Abstract
Background
The traditional exact method for inferring relationships between individuals from genetic data is not easily applicable in all situations that may be encountered in several fields of applied genetics. This study describes an approach that gives affordable results and is easily applicable; it is based on the probabilities that two individuals share 0, 1 or both alleles at a locus identical by state.
Results
We show that these probabilities (zi) depend on locus heterozygosity (H), and are scarcely affected by variation of the distribution of allele frequencies. This allows us to obtain empirical curves relating zi's to H for a series of common relationships, so that the likelihood ratio of a pair of relationships between any two individuals, given their genotypes at a locus, is a function of a single parameter, H. Application to large samples of mother-child and full-sib pairs shows that the statistical power of this method to infer the correct relationship is not much lower than the exact method. Analysis of a large database of STR data proves that locus heterozygosity does not vary significantly among Caucasian populations, apart from special cases, so that the likelihood ratio of the more common relationships between pairs of individuals may be obtained by looking at tabulated zi values.
Conclusions
A simple method is provided, which may be used by any scientist with the help of a calculator or a spreadsheet to compute the likelihood ratios of common alternative relationships between pairs of individuals.
Background
The usual, long-established method of inferring relationships between individuals in forensic genetics is based on the population frequencies of the observed alleles and on the conditional probabilities of the observed genotypes, given two alternative hypothesized relationships [1]. In the more frequent instances, such as paternity testing of trios, or similar cases with deficiencies, well-known formulas have come into common use [2,3]. However, in more complex cases where, for example, the relationship between pairs of individuals from large samples is under investigation, or where the DNA profile of a number of related individuals is known and we want to know the most likely relationships among them, these calculations become exceedingly complex. Each particular problem requires the development of specific formulas, necessitating either the expertise of highly specialized professionals, or recourse to suitable computer programs [4-8] these latter, on the other hand, require trained personnel to be used. In addition, the exact method assumes knowledge of allele frequencies at the marker loci, which often show considerable variability between ethnic groups.
Examples of 'difficult' situations sometimes encountered in forensic science include attribution to missing individuals of one or more body remains [9], identification of the victims of mass disasters [10], validation of large databases of individual genetic profiles [11]. Examples from other fields include linkage analysis (investigators may want to verify the true relationships existing among reported relatives [12,13]), natural and domestic population studies (to resolve kin structures in the wild [14] or confirm the stock source of animal food [15]), and research in physical anthropology (in reconstructing genealogies when there are no civic records [16], or inferring relationships in ancient cemeteries [17,18]).
The increasing availability of highly polymorphic genetic markers and their decreasing cost of typing provide high power of resolving the true biological relationship between individuals even with methods that use only part of the genetic information, being at the same time more easily applicable. The aim of this work is to generalize a method for inferring relationships between pairs of individuals, based on the probabilities (here called z0, z1, z2) that two subjects with a given relationship share 0, 1 or 2 alleles identical by state at a locus. This approach was suggested by Chakraborty and co-authors [19,20], and was subsequently developed by others, generally in the context of genome wide linkage scans [21,22]. We first show that the values of zi for a certain relationship depend on the heterozygosity of a locus (H) and very little on the particular distribution of its allele frequencies; this property allows us to obtain regression equations relating the values of zi to H for the more common relationships; then, we compare the results of our method with those of the conventional exact approach in large samples of mother-child and full-sib pairs. Finally, we examine a large database of gene frequencies of human populations typed for loci commonly used in forensic science. Based on results from this analysis, the zi values of the CODIS and other loci [23] are tabulated for Caucasian populations; these may be directly used by any scientist with the help of a calculator or a spreadsheet to compute the likelihood ratios of common alternative relationships between any pair of individuals. In more general cases, the equations relating zi to H provide an easy way to compute the zi values to be applied to each particular problem.
Results
Relationship between zi and H
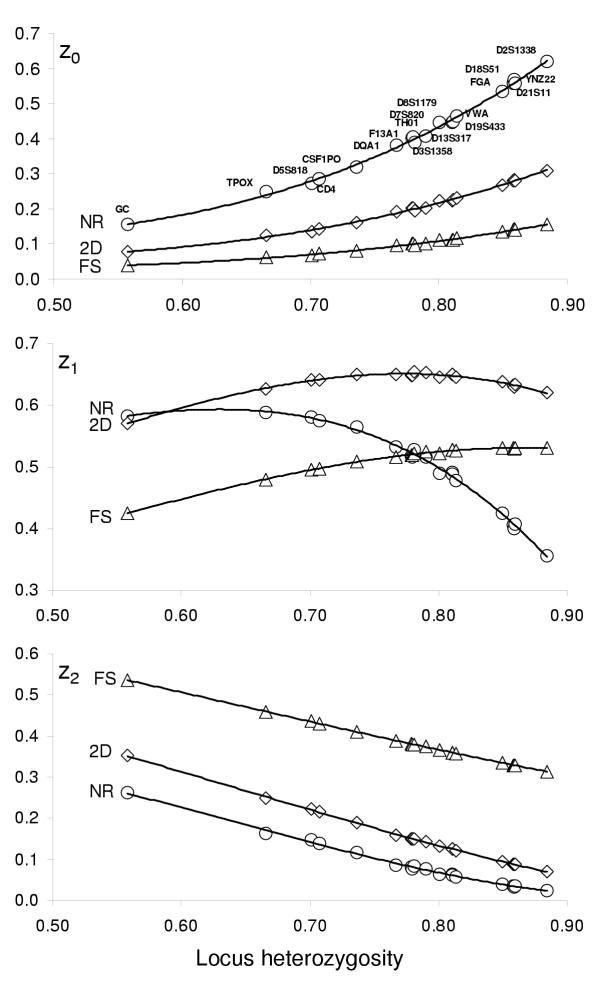
We first worked out approximate equations connecting the probabilities of sharing 0, 1 or 2 alleles (zi) to H for some common relationships and the loci more commonly used in forensic science. The rationale of this task was that these probabilities are exact functions of H when the loci are diallelic; in addition, the relationship between H and zi is linear for parent-child pairs regardless of the number of alleles (namely, z0 = 0, z1 = H, and z2 = 1 - H, see Table 1). However, the dependence of zi on H is not exact for other familial relationships and multi-allelic loci, though it seems reasonable to anticipate that a kind of functional dependence still exists. To investigate this issue, we computed the zi values for many loci with a variable number of alleles covering a wide range of H, and examined their variation among the loci with similar values of heterozygosity. Exact zi values were computed for 19 STR markers commonly used in forensic practice (list in Figure 1); they included the 12 CODIS loci. Marker GC (3 alleles) showed the lowest heterozygosity (H = 0.558), and marker D2S1338 (12 alleles) the highest (H = .885). Fig. 1 shows a plot of the zi values as a function of H for the three relationships FS (full sibs), 2D (any 2.nd degree relationship), and NR (non-relatives). The least-square fitting of these data to a third-order polynomial equation is also shown. It may be seen that H is an excellent predictor of z. There is only minor residual variation of actual zi values around the values of the interpolated equations. In addition, it appears that a third order polynomial is sufficient to obtain adequate approximation over the examined H range. The limit for H → 1 of the zi is of interest. When H = 1, all unrelated individuals are heterozygous for different alleles, and the probability that two siblings share 0, 1 or both alleles coincides with the identity by descent (IBD) probabilities, or 0.25, 0.5 and 0.25, respectively. It may be seen in Fig. 1 that the markers with highest H are in fact approaching these limits. Conversely, the probability of sharing 0 alleles approaches 1 for unrelated individuals, whereas the probability of sharing both alleles vanishes. This is what we see in Fig. 1, particularly in the case of two non-relatives sharing both alleles. All that gives an intuitive justification of the observed strict dependence of the zi values on H.
