off the ventral midline. These streaks represent the first morphological evidence of mammary pattern formation and differentiation.
The second stage occurs around E11 with the definition of the nipple region. Presumptive mammary epithelium forms a lens-shaped disk that becomes associated with underlying condensed mammary mesenchyme. The mammary epithelium continues to grow to form a bulb-shaped mammary bud that elongates and invades the condensed mesenchyme. Mammary epithelial cell identity is firmly established as early as E12.5 (the bud stage), as evidenced by the ability to transplant the presumptive mammary gland into a cleared fat pad and regenerate a ductal tree (CW Daniel, G Robinson, personal communication).
Invasion of the mammary fat pad precursor mesenchyme
As the mammary bud elongates into a mammary sprout, it reaches a second mesenchyme, the fat pad precursor mesenchyme, and undergoes a small amount of branching morphogenesis to form the rudimentary gland of the neonate. Several genes are known to act either at or before this critical transition to allow further growth or to control establishment of sexual dimorphism of the mouse mammary gland (eg Lef1, PThRP, PPR1) [25,26,27]. Tissue recombination experiments demonstrate that mammary mesenchyme and fat pad mesenchyme affect the growth of mammary epithelium in dramatically different ways, suggesting fundamental differences in their identities and biological properties with respect to tissue interactions (see [2]). How these differences are established is unknown.
Growth initiation, cellular differentiation and growth arrest
From birth to puberty, the gland remains rudimentary and relatively growth quiescent. At puberty, ovarian hormones stimulate rapid and invasive ductal elongation driven by growth of a structure called the terminal end bud, which consists of four to six layers of relatively undifferentiated 'body cells' and a surrounding single layer of 'cap cells'. These two populations differentiate into lumenal epithelial cells (also consisting of multiple cell types) and myoepithelial cells, respectively, as the subtending duct is formed [1,28]. Whereas a variety of individual cell types are known to exist and their developmental capacities have begun to be explored, virtually nothing is known about how these cell lineages and fates are established in the first place.
Upon reaching the limits of the fat pad at ductal maturity, ductal elongation ceases and terminal end buds regress to leave a branched system of differentiated ducts. These ducts will remain relatively quiescent as long as the animal remains virgin. How this growth control is achieved and maintained is not known, although it probably involves members of the transforming growth factor-β superfamily.
The cyclical phase: lobuloalveolar differentiation, lactation, involution and gland remodeling
Hormonal changes during pregnancy initiate a cyclical phase of development in which there is a dramatic transition from a predominantly ductal to a predominantly lobuloalveolar gland morphology. Lobuloalveolar progenitor cells located within the ducts proliferate to form alveolar buds, which further differentiate to form the alveoli. Near midpregnancy, the alveolar epithelium acquires the capacity to produce milk proteins (the stage I transition of lactogenesis) but secretory function is inhibited. At parturition, inhibition of secretory function is released and these cells begin to secrete large quantities of milk (the stage II transition of lactogenesis). Upon weaning, milk secretion ceases and the gland involutes. During involution, most alveolar cells undergo apoptosis (programed cell death), while a residual epithelial population remodels itself back into a ductal tree to await the next pregnancy.
Of course, there are known hormonal and growth factor signals that control some of these transitions, but little is known about how epithelial and stromal cells are poised to respond to such signals or how the resultant differentiated state is maintained.
Homeobox genes in embryonic and postnatal mammary gland development
Judging from their properties in the development of other organs, it is likely that homeobox genes may function at many of the transition points already described. The approach that has been most useful thus far in investigating mammary homeobox gene function is to determine which homeobox genes are expressed in the gland, characterize when and where they are expressed throughout development, and assay their function in vitro or, preferably, in vivo (Table 1). Unfortunately, many of the current studies are in their early stages and have been limited to a single detection technique (eg reverse transcription polymerase chain reaction) or to a particular developmental stage with no further analysis. Selected analyses, conveniently divided by gene family, are now highlighted with historical context whenever possible.
Table 1.
