Abstract
Thioredoxin (TRX) is a key protein of the cellular redox metabolism, which expression is increased in several tumors especially gastric tumors. Even though ultraviolet (UV) and hypoxia specifically induce TRX, the mechanisms that lead to increased TRX levels are still ill defined. Here, we show that the heterogenous ribonucleoprotein A18 (hnRNP A18) RNA Binding Domain (RBD) and the arginine, glycine (RGG) rich domain can bind TRX 3′-untranslated region (3′-UTR) independently but both domains are required for maximal binding. Immunoprecipitation (IP) of hnRNP A18-mRNAs complexes and co-localization of hnRNP A18 and TRX transcripts on ribosomal fractions confirm the interaction of hnRNP A18 with TRX transcripts in cells. Moreover, down regulation of hnRNP A18 correlates with a significant reduction of TRX protein levels. In addition, hnRNP A18 increases TRX translation and interacts with the eukaryotic Initiation Factor 4G (eIF4G), a component of the general translational machinery. Furthermore, hnRNP A18 phosphorylation by the hypoxia inducible GSK3β increases hnRNP A18 RNA binding activity in vitro and in RKO cells in response to UV radiation. These data support a regulatory role for hnRNP A18 in TRX post-transcriptional expression possibly through a kissing loop model bridging TRX 3′- and 5′-UTRs through eIF4G.
INTRODUCTION
Thioredoxin (TRX) is a ubiquitous multifunctional protein that has regulatory roles in cellular signaling and gene transcription in addition to cytoprotective activities through the quenching of reactive oxygen species (ROS) (1). The cytoprotective effect of TRX against oxidative stress was best illustrated in experiments where the levels of TRX were reduced with an antisense expression vector (2). These cells became more sensitive to H2O2, a variety of anticancer drugs and ultraviolet (UV) radiation. On the other hand, over-expression of TRX could be detrimental to cellular homeostasis. For example, increased levels of TRX are associated with resistance to chemotherapy and cellular proliferation, probably through inhibition of apoptosis (3). TRX expression is increased in several tumors, especially gastric tumors and colon tumors where TRX mRNA expression is 60 times higher than in normal tissue (4). TRX is also increased in response to a variety of stresses such as X-ray irradiation, UV radiation and other types of oxidative stresses (5). However, the mechanism that leads to increased levels of endogenous TRX in cancer cells is still ill defined (3).
Using a modified Systematic Evolution of Ligands by EXponential enrichment (SELEX) assay, we have identified TRX and 25 other stress-inducible transcripts, as potential targets for the heterogenous ribonucleoprotein A18 (hnRNP A18) (6). hnRNP A18 was the first RNA binding protein (RBP) reported to be inducible by UV radiation (7). hnRNPs are abundant RBPs predominantly found in the nucleus and involved in several cellular activities ranging from transcription and mRNA processing in the nucleus to cytoplasmic mRNA translation and turnover (8). The hnRNPs constitute a family of more than 20 different proteins that are highly conserved (9). hnRNP A18 is different than most hnRNPs in that it contains only one RNA Binding Domain (RBD) and has several arginine and glycine residues (RGG boxes) in its auxiliary domain. In contrast to most hnRNPs that are constitutively expressed, hnRNP A18 mRNA and protein levels can be induced by cellular stress such as UV and hypoxia (10).
The nuclear hnRNP A18 translocates to the cytoplasm in response to UV radiation and binds several stress-responsive mRNA transcripts including TRX (6). hnRNP A18 binds specifically to TRX 3′-UTR and increases translation. Translation initiation can be regulated at both the 5′ and the 3′ end of an mRNA transcript. Modulations of the interactions between RBPs, mRNA transcripts and components of the translational machinery dictate the fate of most transcripts. Proteins that bind to 3′-UTRs can affect both the stability and the translation of a transcript. These proteins can directly contact the basal translational machinery and influence activation or repression of translation (11).