Table 1.
Probabilities of genotype combinations and allele sharing for several common relationships
| Parent-Child | Full sibs | 2.nd degree | Non-relatives | ||
| A) Genotype combination | |||||
| 1 | AA, AA | pA3 | pA2(1+pA)2/4 | pA3(1+pA)/2 | pA4 |
| 2 | AA,AB | 2pA2pB | pA2pB(1+pA) | pA2pB(1+2pA) | 4pA3 pB |
| 3 | AA,BB | 0 | pA2pB2/2 | pA2pB2 | 2pA2pB2 |
| 4 | AB,AB | pApB(pA+pB) | pApB(2pApB+pA+pB+1)/2 | pApB(4pApB+pA+pB)/2 | 4pA2pB2 |
| 5 | AA,BC | 0 | pA2pBpC | 2pA2pBpC | 4pA2pBpC |
| 6 | AB,AC | 2pApBpC | pApBpC(2pA+1) | pApBpC(4pA+1) | 8pA2pBpC |
| 7 | AB,CD | 0 | 2pApBpCpD | 4pApBpCpD | 8pApBpCpD |
| B) Number of shared alleles (Z) | |||||
| 0 | 0 | H2/8 | H2/4 | H2/2 | |
| 1 | H | H(1-H/2) | 3H/2-H2 | 2H-2H2 | |
| 2 | 1-H | 1-H(1-3H/8) | 1-3H/2+3H2/4 | 1-2H+3H2/2 | |
A (top): probabilities of the seven possible combinations of genotypes for multi-allelic loci in pairs of individuals, conditional on their relationship, as functions of allele frequencies. B (bottom): probabilities of sharing 0, 1, or 2 alleles for diallelic loci as functions of locus heterozygosity.
Figure 1.
Relationship between heterozygosity and zi Probabilities of sharing 0, 1 or both alleles at 19 loci as a function of locus heterozygosity for three common relationships (Full sibs, 2nd degree and non-relatives). Lines represent third-order polynomial regression curves.
Table 2 shows the polynomial regression equations fitting the six data series. These equations provide a general way to compute the values of z0, z1, and z2 for any multi-allelic marker based on its heterozygosity. We used these equations to calculate the likelihood ratio (LR) of alternative relationships between individual pairs, in order to compare the statistical power of the IBS method with that of the exact method.
Table 2.
Equations relating heterozygosity to zi
| Full sibs | ||||
| z0 | 0.0035 + | 0.1914 H - | 0.5815 H2 + | 0.6324 H3 |
| z1 | 0.2212 - | 0.2272 H + | 1.7586 H2 - | 1.2504 H3 |
| z2 | 0.7753 + | 0.0358 H - | 1.1771 H2 + | 0.6181 H3 |
| Half sibs | ||||
| z0 | 0.0070 + | 0.3829 H - | 1.1630 H2 + | 1.2647 H3 |
| z1 | 0.4423 - | 0.9544 H + | 3.5173 H2 - | 2.5009 H3 |
| z2 | 0.5507 + | 0.5715 H - | 2.3543 H2 + | 1.2362 H3 |
| Non-relatives | ||||
| z0 | 0.0140 + | 0.7658 H - | 2.3259 H2 + | 2.5295 H3 |
| z1 | 0.8847 - | 2.9088 H + | 7.0345 H2 - | 5.0018 H3 |
| z2 | 0.1013 + | 2.1431 H - | 4.7086 H2 + | 2.4723 H3 |
LRs compared between the IBS method and the exact method
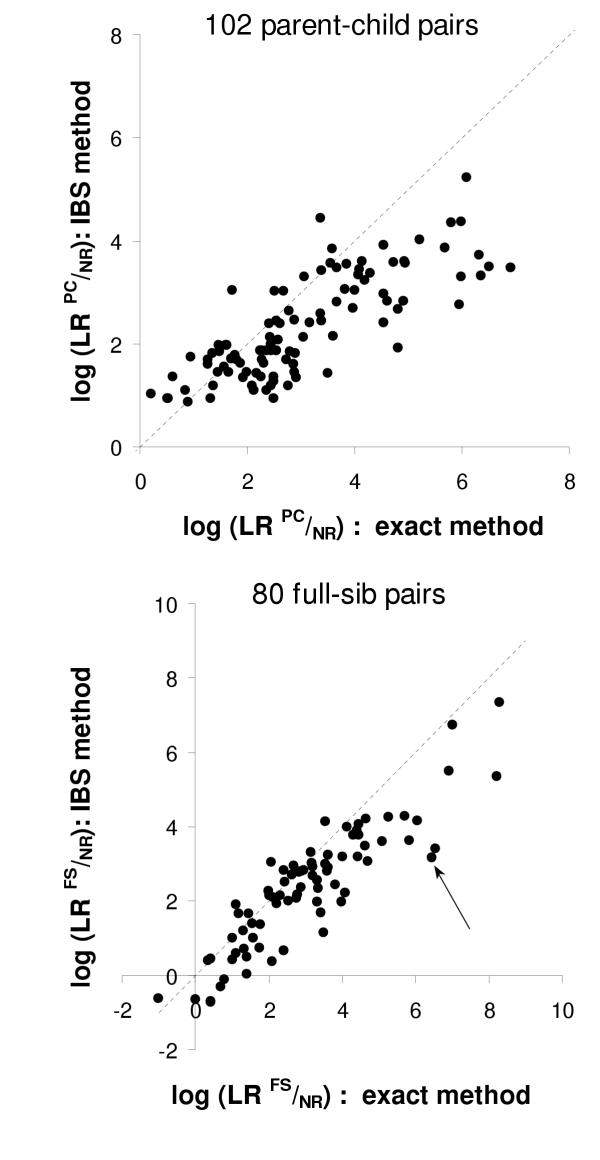
Our historical series of disputed paternity trios (mother/child/alleged father) included 102 cases, typed for a variable number of markers. We selected the mother and the child from each trio, and computed the probabilities that they were true parent-child pairs and non-relative pairs for each locus using both the conventional method and the IBS method (equations from Table 1 and 2). The two resulting LRs were converted to logarithm to base 10, and these were