Summary of homeobox gene expression and function in mammary gland development and neoplasia
| Gene | Gene | Expression | Expression altered in | Mammary or in vitro | |||
| family | namea | detected inb: | Methodc | neoplasias ? | Possible regulation? | phenotype? | Reference |
| Hox/HOX | Hoxa-1 | MG (UD) | RT-PCR; Northern | OE (freq.) | Extracellular matrix | [31*,33*] | |
| Scp2, CID-9 | components | ||||||
| HOXA-1 | MCF7 | RT-PCR | Retinoic acid | [63,30] | |||
| Hoxa-5 | CID-9 | RT-PCR | [33*] | ||||
| Hoxa-7 | MG | RT-PCR | [31*] | ||||
| HOXA-7 | MG | RT-PCR | 1 | ||||
| Hoxa-9 | MG | RT-PCR | KO: lactational defects | [38*] | |||
| HOXA-10 | MCF7 | RT-PCR | Vitamin D | OE in MCF7: promotes cell cycle | [30,64] | ||
| arrest at G1 | |||||||
| Hoxb-6 | MG | RT-PCR; Northern | Lost | [31*] | |||
| HOXB-6 | MCF7 | RT-PCR | [30] | ||||
| Hoxb-7 | MG (E+S) | Northern; in situ | UE | Extracellular matrix | OE in SkBr3 cells: bFGF induction; | [33*,57] | |
| components; developmental | increased proliferation; decreased | ||||||
| growth factor dependency | |||||||
| Hoxb-8 | CID-9 | RT-PCR | [33*] | ||||
| Hoxb-9 | CID-9 E12.5 | RT-PCR; in situ | KO: lactational defects | [33*,38] | |||
| Embryo (MM) | |||||||
| Hoxc-6 | MG | Northern | Lost | Estrogen (-); developmental | [31] | ||
| HOXC-6 | MCF7; MCF7D; | cDNA screen; | Variable | [57,65] | |||
| Hs578Bst; | RT-PCR | ||||||
| MCF10F; T47D | |||||||
| Hoxc-8 | MG | Northern | Lost | Estrogen (-); developmental | [32] | ||
| HOXC-10 | MG | PCR of cDNA library | 2 | ||||
| Hoxd-3 | MG (UD) | Northern | OE (rarely) | [32] | |||
| Hoxd-4 | MG (E) | In situ | Lost | [32] | |||
| Hoxd-8 | MG | Northern | Lost | Developmental | [32] | ||
| Hoxd-9 | MG (E+S); | RT-PCR; | Lost | Developmental | KO: lactational defects | [32,38] | |
| E12.5 Embryo | Northern; in situ | ||||||
| (MM) | |||||||
| Hoxd-10 | MG (E+S) | Northern; in situ | Lost | Developmental | KO: lactational defects | [32,39];2 | |
| Hoxd-11 | MG (UD) | Northern | No | [32] | |||
| Hord-12 | MG | RT-PCR; | OE (myc tumor) | [31*] | |||
| Northern (UD) | |||||||
| Msx | Msx-1 | E13.5 Embryo (E); | Northern; in situ | Maintained but variable | Developmental | KO: none detected in embryos | [40*,41*] |
| MG (E) | |||||||
| Msx-2 | E13.5 Embryo (E); | Northern; in situ | Lost | Estrogen; developmental | KO: MG fail to form | [40*];3 | |
| MG (S) | |||||||
| Iroquois related | IRX-1 | MG | RT-PCR | [48*] | |||
| IRx-2 | MG; primary | RT-PCR; Northern; | Maintained, expression | Developmental | [48*] | ||
| tumors | in situ | levels or splice form | |||||
| usage may be altered | |||||||
| in some tumors | |||||||
| IRX-3 | MG | RT-PCR | [48*] | ||||
| IRX-4 | MG | RT-PCR | [48*] | ||||
| IRX-5 | MG | RT-PCR | [48*] | ||||
| Paired domain | Alx-4 | MG(S) | In situ | Expression associated with | KO: none detected | [45] | |
| actively condensing periductal | |||||||
| stroma | |||||||
| HSIX | HSIX-1 | MCF7 | OE | Expressed in S phase | OE in MCF7 abolishes X-ray induced | [58*] | |
| of cell cycle | cell cycle arrest in G2 phase | ||||||
| POU domain | OCT-1 | MCF7; MCF10; | RT-PCR; | [54] | |||
| SkBr3;MDA- | Western EMSA | ||||||
| MB453 | |||||||
| Oct-1 | MG (lact.) | RT-PCR | [53] | ||||
| OCT-2 | MCF7; MCF10; | ||||||
| SkBr3 | RT-PCR; | [54] | |||||
| Western EMSA | |||||||
| OCT-3 | MG (UD); MCF7; | RT-PCR | OE (freq.) | Form with a 5 amino acid deletion | [54] | ||
| MCF10; SkBr3; | detected om MCF7 cell | ||||||
| primary tumors | |||||||
| Oct-3 | MG (lact.) | RT-PCR | [53] | ||||
| OCT-11 | MG (UD) MCF7; | RT-PCR; EMSA | OE (freq.) | [54] | |||
| skBr3; MDA- | |||||||
| MB453; primary | |||||||
| tumors | |||||||
| Pit-rs 1 | MG (lact.) | RT-PCR | [53] | ||||
| Pou4f1 = Brn-3 | MG (lact.) | RT-PCR | [53] | ||||
| Pou4f-rs1 = | MG (lact.) | RT-PCR | [53] | ||||
| Brn-3R | |||||||
| Pou3f4 = Brn-4 | MG (lact.) | RT-PCR | [53] | ||||
| Pou3f-rs1 = | MG (lact.) | RT-PCR | [53] | ||||
| POU-IIIA | |||||||
| Engrailed related | En-1 | MG | Northern | UE (freq.) | Developmental | [32] | |
| En-2 | MG (UD) | Northern | UD | KO: none detechted | [32] | ||
| MOX | MOX1 | MG | [52] | ||||
| MEIS | MEIS2a | MG | RT-PCR of cDNA library | 4 | |||
| MEIS 2d | MG | RT-PCR of cDNA library | 4 | ||||
| MRG1a | MG | RT-PCR of cDNA library | 4 | ||||
bFGF, basic fibroblast growth factor; E, epithelium; EMSA, electrophoretic mobility shift assay; freq., frequently; KO, knockout mutation; lact., lactating; M, mammary mesenchyme; MG mammary gland; Northern, Northern blot assay; OE, overexpressed; RT-PCR, reverse transcriptase polymerase chain rection; S, periductal stroma; UD, undetectable; UE, underexpressed. aHuman genes are uppercase; mouse genes are lowercase. bCalle lines are listed by their common designation. If expression was detected in the intact mammary gland, the tissue compartment is shown. cNot all detection techniques have been used on all tissue types/cell lines shown. 1Friedmann Y, Daniel CW, unpublished results; 2Lewis MT, Daniel CW, unpublished results; 3Maas R, unpublished results, cited in [41]; 4Lewis MT, unpublished results.