Regulation of protein translation is now recognized as an important component of the cellular response to genotoxic stress (12). More than four decades ago, modification of protein synthesis patterns was recognized as an early phenomenon triggered by cellular stress (13). A typical response was characterized by an immediate arrest of protein synthesis followed by an increased rate after exposure to UV radiation (13). The immediate down regulation of protein synthesis is thought to be an adaptive response triggered to protect the cells and conserve the resources required to survive (14). The increase rate of protein synthesis after cellular stress seems to target specific ribosomal proteins and other proteins important for survival (15). Induction of specific ribosomal proteins in response to stress may indicate the involvement of the translational machinery in sensing and responding to cellular stress (15). The association of several ribosomal proteins with the oxidative stress response (1) is additional evidence that translation regulation is a significant component of the cellular stress response. Several types of stress, including heat shock stress and several chemicals, can induce the synthesis of stress proteins while inhibiting the rate of general protein synthesis (16) (and references within). Thus, regulation of general translation as well as translation of specific transcripts that can confer protective roles are likely to be important component of the stress response in human cells.
In this report we show that hnRNP A18 RBD as well as the RGG domain confer hnRNP A18 RNA binding activity. More importantly, hnRNP A18 and TRX mRNAs form a complex in RKO cells and co-localize on ribosomal fractions. Our data also indicate that levels of hnRNP A18 correlate with TRX protein levels. Furthermore, phosphorylation of hnRNP A18 by the hypoxia inducible kinase GSK3β (17) increases hnRNP A18 RNA binding activity in vitro and in RKO cells in response to UV radiation. Functional assays indicate that hnRNP A18 can increase TRX translation and interacts with eukaryotic Initiation Factor 4G (eIF4G), a component of the general translational machinery. Collectively, these data indicate that hnRNP A18 could mediate post-transcriptional regulation of TRX possibly through a kissing loop model bridging TRX 3′- and 5′-UTRs by binding eIF4G.
MATERIALS AND METHODS
Plasmids
The pcDNA3.1-A18, pcDNA3.1-A18-GFP (green fluorescent protein) and pcDNA3.1-CAT (Chloramphenicol acetyltransferase) expression vectors were constructed as described previously (6). The 3′-UTR of TRX was amplified by PCR from the TRX expression vector pGEM-TRX and cloned into the BamHI/XhoI sites of pcDNA3.1-CAT to generate the plasmid pcDNA3.1-CAT/TRX 3′-UTR. To generate the antisense hnRNP A18 vector, the open reading frame (ORF) of hnRNP A18 was cloned in antisense orientation into the HindIII/XhoI sites of pcDNA3.1. The GST (glutathione S-transferase)-hnRNP A18 expression vectors were constructed with the cDNA of full-length hnRNP A18, RBD with linker (residues 1–92) and RGG domain (residue 84–172) cloned into the BamHI/XhoI site of pGEX-4T-2 vector. These vectors were named pGEX-A18, pGEX-RBD and pGEX-RGG, respectively.
Cell culture and treatment
The rationale for using human colon carcinoma RKO cells is that TRX is over-expressed in colon tumors (4) and RKO cells have a wild-type p53 genotype (18) allowing them to respond normally to genotoxic stress. RKO cells were provided by A. J. Fornace,Jr (National Cancer Institute, National Institutes of Health, Bethesda, MD). The cells were grown in RPMI 1640 containing 10% fetal bovine serum (Invitrogen Corp., Carlsbad, CA). For transient transfection, cells were cultured to 50% confluency, and the indicated plasmids were transfected with Fugene 6 lipid mix (Roche Molecular Biochemicals). For CAT assay, a total amount of 11 µg of plasmids DNA was used and supplement with pcDNA3.1 when necessary. Each dish received 1 µg of CAT or CAT-UTR expression vector and 0 or 10 µg of hnRNP A18. Forty-eight hours after transfection, cells were harvested and lysed for further analyses. Analyses were performed in triplicate. For stable transfection, hnRNP A18-GFP was transfected into 50% confluent RKO cells. Selection was performed with 600 µg/ml hygromycin B for 2 weeks. The RKO antisense cell line was established as described earlier (6). The UVC source was a Philips 30 W germicidal lamp emitting at 253.7 nm and the intensities of the UV lamp were determined with a UVX Radiometer (UVP Inc., Upland, CA).