summed over all loci. The final values produced by the two methods were thus directly comparable. Fig. 2A shows the results of this analysis. Each point represents a parent-child pair, where the X axis is the log(LR) computed by the exact method and the Y axis is the log(LR) computed by the IBS method. In case of perfect correspondence between the two methods, the points would lay on the diagonal line. Fig. 2B shows the same analysis applied to full-sib data.
Figure 2.
Contrasting likelihood ratios between the exact and the IBS method Likelihood ratios that 102 true parent-child pairs (top) are parent-child pairs rather than non-relatives, conditional on their genotypes at multiple loci, calculated by the exact method (X axis) and by the IBS method (Y axis). Bottom: same analysis applied to 80 true full-sib pairs.
In both cases, the majority of points were located below the diagonal line. This means that the exact method is generally more powerful than the IBS method. Table 3 shows, for increasing values of log(LR), the percentage of pairs with values higher than that value, for both the PC and the FS pairs. The table also report the LRs in linear scale and the associated probabilities. For example, 85.3% of PC pairs get a probability >95% of being PC rather than unrelated using the exact method, versus 75.5% using the IBS method; for the FS pairs, the corresponding percentages are 77.5 and 72.5. At the probability level of 99%, the percentages are 74.5 vs. 50.0 for PC pairs and 71.3 vs 63.8 for FS pairs. It appears that a decreasing fraction of pairs get very high probability values using the IBS method. In other words, the exact method produces LRs comparably higher when the available information to infer relationships is very high. However, the two methods produce comparable results at the probability levels usually considered in the scientific work (95% or 99%). It is also noteworthy that in a relevant percentage of cases (26 out of 102 PC pairs, or 25%, and 18 out of 80 FS pairs, or 23%) the IBS method provided higher evidence for the correct relationship. This means that it may be particularly useful to apply both methods to borderline cases. In addition, the two methods produced highly correlated values. The values of Pearson correlation coefficients were 0.789 for PC pairs and 0.892 for FS pairs. We computed the LRs that the true FS pairs were HS pairs by the same approach (data not shown), and the correlation between the two methods was still higher (r = 0.962). This means that the values produced by the exact method can be predicted using the IBS method with good confidence, albeit after taking into account that the values of the exact method are on average higher than those produced by the IBS method. This may help deciding, for example, when the collected evidence for a certain problem is sufficient for the given purposes, or it is advisable to type additional loci, before embarking in complex calculations.
Table 3.