Hox/HOX
In the mouse (and human), 39 known Hox (HOX) complex genes are arranged in four paralogous gene clusters, one on each of four different chromosomes. These genes are primarily responsible for anterior-posterior patterning of the body and limbs but organ-specific functions are also known. Targeted disruption (knockout) strains exist for most Hox genes, as do a limited number of overexpressing transgenic strains. Both types of mutants have rarely been examined for mammary defects and tumors, and overall phenotypic analysis of single gene mutations has been somewhat hampered by functional compensation from paralogous genes within the complex itself. Nevertheless, it is already clear that some Hox genes are critically important for proper mammary gland development and function (Table 1).
Early immunohistochemical studies and screens based on polymerase chain reaction first detected homeobox gene expression in mammary epithelial cell lines and tumors [29,30]. Subsequently, Friedmann and coworkers identified several Hox genes that were expressed in the normal mouse mammary gland and associated neoplasias (Table 1) [31,32]. In situ hybridization demonstrated that many of these genes are expressed either in the mammary epithelium or in the periductal stroma, or both (Fig. 2).
Figure 2.
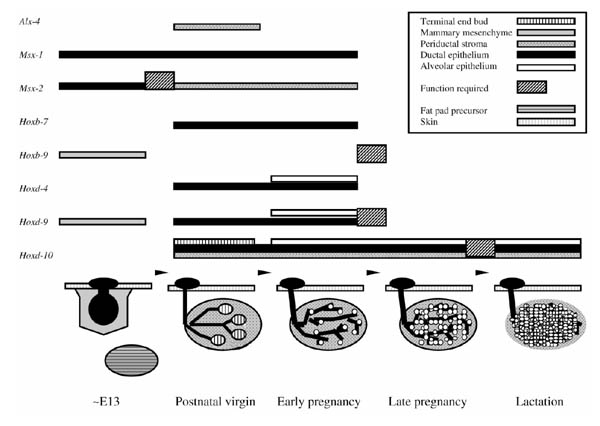
Tissue distribution of homeobox gene expression through mouse mammary gland development. Selected stages of mammary gland development are depicted with reference to the expression patterns of several homeobox genes as demonstrated by in situ hybridization. Expression at a given stage is shown by a bar above the stage. Bars are pattern coded to represent a unique tissue compartment or epithelial structure. Transition points affected by a given homeobox gene mutation are denoted by a hatched box above the arrow representing the transition.
Several genes, including Hoxc-6, Hoxc-8, Hoxd-8, Hoxd-9 and Hoxd-10, appeared to be regulated developmentally. Most intriguing was the demonstration that expression of some homeobox genes was sensitive to manipulation of the estrogen level. Together, these data provided the first substantive clues that homeobox genes are important for normal mammary development.
Soon after, Srebrow et al [33] identified five Hox genes expressed in CID-9 mouse mammary epithelial cells. When these cells are cultured on reconstituted basement membrane in the presence of lactogenic hormones, they form alveolar-like hollow spheres and differentiate to express milk proteins. Expression of two genes, Hoxa-1 and Hoxb-7, was shown to be downregulated by exogenous basement membrane, suggesting that homeobox gene expression may be modulated in vivo by interactions between epithelial cells and components of the basement membrane. Such interactions have been shown to be critical for mammary differentiation and function, and tend to be altered on neoplastic progression [34,35,36,37].
Taking cue from these studies, Chen and Cappechi [38] demonstrated compromised mammary function in mutant mice carrying various deletions of paralogous genes Hoxa-9, Hoxb-9 and Hoxd-9. Single mutant lines disrupted for either Hoxa-9 or Hoxb-9 (heterozygous at the remaining relevant Hox loci) showed only a small decrease in newborn survival, while the Hoxd-9 disruption alone reduced survival to below 50%. Double mutant combinations showed synergistic interaction that reduced newborn survival well below the additive expected values for each single mutation, suggesting functional cooperativity. Dams with the most severely affected genotypes (Aabbdd, aaBbdd and aabbdd) demonstrated marked hypoplasia of the mammary gland after parturition with gland morphology resembling that of a midpregnant animal.
During embryogenesis, Hoxb-9 and Hoxd-9 are each expressed in condensed mammary mesenchyme surrounding the epithelial bulb, suggesting roles in mesenchyme condensation or in modulation of epithelial-mesenchymal interactions during early mammary gland development [38]. Unfortunately, in situ hybridization against Hoxa-9 and Hoxb-9 using adult tissues has not been performed. However, Friedmann has examined expression of Hoxd-9 throughout postnatal development and showed it to be expressed highly in periductal fibroblasts and ductal epithelium in the virgin, but only expressed weakly in these cell types during pregnancy. Interestingly, Hoxd-9 showed enhanced expression in developing alveolar epithelium relative to subtending ducts, consistent with its apparent role in alveolar differentiation during pregnancy.
In situ hybridization has also demonstrated expression of Hoxd-10 in both epithelium and stroma with elevated levels in the epithelium of developing and secreting alveoli. These observations suggested a role in lobuloalveolar differentiation or the transition from pregnancy to lactation. Consistent with this hypothesis, targeted disruption of Hoxd-10 led to lactation failure in a significant percentage of homozygous mutant animals ([39]; Lewis Mt, Daniel CW, unpublished results). In glands of affected animals, alveolar development progressed through late pregnancy but failed, in whole or in part, to make the transition to lactation. In severely affected animals, alveoli failed to expand and increased pup mortality was observed. Together with the data for the group 9 paralogs, these observations firmly establish a functional role for Hox genes in alveolar differentiation and function.
Msx
Msx is a small family of three related genes. In the mouse, at least two members (Msx-1 and Msx-2) appear to play roles in mediating inductive tissue interactions during organogenesis. Only Msx-1 and Msx-2 have been examined in the mouse mammary gland [40,41].