Protein purification
Expression of GST fusion hnRNP A18 full-length, hnRNP A18 RBD and hnRNP A18 RGG were achieved in Escherichia coli BL21(DE3) as recommended by the manufacturer (Novagen) except that the bacteria were grown at 30°C. Glutathione Sepharose 4B was purchased from Amersham Biosciences and the fusion proteins were purified according to the protocol provided by the manufacturer.
RNA binding activity in vitro
RNA band shift and Northwestern assay
3′-UTR of TRX cDNAs plus polyA tail (319–485) was amplified by PCR with a T7 promoter (5′-AGTGAATTGTAATACGACTCACTATAGGGC-3′) engineered at their 5′ ends and in vitro transcribed with MAXI-scripts (Ambion Inc.). 32P-radiolabeled TRX 3′-UTR transcripts were gel purified. Binding reactions for gel mobility shift assays were performed with a range of GST–hnRNP A18 fusion protein concentrations and 0.15 nM 32P-labeled RNA in a final volume of 25 µl containing 10 mM HEPES (pH 7.9), 150 mM KCl, 5 mM MgCl2, 0.2 mM DTT, 10 U of RNasin (Invitrogen), 8% glycerol and 50 µg/ml yeast tRNA. Reactions were incubated for 20 min at room temperature and immediately fractionated through 1% agarose gel. For Northwestern assay, the RNA–protein binding condition was the same as described above except that the recombinant GST or GST–hnRNP A18 fusion proteins were transferred to a nitrocellulose membrane first and the proteins were renatured over-night in a buffer containing 10 mM HEPES (pH 7.9), 40 mM KCl, 3 mM MgCl2, 5 mg/ml BSA, 5% glycerol, 0.2% non-ionic detergent P-40 (NP-40), 0.1 mM EDTA, 1 mM DTT before incubating the membrane with 1 × 106 CPM/ml of 32P-labeled RNA probes.
Small interference RNA (siRNA)
The 21 nt double-stranded RNA was synthesized using Ambion RNAi construction kit. The target sequence (5′-GAGACAGTTACGACAGTTA-3′) corresponds to 19 nt from the hnRNP A18 ORF and a two-dT overhang. The scrambled RNA is composed of four random sequences of 19 nt with a two-UU overhang on each side (nonspecific control duplexes XIII, pool of four; Dharmacon, Inc., Boulder, CO). The siRNAs (20 nM) were transfected in RKO cells with TransMesenger transfecton reagent (Qiagen) according to the manufacturer's recommendations. The cells were exposed to UV radiation 24 h after transfection.
Western blots
The western blot analyses were performed, as described previously (6) on different amount of protein extracts (indicated in the figure legends). The cells were lysed in radioimmune precipitation assay (RIPA) buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 µg/ml leupeptin and 5 mM DTT]. The proteins were run on a 15% polyacrylamide gel, transferred on to nitrocellulose, and reacted with either TRX mouse monoclonal antibody (BD) at 1:5000 dilution, hnRNP A18 rabbit polyclonal antibody (6) at 1:200 dilution, eIF4G rabbit monoclonal antibody (Bethyl) at 1:1000, GSK3β mouse monoclonal antibody (Upstate) at 1:1000 dilution, or actin mouse monoclonal antibody (Oncogene Research Products) at 1:5000 dilution to control for even protein loading. The blots were then reacted with their respective secondary antibodies conjugated to horseradish peroxidase (HRP) and reacted with a chemiluminescence substrate (ECL, Amersham Biosciences) as recommended by the manufacturer.