Percentages of pairs with LR higher than given cut-off values
| Parent-child | Full sibs | |||||
| Log(LR) | LR | Probablity | Exact method | IBS method | Exact method | IBS method |
| >0 | 1 | 0.500 | 100.0% | 100.0% | 98.8% | 92.5% |
| >0.5 | 3.2 | 0.760 | 98.0% | 100.0% | 92.5% | 86.3% |
| >1 | 10 | 0.909 | 93.1% | 95.1% | 87.5% | 80.0% |
| >1.5 | 31.6 | 0.969 | 85.3% | 75.5% | 77.5% | 72.5% |
| >2 | 100 | 0.990 | 74.5% | 50.0% | 71.3% | 63.8% |
| >2.5 | 316 | 0.997 | 56.9% | 39.2% | 60.0% | 50.0% |
| >3 | 1000 | 0.999 | 42.2% | 30.4% | 47.5% | 32.5% |
| >3.5 | 3162 | 0.9997 | 34.3% | 14.7% | 36.3% | 20.0% |
| >4 | 10000 | 0.9999 | 26.5% | 4.9% | 27.5% | 12.5% |
| >4.5 | 31622 | 0.99997 | 20.6% | 1.0% | 17.5% | 5.0% |
| >5 | 1.0E+05 | 0.99999 | 10.8% | 1.0% | 13.8% | 5.0% |
| >5.5 | 3.2E+05 | 0.999997 | 9.8% | – | 11.3% | 3.8% |
| >6 | 1.0E+06 | 0.999999 | 4.9% | – | 8.8% | 2.5% |
| >6.5 | 3.2E+06 | 0.9999997 | 2.0% | – | 6.3% | 2.5% |
| >7 | 1.0E+07 | 0.9999999 | – | – | 2.5% | 1.3% |
| >7.5 | 3.2E+07 | 0.99999997 | – | – | 2.5% | – |
| >8 | 1.0E+08 | 0.99999999 | – | – | 2.5% | – |
Variation of STR heterozygosity among populations
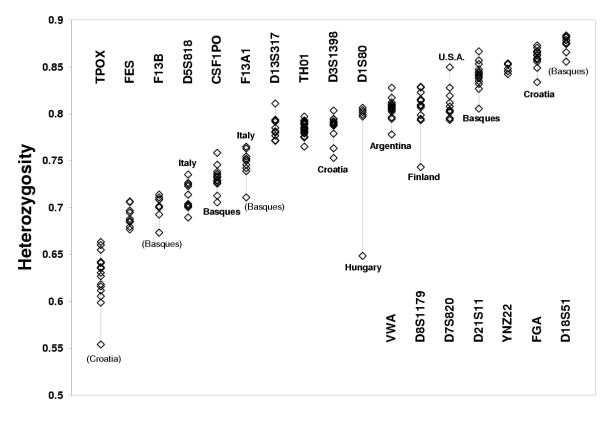
As locus heterozygosities are critical in determining the values of z0, z1, z2, we investigated its variation among human populations. We extracted the allele frequency data from "The Distribution of the Human DNA-PCR Polymorphisms" on-line database, and computed heterozygosities and their standard deviations of 122 different population samples. These included a total of 452 values of H from 17 loci (the D16S539 locus was added to the database later, and it was not considered in this analysis). After merging data that obviously referred to the same population, we ended up with 74 different populations typed for 1 to 17 markers (373 values of H). The Caucasoid group of populations included 56% of all data, and consisted of k = 27 different populations, typed for one (Cyprus, Slovakia, Albania, Australia) to 17 loci (Germany, Italy). By pairing all populations to each other, separately for all loci, and applying Tukey's multiple comparison method, we found the following results. Seven loci did not show any pair comparison of H exceeding the 0.01 critical value of the studentized range distribution (D13S317, k = 11; D18S51, k = 11; F13B, k = 8; FES, k = 9; TPOX, k = 15; TH01, k = 23; YNZ22, k = 5). The remaining ten loci showed a single outlier population, as follows: D1S80 (k = 6, Hungary was outlier with lowest H); CSF1PO (k = 18, Basques outlier with lowest H), D21S11 (k = 14, Basques outlier with lowest H), VWA (k = 15, Argentina outlier with lowest H), D3S1358 (k = 12, Croatia outlier with lowest H), D5S818 (k = 10, Italy outlier with highest H), F13A1 (k = 10, Italy outlier with highest H), FGA (k = 16, Croatia outlier with lowest H), D7S820 (k = 11, USA outlier with highest H), D8S1179 (k = 14, Finland outlier with lowest H). In conclusion, the Basques, the Croatians and Italians were outliers for two loci each (the first two with low H, the last with high H), and Argentina, Finland, Hungary and USA were outlier for a single locus each. Figure 3 shows a plot of H of all tested populations; loci are ordered by increasing value of the weighted mean of H. The populations that had outlier results at the test are indicated; in addition, it may be noted that the Basques scored lowest in H for three additional loci (F13B, F13A1, and D18S51), though the test was non-significant in these cases; similarly, the sample from Croatia showed a markedly lower heterozygosity than the average for a locus (TPOX) that was not significant at the test. These analyses show that marker heterozygosity is sufficiently stable across a major ethnic group as to permit in most cases the use of a single averaged value for its sub-populations, apart from special cases.
Figure 3.
Heterozygosities of 17 loci in samples from 27 Caucasian populations For each locus, the populations that resulted outliers at the Tukey multiple comparison test are indicated (bold); populations in brackets were non-significant at the test, though they point to samples with heterozygosity consistently lower than average (Basques and Croatians).
Table 4 shows the values of the zi values computed by the equations of Table 2 for PC, FS, 2D, and NR pairs for the Caucasian populations and for 18 markers, including CODIS loci and other commonly used STRs. These values may be directly used to infer the relationship existing between pairs of individuals, when they are assumed to come from a Caucasian population that has no particular reasons of being more inbred than average. In other cases, equations from table 2 may be used to calculate the appropriate zi values by supplying a population-specific value for H.
Table 4.