Expression of Msx-1 and Msx-2 has been demonstrated in mammary buds during embryogenesis, with both genes expressed in the epithelium [41]. In contrast, in postnatal mice, Msx-1 and Msx-2 are expressed in reciprocal tissue compartments, with Msx-1 expressed in the epithelium and Msx-2 expressed in the periductal stroma (Fig. 2). The expression of Msx-2 in the fat pad was dependent on the presence of mamary epithelium. Results are suggestive of a fundamental transition in epithelial-stromal signaling that leads to tissue compartment switching of Msx-2 expression during organogenesis.
Expression studies also suggested a potential role in mediation of hormone responses. The Msx-2 mRNA was down-regulated in glands of ovariectomized and antiestrogen treated animals, and upregulated by estrogen replacement in ovariectomized animals. So far, Msx-1, Msx-2 and Msx-1/Msx-2 targeted disruption strains have been examined for mammary defects only during embryogenesis. No defects in embryonic mammary gland development were observed in Msx-1 mutants but Msx-2 and Msx-1/Msx-2 animals reportedly showed developmental defects as early as E13.5 ([41]; Maas R, unpublished results cited in [41]).
As with other organs, data from the mammary gland are consistent with a role in mediation of tissue interactions. Because homozygous Msx-1 and Msx-2 mutations are embryonic lethal, analyses of Msx gene function in the adult mammary gland are largely absent. Transplantation rescue experiments and gland reconstitution assays using embryonic tissue should be extremely useful in further examining Msx gene function in the adult mammary gland [42,43,44].
Paired domain class
Aristaless-like
The Aristaless-like gene Alx-4 is expressed in mesenchymal condensations of tissues whose development is dependent on epithelial-mesenchymal interactions [45]. In the mammary gland, Alx-4 expression was associated primarily with actively condensing stroma at the neck of terminal end buds (Fig. 2). Alx4 mutants failed to complement Strong's luxoid (lst) mutants and the two strains show similar defects [46,47]. Interestingly, mammary gland of lst mice appear normal, as do other ventral (but not dorsal) structures in which Alx-4 is expressed [45,46,47], although it is not clear to what extent the mammary glands from these animals were analyzed.
Iroquois-related
At least five members of the Iroquois-related homeobox (IRX) gene family are expressed in the human mammary gland [48]. One member, IRX-2, was shown to be expressed in discrete epithelial cell lineages being found in lumenal epithelium of both ducts and alveoli, but not myoepithelium. IRX-2 also showed developmental regulation: expression was enhanced in terminal buds and terminal lobules of the immature gland but uniformly distributed in mature glands. During lactation, some alveolar epithelial cells (~18%) showed reduced levels of mRNA relative to adjacent strongly expressing cells. Uniform epithelial expression was reestablished upon involution. Mutational analyses of these genes are underway in several laboratories.
Engrailed-related
Both Engrailed-related (En) En-1 and En-2 have been examined in the mouse mammary gland by Northern hybridization [32]. En-2 was not detected in any tissue examined; in contrast, En-1 was expressed at virtually constant levels through early pregnancy but was not detectable during late pregnancy or lactation. Homozygous En-1 mutations are embryonic lethal. Because neither embryonic glands nor epithelial transplants have been examined, it remains possible that En-1 serves a function in the mammary gland near the early→late pregnancy transition [49,50,51].
Other homeobox genes
Several other homeobox genes have been identified either in intact mammary glands of from cDNA library screens. These include members of the POU domain containing, MOX, and MEIS gene families ([52,53,54]; Lewis MT, Daniel CW, unpublished results). No functional or detailed expression data are yet available. Given the known importance of the POU and MEIS gene families in development and cancer, these should be among the first to be evaluated for mammary function.
Intrinsic versus extrinsic function: the Forkhead transcription factor Fkh-5/Mf3
Forkhead-related genes encode transcription factors containing a divergent 'winged helix' homeodomain. One of these genes, Fkh-5/Mf3, was shown to be required for brain development and postnatal growth with most homozygous null animals dead at birth. Surviving adult homozygous females were unable to feed their pups, yet did not show overt defects in mammary gland morphology or secretory function. Further analysis showed a defect in the milk letdown response. This defect was alleviated by injection with oxytocin and was probably caused by improper hypothalamic development [55,56].
This example illustrates the important point that the mammary gland cannot be considered an isolated organ. Rather, it must be considered an organ profoundly influenced by, and dependent upon, proper neuroendocrine function. Given that many homeobox genes affect development of the central nervous system and endocrine organs, it becomes imperative to demonstrate experimentally (eg via transplantation) that observed mammary defects are intrinsic to the gland and are not a downstream consequence of an extrinsic physiologic dysfunction.
Homeobox genes in mammary neoplasia
Embryogenesis and oncogenesis share several common features. Among them, both processes require cell proliferation, modulation of cell death (apoptosis), cell motility, invasion of surrounding tissue and neovascularization. In embryogenesis, the cells that accomplish these tasks are generally relatively undifferentiated. Similarly, in cancer progression, cells that contribute to neoplasias tend to appear relatively undifferentiated, or dedifferentiated, as the case may be.
This consideration leads to the following hypotheses. If cells require accurate spatial and temporal regulation of homeobox gene expression during embryogenesis to acquire the proper differentiated state (identity), misexpression of homeobox genes could lead to failure of differentiation, loss of the differentiated state or adoption of an alternative cell identity (a homeotic transformation). The expression profile of homeobox genes (and their targets) may then be taken as a crude measure of the identity or functional state of a given cell. If so, misregulation of homeobox genes during neoplastic progression may be indicative of progressive alteration of epithelial cell identity. If such cells consequently possessed inappropriate characteristics (eg loss of cell cycle control, decreased apoptosis, altered cell-cell adhesions, altered hormone and growth factor responses or increased protease expression, to name a few), misexpression of homeobox genes could easily contribute to cancer initiation or progression.
While evidence has accumulated that altered homeobox gene function plays a causal role in development of other types of cancers, particularly leukemias, there is only circumstantial evidence for their involvement in mammary neoplasia. However, when taken in aggregate and in the context of cancer in other organs, a number of observation suggest a contributory role of homeobox genes in the initiation or progression of mammary cancer. Again, the more suggestive existing data can be easily recounted by gene family and are now summarized.
Hox/HOX
Expression of all eight normally expressed homeobox genes examined (Hoxb-6, Hoxb-7, Hoxc-6, Hoxc-8, Hoxd-4, Hoxd-8, Hoxd-9 and Hoxd-10) was lost on neoplastic progression in a selected population of mouse hyperplasias and tumors [32]. Conversely, three genes not normally expressed were activated in subsets of tumors (Hoxa-1, Hoxd-3 and Hoxd-12).
Thus far, it appears that loss of function mutations simply affect developmental progression but that gain of function mutations impact neoplastic progression unpredicatable ways. For example, loss of function mutations of Hoxa-9/Hoxb-9/Hoxd-9 or of Hoxd-10 resulted in developmental failure, rather than tumor formation. As for gain of function mutations, whereas overexpression of human HOXA-10 in MCF7 cells promoted cell cycle arrest in G1, overexpression of murine Hoxb-7 in SkBr3 mammary cancer cells caused increased proliferation and decreased growth factor dependency [57]. These observations suggest that the consequences of a given Hox gene mutation cannot be predicted a priori, but will depend entirely on the nature of the mutation and on the battery of downstream genes misregulated as a result.
HSIX1
HSIX1 is expressed toward the end of S phase of the cell cycle in MCF7 cells [22,58]. Overexpression of HSIX1 in these cells abolished the G2 cell cycle checkpoint in response to irradiation, leading to inappropriate entry into mitosis. Interestingly, very low expression of HSIX1 was observed in the normal mammary gland, but elevated expression was observed in 44% of primary breast cancers and in an astonishing 90% of metastatic lesions. These data represent the only known involvement of homeobox genes in mammary cell cycle control at the G2 checkpoint. Given that loss of this and other checkpoints is associated with mammary cancer [59,60,61,62], these data represent the strongest indication to date that homeobox genes may contribute to neoplastic progression in the mammary gland.
Teasers
Msx
Msx-1 expression was maintained in all tumors examined [40]. In contrast, Msx-2 became undetectable in neoplasias. Given its reportedly critical role in embryonic mammary gland development, loss or alteration of stromal Msx-2 function in the adult mammary gland might be expected to have profound effects on interactions between the neoplastic epithelium and its associated stroma.
IRX
Similar to the expression pattern observed for Msx-1, IRX-2 expression was also maintained in all tumor types examined [48]. However, in some tumors, there was evidence of either increased expression or altered ratios of the two known transcripts. Since the IRX-2b transcript lacks the homeobox found in the IRX-2a transcript, target gene regulation may be expected to be altered.
POU domain
At least four POU class homeobox genes have been shown to be expressed in various human breast cancer cell lines, including OCT1, OCT2, OCT3 and OCT11 [54]. OCT3 and OCT11 were not detected in the normal human breast samples examined. Since OCT3 is generally considered to be an embryonic transcription factor, it is conceivable that these tumor cells adopted a more embryonic cell identity. Interestingly, Oct3 has also been detected in the normal mouse mammary gland during lactation [53], but the significance of this observation is not known.
Conclusion
Important questions remain with respect to homeobox gene function in the normal and neoplastic mammary gland. The studies summarized in this review represent only a small fraction of the homeobox genes likely to be involved. What has become evident from the more extensive studies is that, whereas a given homeobox gene may be expressed in regulated patterns throughout development (eg Msx-2, Hoxd-9, Hoxd-10), mutation of that gene may overtly affect only a single transition point or developmental stage (Fig. 2). If this general observation holds for other homeobox genes, functional analysis will require in vivo phenotypic evaluation of mutant animals throughout mammary organogenesis, including embryogenesis.
Despite the temptation to try, a unified model for homeobox gene function in the mammary gland is unlikely to be developed given the diverse functions and complex interactions known for many of these genes. As a consequence, the mechanistic details of how the various homeobox genes dictate mammary gland identity, form and function will require years of effort and careful attention to information derived from other model systems, particularly Drosophila. However, a general frame work can be postulated based on data gathered thus far and by analogy with known roles in other organs. In all likelihood, homeobox genes control mammary gland development and function by sequential activation or inactivation of specific sets of homeobox genes at specific developmental transition points. For example, Hoxa9, Hoxb9 and Hoxd9 appear to be important for alveolar development near midpregnancy, while Hoxd10 appears to be required at the next major transition point, the onset of lactation. As already suggested by currently available expression data, layered on top of this temporal regulation will no doubt be tissue specific and cell type specific regulation of gene function (Fig. 2).
With respect to normal development, it is interesting to recount what homeobox gene mutations have not been reported to cause in the mammary gland (yet?). So far, no alterations in nipple placement (patterning and cell identity), no alterations in mammary gland number (patterning and cell identity), no homeotic transformation of mammary tissues to other ectodermally derived tissues (eg those of the sweat or sebaceous glands, skin or hair follicle) and no demonstrated function during postnatal virgin development or early pregnancy have been observed.