Probabilities of zi for 18 loci and four common relationships
| PARENT-CHILD | FULL SIB | SECOND DEGREE | NON RELATIVES | |||||||||
| MARKER |
z0 |
z1 | z2 | z0 | z1 | z2 | z0 | z1 | z2 | z0 | z1 | z2 |
| TPOX | 0 | 0.632 | 0.368 | 0.052 | 0.465 | 0.483 | 0.104 | 0.613 | 0.283 | 0.208 | 0.593 | 0.199 |
| FES | 0 | 0.690 | 0.310 | 0.067 | 0.491 | 0.442 | 0.133 | 0.637 | 0.230 | 0.266 | 0.584 | 0.150 |
| F13B | 0 | 0.708 | 0.292 | 0.072 | 0.498 | 0.430 | 0.144 | 0.642 | 0.214 | 0.288 | 0.576 | 0.136 |
| D5S818 | 0 | 0.719 | 0.281 | 0.076 | 0.502 | 0.422 | 0.151 | 0.645 | 0.204 | 0.303 | 0.571 | 0.127 |
| CSF1PO | 0 | 0.731 | 0.269 | 0.080 | 0.506 | 0.414 | 0.160 | 0.647 | 0.193 | 0.319 | 0.563 | 0.117 |
| F13A1 | 0 | 0.747 | 0.253 | 0.086 | 0.512 | 0.403 | 0.172 | 0.650 | 0.179 | 0.343 | 0.552 | 0.105 |
| D16S539 | 0 | 0.772 | 0.228 | 0.096 | 0.519 | 0.386 | 0.192 | 0.651 | 0.157 | 0.384 | 0.530 | 0.087 |
| D13S317 | 0 | 0.786 | 0.214 | 0.102 | 0.522 | 0.376 | 0.204 | 0.651 | 0.145 | 0.408 | 0.515 | 0.077 |
| THO1 | 0 | 0.787 | 0.213 | 0.102 | 0.522 | 0.376 | 0.205 | 0.651 | 0.145 | 0.410 | 0.514 | 0.077 |
| D3S1358 | 0 | 0.790 | 0.210 | 0.104 | 0.523 | 0.374 | 0.207 | 0.650 | 0.142 | 0.414 | 0.511 | 0.075 |
| D1S80 | 0 | 0.799 | 0.201 | 0.108 | 0.525 | 0.368 | 0.216 | 0.649 | 0.135 | 0.432 | 0.500 | 0.069 |
| VWA | 0 | 0.807 | 0.193 | 0.112 | 0.526 | 0.362 | 0.224 | 0.648 | 0.128 | 0.447 | 0.489 | 0.063 |
| D8S1179 | 0 | 0.814 | 0.186 | 0.115 | 0.527 | 0.358 | 0.231 | 0.647 | 0.122 | 0.461 | 0.480 | 0.059 |
| D7S820 | 0 | 0.817 | 0.183 | 0.116 | 0.527 | 0.356 | 0.233 | 0.647 | 0.121 | 0.466 | 0.477 | 0.058 |
| D21S11 | 0 | 0.848 | 0.152 | 0.133 | 0.531 | 0.336 | 0.267 | 0.637 | 0.096 | 0.534 | 0.426 | 0.040 |
| YNZ22 | 0 | 0.849 | 0.151 | 0.134 | 0.531 | 0.335 | 0.268 | 0.637 | 0.095 | 0.536 | 0.424 | 0.040 |
| FGA | 0 | 0.861 | 0.139 | 0.141 | 0.531 | 0.328 | 0.282 | 0.632 | 0.086 | 0.565 | 0.402 | 0.034 |
| D18S51 | 0 | 0.878 | 0.122 | 0.151 | 0.531 | 0.318 | 0.303 | 0.623 | 0.074 | 0.606 | 0.368 | 0.026 |
Probabilities of sharing 0, 1, or 2 alleles at 18 loci commonly used in the forensic practice, and for the indicated relationships, ordered by increasing value of heterozygosity (corresponding to parent-child z1 values); heterozygosity values were computed as weighted means among Caucasian populations.
Discussion
Inferring the biological relationships existing between two or more specimens using genetic polymorphisms is a cornerstone task in forensic science, which is also encountered in a variety of problems of applied genetics. Application of the conventional exact method requires three critical steps: 1) identification of the correct genotype combination for each locus (among seven possible types); 2) identification of the allele(s) whose frequency must be entered into the appropriate formula; 3) identification of the proper allele frequencies to be used in the calculation. Steps 1 and 2 are computationally tricky; if large databases have to be examined to identify first-degree relatives among unrelated individuals, one must recourse to programs developed by experts. Point 3 is also computationally not trivial (it requires to look up values in external tables to be imported into the specific formula appropriate for each pair), but it is also critical from a conceptual point of view, as sometimes we are uncertain about the allele frequencies that are appropriate for a given problem. This latter problem is particularly important, for example, in mass disasters, when large numbers of ethnically diverse victims must be screened against large numbers of possible relative matches. In such a situation, use of H in the IBS method may be much more appropriate than using incorrect "average" values of allele frequencies in the exact method. Once a match has been made for a pair of relatives, then its exact probabilities can be computed with the exact method using the ethnically appropriate allele frequencies.
In this study, we have shown that an approach based on the number of alleles shared IBS at each locus may be conveniently used for the purpose of inferring relationships. In this method, the probability of a certain relationship, given the genotypes observed in a pair, depends on a single parameter, the locus heterozygosity (H). This makes it easy to handle even large volume of data in a single spreadsheet, since these probabilities depend on the number of shared alleles (0, 1 or 2) and on a constant (H). For example, Presciuttini et al. [17] were interested in the relationships connecting 26 individuals buried in a 18th century cemetery; application of the IBS method to all possible pairs of this sample was not only computationally easier than the exact method, but it was also theoretically more robust, as it did not require making inferences about the allele frequencies that characterized the population, but only required assumptions about H. The functional dependence on H is one of the main advantages of the IBS approach. Heterozygosity, being a composite parameter, is inherently less variable among populations than individual allele frequencies. To examine this issue in real data, we analysed the sampling variance of H in a large database of allele frequencies, and concluded that heterozygosity is sufficiently homogeneous, at least among Caucasian populations, as to justify the adoption of a single common mean value, apart from special cases of historically isolated groups. Based on this observation, we tabulated the values of the zi for the more common relationships and the more frequently used loci. The values of H of these 18 loci span from 0.632 (TPOX) to 0.878 (D18S51), thus covering a wide range of values. This table has two main applications. The first concerns all those studies in which the individuals belong to a population whose allele frequencies are unknown (e.g., immigrants from poorly investigated ethnic groups or large samples of ethnically mixed victims and putative relatives); in this situation, applying the exact method is problematic, due to the uncertainty about the proper allele frequencies to be used, whereas the IBS method is straightforward. The second application concerns the cases where a new locus has been typed in a certain population, maybe of an animal species, and its heterozygosity is known; in this case, one may use the tabulated values of a locus with a similar heterozygosity, or may interpolate them (for out-of-range loci, or for more precise results, the regression equations may be used). More generally, using the tabulated zi values makes it easy to obtain a first-hand inference about the relationship between any two samples by simple inspection of genotype data. One may write down, given the number of alleles the pair shares at a locus, the corresponding probabilities of any two relationships to be tested, and then multiply the ratios of the two probabilities across all typed loci. The exact approach is more cumbersome and error prone, as it requires a table of formulas to be applied to each genotype combination and a table of allele frequencies from which one obtains the correct frequencies.