As for a possible role in the development of mammary cancer, the initial case is far weaker than one might hope. However, lack of solid evidence at this early stage should not be taken as an indication that the hypothesis is fundamentally flawed. There are still a huge number of targeted disruption strains as well as appropriately engineered strains that overexpress candidate oncogenic homeobox genes to be made and examined. For example, notably absent from current studies, or poorly understood, are those gene families that have been implicated causally in development of other types of cancer, including PBX, PAX and MEIS.
Homeobox genes clearly represent attractive candidates for the control of mammary development and neoplastic progression. Unfortunately, given the limitations recounted in the introduction of this review, analysis of homeobox gene expression and function in mammary gland development and neoplasia will be an exceptional undertaking. Fortunately, the mammary gland has several advantages over many other developmental models that bypass many of these potential problems. Identification of target genes, upstream regulators and cofactors will be enhanced significantly by the coupling of mouse developmental genetics with powerful gene expression analysis techniques such as DNA microarray and real-time reverse transcription polymerase chain reaction, for which the mammary gland provides ample material. The problem of embryonic or perinatal lethality, particularly with animals carrying combinatorial mutations, can be addressed using two powerful approaches. First, engineered mouse strains carrying tissue-restricted gene disruptions or overexpressing transgenes can be generated, which should allow genetic analyses of otherwise lethal or detrimental mutant combinations. Second, transplantation techniques have been developed that allow transplantation of mammary epithelium or intact mammary glands from embryonic and postnatal sources into wild type host animals. Such manipulations allow development of otherwise moribund mammary tissue in a physiologically relevant in vivo setting.
The study of homeobox gene function in the mammary gland is truly a daunting task but, given what we already know, it should be well worth the effort.
References
- Daniel CW, Silberstein GB. Developmental biology of the mammary gland. . The Mammary Gland. 1987. pp. 3–36.
- Sakakura T. Mammary embryogenesis. The Mammary Gland. 1987. pp. 37–66.
- Russo J, Russo IH. Development of the human mammary gland. The Mammary Gland. 1987. pp. 67–93.
- Manak JR, Scott MP. A class act: conservation of homeodomain protein functions. Dev Suppl. 1994: 61–77. [PubMed] [Google Scholar]
- Thesleff I, Vaahtokari A, Partanen AM. Regulation of organogenesis. Common molecular mechanisms regulating the development of teeth and other organs. . Int J Dev Biol. 1995;39:35–50. [PubMed] [Google Scholar]
- Capecchi MR. Hox genes and mammalian development. Cold Spring Harb Symp Quant Biol. 1997;62:273–281. [PubMed] [Google Scholar]
- Mark M, Rijli FM, Chambon P. Homeobox genes in embryogenesis and pathogenesis. Pediatr Res. 1997;42:421–429. doi: 10.1203/00006450-199710000-00001. [DOI] [PubMed] [Google Scholar]
- Burglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucl Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MP, Tamkun JW, Hartzell GWd. The structure and function of the homeodomain. Biochim Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Stein S, Fritsch R, Lemaire L, Kessel M. Checklist: vertebrate homeobox genes. Mech Dev. 1996;55:91–108. doi: 10.1016/0925-4773(95)00494-7. [DOI] [PubMed] [Google Scholar]
- Mannervik M. Target genes of homeodomain proteins. Bioessays. 1999;21:267–270. doi: 10.1002/(SICI)1521-1878(199904)21:4<267::AID-BIES1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Schumacher A, Magnuson T. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- Kennison JA, Tamkun JW. Trans-regulation of homeotic genes in Drosophila. New Biol. 1992;4:91–96. [PubMed] [Google Scholar]
- Kennison JA. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- Hanson RD, Hess JL, Yu BD, et al. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. . Proc Natl Acad Sci USA. 1999;96:14372–14377. doi: 10.1073/pnas.96.25.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabin CJ. Why we have (only) five fingers per hand: hox genes and the evolution of paired limbs. Development. 1992;116:289–296. doi: 10.1242/dev.116.2.289. [DOI] [PubMed] [Google Scholar]
- Zakany J, Fromental-Ramain C, Warot X, Duboule D. Regulation of number and size of digits by posterior Hox genes: a dose-dependent mechanism with potential evolutionary implications. Proc Natl Acad Sci USA. 1997;94:13695–13700. doi: 10.1073/pnas.94.25.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Jacobs Y, Nakamura T, Jenkins NA, Copeland NG, Cleary ML. Meis proteins are major in vivo DNA binding partners for wildtype but not chimeric Pbx proteins. . Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan ML, Rambaldi I, Featherstone MS. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS. The specificity of homeotic gene function. Bioessays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- Stuart ET, Yokota Y, Gruss P. PAX and HOX in neoplasia. Adv Genet. 1995;33:255–274. doi: 10.1016/s0065-2660(08)60336-3. [DOI] [PubMed] [Google Scholar]