The main disadvantage of the IBS method is, of course, its reduced power. If the results are part of a legal case, all available information must be used to support a given hypothesis, and the exact method must always be used if it is applicable. However, there are many instances in which a standard significance level (0.05 or 0.01) is acceptable for screening a large database or to draw provisional scientific conclusions, and the IBS method may reach these limits even with a small number of typed loci, at least for discriminating first-degree relatives from non-relatives.
A worked example may be useful. Table 5 shows the genotypes at all 13 typed markers for a particular sib pair of our series (marked with an arrow in Fig. 2). This pair was chosen for being the more discordant full sib pair, with a LR much higher under the conventional method (log(LR) = 6.4 vs 2.9, respectively). Table 5 compares, locus by locus, the results of the exact calculation (columns 4–7) with the results of the IBS method (columns 8–10). For the exact calculation, the genotype combinations from Table 1A are listed, then the alleles whose frequency must be entered in the appropriate equations are shown, and the likelihood ratios that these two subjects were full sibs rather than non-relatives are calculated. The logarithm of the product of all LRs and corresponding probability are indicated in the last rows. The next three columns show the likelihood ratios computed according to the IBS method. Given the observed number of shared alleles between the two subjects, the LRs of these relationships were obtained by looking for the appropriate values in Table 4. For the two markers LPL and D19S253 (not included in Table 4), the probabilities of sharing the observed number of alleles and the corresponding LRs were obtained using the equations of Table 2, with heterozygosities computed from sample data.
Table 5.
Comparison of exact and IBS methods in a particular case
| Genotype data | Exact calculation | IBS method | Ratio LR1/LR2 | |||||||
| Marker | Sib1 | Sib2 | Genotype combination(1) | Allele frequencies(2) | Likelihood ratio(3) | Shared alleles | H(4) | Likelihood ratio(5) | ||
| CSF1PO | 9–13 | 9–13 | 4 | p9 = 0.044 | p13 = 0.062 | 50.93 | 2 | 3.52 | 14.5 | |
| FGA | 20–24.2 | 20–24.2 | 4 | p20 = 0.156 | p24.2 = 0.009 | 103.97 | 2 | 9.78 | 10.6 | |
| FES | 12–13 | 12–13 | 4 | p12 = 0.266 | p13 = 0.047 | 13.38 | 2 | 2.94 | 4.6 | |
| F13A1 | 5–5 | 5–5 | 1 | p5 = 0.184 | 10.32 | 2 | 3.83 | 2.7 | ||
| D19S253 | 8–12 | 12–12 | 2 | p12 = 0.369 | 0.93 | 1 | 0.760 | 0.35 | 2.7 | |
| F13B | 6–8 | 6–10 | 6 | p6 = 0.080 | 1.81 | 1 | 0.86 | 2.1 | ||
| LPL | 10–12 | 10–10 | 2 | p10 = 0.275 | 1.16 | 1 | 0.723 | 0.89 | 1.3 | |
| TH01 | 9.3–9.3 | 9–9.3 | 2 | p9.3 = 0.241 | 1.29 | 1 | 1.02 | 1.3 | ||
| D18S51 | 13–19 | 14–17 | 7 | 0.25 | 0 | 0.25 | 1.0 | |||
| D8S1179 | 13–15 | 13–15 | 4 | p13 = 0.344 | p15 = 0.116 | 4.82 | 2 | 6.07 | 0.8 | |
| TPOX | 8–11 | 8–10 | 6 | p8 = 0.512 | 0.49 | 1 | 0.78 | 0.6 | ||
| VWA | 17–18 | 17–18 | 4 | p17 = 0.275 | p18 = 0.206 | 3.52 | 2 | 5.71 | 0.6 | |
| D21S11 | 63–67 | 63–65 | 6 | p63 = 0.244 | 0.76 | 1 | 1.17 | 0.6 | ||
| Cumulative log(LR) | 6.47 | 2.92 | ||||||||
| Probability | >99.999% | 99.9% | ||||||||
Calculations applied to the most discordant pair from Fig. 2 are fully displayed. (1)from Table 1A; (2)calculated from full-sib sample data; (3)FS rather than NR, computed using formulas from Table 1A; (4)computed from sample data; (5)FS rather than NR, computed from values in Table 3 or applying formulas from Table 2 to the displayed H values.
In Table 5, markers are arranged in decreasing order of the ratio between the two LRs (last column). It may be seen that the three topmost markers provide most of the bias in favor of the exact method, and this is clearly the consequence of the occurrence, in this particular pair, of rare alleles. This highlights the major difference between the two methods. In the exact method, the frequencies of the observed alleles are both necessary for the calculation and critical. They are necessary because they contain all information we can use for inference, and they are critical because small changes of their values may cause large variations of the resulting likelihood ratios. The exact method assumes the allele frequencies are known without error; if they are misspecified (because of poor quality of published estimates, inadequate information about the ethnicity of the members of the putative pair, etc.), then the results of the exact method will be incorrect. When the alleles shared by any two individuals are rare, the LR that they are related may reach high values. In the IBS method, the frequencies of the observed alleles are irrelevant, so that we do not expect to find high peaks of LR in any pair. However, the occurrence of rare alleles in random pairs of individuals is also rare, so that, on the average, the power of the IBS method is not much lower than that of the exact method. This was apparent in our analysis of true parent-child and full-sib pairs; the exact method produced a tail of pairs with very high LR, whereas the IBS method appeared to be more constrained in the upper bound.
Conclusions
In conclusion, the IBS method presented here may be conveniently used as a preliminary approach to investigate the relationship existing between any pair of individuals. It can be applied by anybody using a desk calculator or a spreadsheet. Future work based on extensive computer simulations will address issues that we have not examined here. These include analysis of statistical power (which requires considering the distribution of LR when assumed relationships are false), the effects of typing errors and gene mutations, the robustness of the method to deviations in any of the assumptions (such as taking average H values).
Using the IBS method may help deciding when the collected evidence for a certain problem is sufficient for the purposes, or it is advisable to type additional loci, before embarking in exact calculations. In addition, the IBS method's using of estimates of H rather than of allele frequencies makes the IBS method particularly attractive in all those cases where ethnicity pose a problem, since H varies less across ethnicities. Furthermore, the results of the IBS method may even be accepted without further analyses in certain circumstances, since the LRs are highly correlated with those calculated by the exact method. Of course, the exact method should always follow IBS analysis when the results are critical to living human subjects.
Methods
Methodology outline
The conventional approach to determine the biological relationship existing between pairs of individuals is based on the probability P(X|R) of the observed marker genotypes (X), conditional on a certain relationship R. Here, X may be a multi-locus genotype. The collected evidence is then summarized in the form of a likelihood ratio of two alternative hypotheses, the probability of observation X given the relationship R1 and the probability of the same observation X given the relationship R2. Seven possible configurations of genotypes (regardless of order) are generally possible for two individuals and a multi-allelic locus [24]; Table 1A shows the formulas expressing P(X|R) for the following four relationships: 1) parent-child (PC), 2) full sibs (FS), 3) second degree relationships, including half sibs, avuncular pairs, and grandparent-grandchild pairs (2D), and 4) non-relatives (NR), as a function of the allele frequencies at a single locus.
In the IBS method, we consider the probabilities P(Z|R) that two individuals share 0, 1 or 2 alleles identical by state (Z) at a locus (again, Z may be a multi-locus vector), given a certain relationship R. Thus, the particular genotypes observed in each individual are irrelevant, as the observed variable Z is the number of alleles they have in common. In the case of a diallelic locus, these probabilities (z0, z1, z2, for Z = 0, Z = 1, and Z = 2, respectively) were easily obtained for the most common relationships as simple functions of the locus heterozygosity H (Table 1B). As the number of alleles increases, the values of z0, z1, z2 show increasing departures from those predicted by the diallelic formulas; only the linear relation of parent-child pairs remains valid for any value of H and for any number of alleles. For example, z0 is still H2/2 in the case of a tri-allelic locus and a pair of non-relatives (Table 1), whereas z2 = 1 - 2H + H2 + 2(p2q2 + p2r2 + q2r2). If the last term of this equation were = H2/2, the equation would have been identical to that shown in Table 1; in contrast, this term is smaller than H2/2 by three cross-product terms (4p2qr, 4pq2r, and 4pqr2, respectively), so that z2 cannot be expressed as a simple function of H. Of course, the value of z1 is higher than that predicted by the diallelic formula of the same amount. In the case of a locus with four alleles, even the value of z0 is different from H2/2. This suggests that the zi values of multi-allelic loci, albeit being related to H, are not exact functions of it.
Computation of exact zi values for markers with arbitrary numbers of alleles and for the four above relationships was obtained by first determining the population probability of all possible genotype pairs for each locus; this was accomplished by listing all possible genotype pairs for each locus and then applying to each pair the exact formulas of Table 1A. The number of shared alleles (Z) was also determined for each pair, and the values of zi were simply calculated by summing together the probabilities of all pairs for the three different values of Z. In this procedure, we used allele frequencies from the databases currently used in our forensic casework studies; these values were also used in computing likelihood ratios of different relationships for parent-child pairs. The obtained exact zi values were fitted to third-order polynomial equations, where the independent variable was the locus heterozygosity.
Data analysis
In the analysis of true familial data, two independent samples were used: i) a series of 102 mother-child pairs from disputed paternity studies, typed for a variable number of markers (5 to 17, out of 26 codominant loci), and, ii) a sample of 80 sib pairs, the bone marrow transplant recipients and donors [25] typed for 13 loci (list is shown in Table 5). These siblings were identical for haplotypes of both HLA class I and class II loci, making it unlikely that any of these pairs were actually biologically unrelated. In the analysis of interpopulation variation of H, allele frequency data of STR markers and sample sizes were extracted from the on-line database "The Distribution of the Human DNA-PCR Polymorphisms" http://www.uni-duesseldorf.de/WWW/MedFak/Serology/dna.html[26]. Heterozygosities and their sampling variance were computed by formulas 8.3 and 8.13 in Nei [27], respectively. Tukey's multiple comparison procedure [28] was used to test differences in single-locus heterozygosities between populations. This test is essentially a t-test applied to multiple means, and uses an appropriate and controlled significance level; it is designed to recognize the mean(s) that are significantly different from one or more other means in a given group. In applying this test to our data, we formed all possible pairs of the population to be tested and calculated the test statistics q = (2d)1/2/sd, d being the difference in H between two populations and sd being the standard error of this difference. The q's critical values are tabulated (the Tukey's studentized range distribution tables, see e.g. http://cse.niaes.affrc.go.jp/miwa/probcalc/s-range/).
Authors' contributions
SP conceived of the study, coordinated all phases of statistical analyses, and drafted the manuscript. CT participated in the design of the study, collected the parent-child data and carried out related statistical analysis. ET performed the analysis of inter-population variation of heterozygosity. SV and LC produced and organized the genetic data on sib pairs. IS participated in the production of parent-child data. FdS and RD participated in the design of the study and coordinated the experimental work. JEBW participated in the study conception and interpretation of data, and provided critical revision of the manuscript for important intellectual content.
Contributor Information
Silvano Presciuttini, Email: sprex@cidr.nhgri.nih.gov.
Chiara Toni, Email: chiara@hint.it.
Elena Tempestini, Email: e.tempestini@biomed.unipi.it.
Simonetta Verdiani, Email: labgenfor@libero.it.
Lucia Casarino, Email: destefan@unige.it.
Isabella Spinetti, Email: spinetti@biomed.unipi.it.
Francesco De Stefano, Email: destefan@pacs.unica.it.
Ranieri Domenici, Email: ranieri@biomed.unipi.it.
Joan E Bailey-Wilson, Email: jebw@cidr.nhgri.nih.gov.
References
- Evett IW, Weir BS. Interpreting DNA evidence. Sunderland, Sinauer Associated Inc. 1998.