- Ford HL. Homeobox genes: a link between development, cell cycle, and cancer? Cell Biol Int. 1998;22:397–400. doi: 10.1006/cbir.1998.0329. [DOI] [PubMed] [Google Scholar]
- Cillo C, Faiella A, Cantile M, Boncinelli E. Homeobox genes and cancer. Exp Cell Res. 1999;248:1–9. doi: 10.1006/excr.1999.4451. [DOI] [PubMed] [Google Scholar]
- Deschamps J, Meijlink F. Mammalian homeobox genes in normal development and neoplasia. Crit Rev Oncog. 1992;3:117–173. [PubMed] [Google Scholar]
- van Genderen C, Okamura RM, Farinas I, et al. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- Dunbar ME, Wysolmerski JJ. Parathyroid hormone-related protein: a developmental regulatory molecule necessary for mammary gland development. . J Mammary Gland Biol Neoplasia. 1999;4:21–34. doi: 10.1023/a:1018700502518. [DOI] [PubMed] [Google Scholar]
- Dunbar ME, Dann PR, Robinson GW, Hennighausen L, Zhang JP, Wysolmerski JJ. Parathyroid hormone-related protein signaling is necessary for sexual dimorphism during embryonic mammary development. Development. 1999;126:3485–3493. doi: 10.1242/dev.126.16.3485. [DOI] [PubMed] [Google Scholar]
- Chepko G, Smith GH. Mammary epithelial stem cells: our current understanding. J Mammary Gland Biol Neoplasia. 1999;4:35–52. doi: 10.1023/a:1018752519356. [DOI] [PubMed] [Google Scholar]
- Wewer UM, Mercurio AM, Chung SY, Albrechtsen R. Deoxyribonucleic-binding homeobox proteins are augmented in human cancer. Lab Invest. 1990;63:447–454. [PubMed] [Google Scholar]
- Castronovo V, Kusaka M, Chariot A, Gielen J, Sobel M. Homeobox genes: potential candidates for the transcriptional control of the transformed and invasive phenotype. Biochem Pharmacol. 1994;47:137–143. doi: 10.1016/0006-2952(94)90447-2. [DOI] [PubMed] [Google Scholar]
- Friedmann Y, Daniel CA, Strickland P, Daniel CW. Hox genes in normal and neoplastic mouse mammary gland. Cancer Res. 1994;54:5981–5985. [PubMed] [Google Scholar]
- Friedmann Y. Expression and developmental role of homeobox containing genes during mouse mammary gland morphogenesis. . In Biology. 1995.
- Srebrow A, Friedmann Y, Ravanpay A, Daniel CW, Bissell MJ. Expression of Hoxa-1 and Hoxb-7 is regulated by extracellular matrix-dependent signals in mammary epithelial cells. J Cell Biochem. 1998;69:377–391. doi: 10.1002/(SICI)1097-4644(19980615)69:4<377::AID-JCB1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Robinson GW, Karpf AB, Kratochwil K. Regulation of mammary gland development by tissue interaction. J Mammary Gland Biol Neoplasia. 1999;4:9–19. doi: 10.1023/a:1018748418447. [DOI] [PubMed] [Google Scholar]
- Rosen JM, Wyszomierski SL, Hadsell D. Regulation of milk protein gene expression. Annu Rev Nutr. 1999;19:407–436. doi: 10.1146/annurev.nutr.19.1.407. [DOI] [PubMed] [Google Scholar]
- Streuli CH, Gilmore AP. Adhesion-mediated signaling in the regulation of mammary epithelial cell survival. J Mammary Gland Biol Neoplasia. 1999;4:183–191. doi: 10.1023/a:1018729308878. [DOI] [PubMed] [Google Scholar]
- Weaver VM, Bissell MJ. Functional culture models to study mechanisms governing apoptosis in normal and malignant mammary epithelial cells. J Mammary Gland Biol Neoplasia. 1999;4:193–201. doi: 10.1023/a:1018781325716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Capecchi MR. Paralogous mouse Hox genes, Hoxa9, Hoxb9, and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proc Natl Acad Sci USA. 1999;96:541–546. doi: 10.1073/pnas.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter EM, Goddard JM, Davis AP, Nguyen TP, Capecchi MR. Targeted disruption of Hoxd-10 affects mouse hindlimb development. Development. 1997;124:4505–4514. doi: 10.1242/dev.124.22.4505. [DOI] [PubMed] [Google Scholar]
- Friedmann Y, Daniel CW. Regulated expression of homeobox genes Msx-1 and Msx-2 in mouse mammary gland development suggests a role in hormone action and epithelial-stromal interactions. Dev Biol. 1996;177:347–355. doi: 10.1006/dbio.1996.0168. [DOI] [PubMed] [Google Scholar]
- Phippard DJ, Weber-Hall SJ, Sharpe PT, et al. Regulation of Msx-1, Msx-2, Bmp-2 and Bmp-4 during foetal and postnatal mammary gland development. Development. 1996;122:2729–2737. doi: 10.1242/dev.122.9.2729. [DOI] [PubMed] [Google Scholar]
- Brisken C, Park S, Vass T, Lydon JP, O'Malley BW, Weinberg RA. A paracrine role for the epithelial progesterone receptor in mammary gland development. Proc Natl Acad Sci USA. 1998;95:5076–5081. doi: 10.1073/pnas.95.9.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MT, Ross S, Strickland PA, et al. Defects in mouse mammary gland development caused by conditional haploinsufficiency of Patched-1 (Ptc-1). Development. 1999;126:5181–5193. doi: 10.1242/dev.126.22.5181. [DOI] [PubMed] [Google Scholar]
- Wiesen JF, Young P, Werb Z, Cunha GR. Signaling through the stromal epidermal growth factor receptor is necessary for mammary ductal development. . Development. 1999;126:335–344. doi: 10.1242/dev.126.2.335. [DOI] [PubMed] [Google Scholar]