- Lee HS, Lee JW, Han GR, Hwang JJ. Motherless case in paternity testing. Forensic Sci Int. 2000;114:57–65. doi: 10.1016/S0379-0738(00)00293-0. [DOI] [PubMed] [Google Scholar]
- Ayres KL. Relatedness testing in subdivided populations. Forensic Sci Int. 2000;114:107–115. doi: 10.1016/S0379-0738(00)00289-9. [DOI] [PubMed] [Google Scholar]
- Egeland T, Mostad PF, Mevag B, Stenersen M. Beyond traditional paternity and identification cases. Selecting the most probable pedigree. Forensic Sci Int. 2000;110:47–59. doi: 10.1016/S0379-0738(00)00147-X. [DOI] [PubMed] [Google Scholar]
- Brenner CH. Symbolic kinship program. Genetics. 1997;145:535–42. doi: 10.1093/genetics/145.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate J, Marshall TC, Pemberton JM. A retrospective assessment of the accuracy of the paternity inference program CERVUS. Mol Ecol. 2000;9:801–808. doi: 10.1046/j.1365-294x.2000.00930.x. [DOI] [PubMed] [Google Scholar]
- Maviglia R, Mortera J, Dobosz M, Caglià A, Pascali VL, van Boxel DW, Dawid AP. Forensic inference from incomplete pedigrees by probabilistic expert systems. Progress in Forensic Genetics. 2000;8:399–401. [Google Scholar]
- Epstein MP, Duren WL, Boehnke M. Improved inference of relationship for pairs of individuals. Am J Hum Genet. 2000;67:1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corach D, Sala A, Penacino G, Iannucci N, Bernardi P, Doretti M, Fondebrider L, Ginarte A, Inchaurregui A, Somigliana C, Turner S, Hagelberg E. Additional approaches to DNA typing of skeletal remains: the search for "missing" persons killed during the last dictatorship in Argentina. Electrophoresis. 1997;18:1608–1612. doi: 10.1002/elps.1150180921. [DOI] [PubMed] [Google Scholar]
- Hsu CM, Huang NE, Tsai LC, Kao LG, Chao CH, Linacre A, Lee JC. Identification of victims of the 1998 Taoyuan Airbus crash accident using DNA analysis. Int J Legal Med. 1999;113:43–46. doi: 10.1007/s004140050277. [DOI] [PubMed] [Google Scholar]
- Ruitberg CM, Reeder DJ, Butler JM. STRBase: a short tandem repeat DNA database for the human identity testing community. Nucleic Acids Res. 2001;29:320–322. doi: 10.1093/nar/29.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Abney M, McPeek MS. Detection of mis-specified relationships in inbred and outbred pedigrees. Genet Epidemiol. 2001;21:S36–41. doi: 10.1002/gepi.2001.21.s1.s36. [DOI] [PubMed] [Google Scholar]
- Sieberts SK, Wijsman EM, Thompson EA. Relationship inference from trios of individuals, in the presence of typing error. Am J Hum Genet. 2002;70:170–80. doi: 10.1086/338444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderson GW, Gibbs HL, Sealy SG. Parentage and kinship studies in an obligate brood parasitic bird, the brown-headed cowbird (Molothrus ater), using microsatellite DNA markers. J Hered. 1999;90:182–90. doi: 10.1093/jhered/90.1.182. [DOI] [PubMed] [Google Scholar]
- Ciampolini R, Moazami-Goudarzi A, Vaiman D, Dillmann C, Mazzanti E, Foulley JL, Leveziel H, Cianci D. Individual multilocus genotypes using microsatellite polymorphisms to permit the analysis of the genetic variability within and between Italian beef cattle breeds. J Anim Sci. 1995;73:3259–3268. doi: 10.2527/1995.73113259x. [DOI] [PubMed] [Google Scholar]
- Calafell F, Shuster A, Speed WC, Kidd JR, Black FL, Kidd KK. Genealogy reconstruction from short tandem repeat genotypes in an Amazonian population. Am J Phys Anthropol. 1999;108:137–146. doi: 10.1002/(SICI)1096-8644(199902)108:2<137::AID-AJPA1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Presciuttini S, Bramanti B, Hummel S, Herrmann B. Assessing relationships in an ancient skeletal collection by the number of alleles shared identical by state (IBS) among pairs of individuals. Progress in Forensic Genetics. 2002.
- Shinoda K, Kanai S. Intracemetery genetic analysis at the Nakazuma Jomon site in Japan by mitochondrial DNA sequencing. Anthropol Sci. 1999;107:129–140. [Google Scholar]
- Chakraborty R, Jin L. Determination of relatedness between individuals using DNA fingerprinting. Hum Biol. 1993;65:875–895. [PubMed] [Google Scholar]
- Stivers DN, Zhong Y, Hanis CL, Chakraborty R. RELTYPE: a computer program for determining biological relatedness between individuals based on allele sharing at microsatellite loci. Am J Hum Genet. 1995;suppl 59:A190. [Google Scholar]
- Ehm MG, Wagner M. A test statistic to detect errors in sib-pair relationship. Am J Hum Genet. 1998;62:181–188. doi: 10.1086/301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of mispecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JM. Forensic DNA typing. London San Diego, Academic Press. 2001.
- Lange K. A test statistic for the affected-sib-set method. Ann Hum Genet. 1986;50:283–290. doi: 10.1111/j.1469-1809.1986.tb01049.x. [DOI] [PubMed] [Google Scholar]
- Bacigalupo A, van Lint MT, Valbonesi M, Lercari G, Carlier P, Lamparelli T. Thiotepa cyclophosphamide followed by granulocyte colony-stimulating factor mobilized allogenic peripheral blood cells in adults with advanced leukemia. Blood. 1996;88:353–357. [PubMed] [Google Scholar]
- Huckenbeck W, Kuntze K, Scheil HG. The distribution of the human dna-PCR polymorphisms. Berlin, Verlag Köster. 2002.
- Nei M. Molecular evolutionary genetics. New York, Columbia University Press. 1987.
- Kirk RE. In Experimental Design, Belmont, Brooks/Cole. 2. 1982. Multiple comparison tests. pp. 90–126. [Google Scholar]