- Hudson R, Taniguchi-Sidle A, Boras K, Wiggan O, Hamel PA. Alx-4, a transcriptional activator whose expression is restricted to sites of epithelial-mesenchymal interactions. Dev Dyn. 1998;213:159–169. doi: 10.1002/(SICI)1097-0177(199810)213:2<159::AID-AJA1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Qu S, Niswender KD, Ji Q, et al. Polydactyly and ectopic ZPA formation in Alx-4 mutant mice. Development. 1997;124:3999–4008. doi: 10.1242/dev.124.20.3999. [DOI] [PubMed] [Google Scholar]
- Qu S., Tucker SC, Ehrlich JS, et al. Mutations in mouse Aristaless-like4 cause Strong's luxoid polydactyly. Development. 1998;125:2711–2721. doi: 10.1242/dev.125.14.2711. [DOI] [PubMed] [Google Scholar]
- Lewis MT, Ross S, Strickland PA, Snyder CJ, Daniel CW. Regulated expression patterns of IRX-2, an Iroquois-class homeobox gene, in the human breast. Cell Tissue Res. 1999;296:549–554. doi: 10.1007/s004410051316. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Herrup K, Auerbach BA, Davis CA, Rossant J. Subtle cerebellar phenotype in mice homozygous for a targeted deletion of the En-2 homeobox. . Science. 1991;251:1239–1243. doi: 10.1126/science.1672471. [DOI] [PubMed] [Google Scholar]
- Hanks M, Wurst W, Anson-Cartwright L, Auerbach AB, Joyner AL. Rescue of the En-1 mutant phenotype by replacement of En-1 with en-2. Science. 1995;269:679–682. doi: 10.1126/science.7624797. [DOI] [PubMed] [Google Scholar]
- Loomis CA, Harris E, Michaud J, Wurst W, Hanks M, Joyner AL. The mouse Engrailed-1 gene and ventral limb patterning. Nature. 1996;382:360–363. doi: 10.1038/382360a0. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Cochran C, Rosenthal J, et al. Isolation of a diverged homeobox gene, MOX1, from the BRCA1 region on 17q21 by solution hybrid capture. Hum Mol Genet. 1994;3:1359–1364. doi: 10.1093/hmg/3.8.1359. [DOI] [PubMed] [Google Scholar]
- Jehn B, Chicaiza G, Martin F, Jaggi R. Isolation of three novel POU-domain containing cDNA clones from lactating mouse mammary gland. Biochem Biophys Res Commun. 1994;200:156–162. doi: 10.1006/bbrc.1994.1428. [DOI] [PubMed] [Google Scholar]
- Jin T, Branch DR, Zhang X, Qi S, Youngson B, Goss PE. Examination of POU homeobox gene expression in human breast cancer cells. Int J Cancer. 1999;81:104–112. doi: 10.1002/(SICI)1097-0215(19990331)81:1<104::AID-IJC18>3.3.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Labosky PA, Winnier GE, Jetton TL, et al. The winged helix gene, Mf3, is required for normal development of the diencephalon and midbrain, postnatal growth and the milk-ejection reflex. Development. 1997;124:1263–1274. doi: 10.1242/dev.124.7.1263. [DOI] [PubMed] [Google Scholar]
- Wehr R, Mansouri A, de Maeyer T, Gruss P. Fkh5-deficient mice show dysgenesis in the caudal midbrain and hypothalamic mammillary body. Development. 1997;124:4447–4456. doi: 10.1242/dev.124.22.4447. [DOI] [PubMed] [Google Scholar]
- Care A, Silvani A, Meccia E, Mattia G, Peschle C., Colombo MP. Transduction of the SkBr3 breast carcinoma cell line with the HOXB7 gene induces bFGF expression, increases cell proliferation and reduces growth factor dependence. Oncogene. 1998;16:3285–3289. doi: 10.1038/sj/onc/1201875. [DOI] [PubMed] [Google Scholar]
- Ford HL, Kabingu EN, Bump EA, Mutter GL, Pardee AB. Abrogation of the G2 cell cycle checkpoint associated with overexpression of HSIXI: a possible mechanism of breast carcinogenesis. Proc Natl Acad Sci USA. 1998;95:12608–12613. doi: 10.1073/pnas.95.21.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lowe M, Keyomarsi K. UCN-01-mediated G1 arrest in normal but not tumor breast cells is pRb-dependent and p53-independent. Oncogene. 1999;18:5691–5702. doi: 10.1038/sj/onc/1202948. [DOI] [PubMed] [Google Scholar]
- Jones JM, Cui XS, Medina D, Donehower LA. Heterozygosity of p21WAF1/CIP1 enhances tumor cell proliferation and cyclin D1-associated kinase activity in a murine mammary cancer model. Cell Growth Diff. 1999;10:213–222. [PubMed] [Google Scholar]
- Larson JS, Tonkinson JL, Lai MT. A BRCA1 mutant alters G2-M cell cycle control in human mammary epithelial cells. Cancer Res. 1997;57:3351–3355. [PubMed] [Google Scholar]
- Kuerbitz SJ, Plunkett BS, Walsh WV, Kastan MB. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. . Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chariot A, Moreau L, Senterre G, Sobel ME, Castronovo V. Retinoic acid induces three newly cloned HOXA1 transcripts in MCF7 breast cancer cells. Biochem Biophys Res Commun. 1995;215:713–720. doi: 10.1006/bbrc.1995.2522. [DOI] [PubMed] [Google Scholar]
- Rots NY, Liu M, Anderson EC, Freedman LP. A differential screen for ligand-regulated genes: identification of HoxA10 as a target of vitamin D3 induction in myeloid leukemic cells. Mol Cell Biol. 1998;18:1911–1918. doi: 10.1128/mcb.18.4.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chariot A, Castronovo V, Le P, Gillet C, Sobel ME, Gielen J. Cloning and expression of a new HOXC6 transcript encoding a repressing protein. Biochem J. 1996;319:91–97. doi: 10.1042/bj3190091. [DOI] [PMC free article] [PubMed] [Google Scholar]