Linear sucrose gradient fractionation
RKO cells (40 × 106) exposed or not to UV radiation were treated for 15 min with cycloheximide (100 µg/ml) and then harvested. The cytoplasmic extracts were prepared by resuspention of the cell pellets in polysome lysis buffer [10 mM HEPES (pH 7.0), 100 mM KCl, 5 mM MgCl2, 0.5% NP-40] containing 1 mM DTT, 100 U/ml RNase inhibitor (Gibco-BRL) and protease inhibitor cocktail (Roche). The cells were homogenized with a Dounce's homogenizer and centrifuged at 14 000 g. The supernatants (800 µl) were loaded on to sucrose gradients (10–50% w/v) in polysome lysis buffer without NP-40. After centrifugation (Sorval AH-629, 25 000 r.p.m., 3 h, 4°C), the samples were fractionated from the bottom into ten 1.5 ml aliquots. For western blot, 50 µl from each fraction was loaded on to SDS–PAGE gel. For RNA analysis, half of each fraction (750 µl) was diluted with an equal volume of water and RNA was isolated by phenol–chloroform followed by isopropanol precipitation. For total RNA analysis, ∼40% of the total RNA isolated from each gradient fraction was fractionated on 1.2% agarose gel. For RT–PCR, 5% of isolated total RNA (1 µl) from each fraction was used.
In vitro phosphorylation
FLAG-ATM (Ataxia Telangiectasia mutant) and ATR expression vectors were obtained from Dr Mike Kastan (St. Jude Children's Research Hospital, Memphis, TN) and Aziz Sancar (University of North Carolina, Chapel Hill, NC), respectively. The FLAG-SMG1ATX was obtained from Dr Alan Fields (University of Texas, Galveston, TX). The vectors were stably transfected in RKO cells and the respective kinases were immunoprecipitated with a FLAG antibody after UV radiation. The recombinant CK2, GSK3α and GSK3β kinase were purchased from Upstate Biotechnology. The kinase reaction was performed with 1 mCi of [32P]ATP in 15 µl of 10 mM Tris–HCl (pH 7.4), 150 mM NaCl, 10 mM MgCl2, 0.5 mM DTT (kinase reaction buffer) for 30 min at 37°C. Samples were run on an SDS–12% polyacrylamide gel. The gel was dried and exposed to X-ray sensitive film.
RNA binding activity in vivo
Northwestern and GSK3β siRNA treatment
RKO cells (30% confluency) stably transfetced with GFP–hnRNP A18 were transiently transfected with either 20 nM of GSK3β siRNA (SMRTpool GSK3β, Upstate) or scrambled siRNA (Ambion). Forty-eight hours after transfection, the cells were exposed or not to UV (14 Jm−2) radiation and harvested 4 h later. Cell lysis were performed as described above and GFP–hnRNP A18 pulled down is described below. One mg of total protein from each sample was used to pull down GFP-hnRNP A18. The pulled down GFP–hnRNP A18 was loaded on a 12% SDS–PAGE gel and transferred to a nitrocellulose membrane. Northwestern was performed as described above.
Co-immunoprecipatation of hnRNP A18 mRNA complexes and hnRNP A18 protein complexes
RKO cells and RKO cells stably transfected with GFP–hnRNP A18 were exposed to 20 Jm−2 UV radiation and harvested 4 h later. Immunoprecipitation (IP) of endogenous hnRNP A18 mRNA complexes were performed essentially as described in (19) except that the cell lysate was separated into two samples; sample (a) was used to immunoprecipitate mRNAs bound to hnRNP A18 and sample (b) was used to immunoprecipitate proteins associated with hnRNP A18. Briefly, the cells were lysed with polysome lysis buffer [10 mM HEPES (pH 7.0), 100 mM KCl, 5 mM MgCl2, 0.5% NP-40] and total lysate (1.5 mg) were incubated in the presence of EDTA (20 mM) with protein A-Sepharose CL-4B beads (Sigma) that had been pre-coated with 30 µg of either anti-GFP (Santa Cruz Biotech.) or anti-IgG1 (Calbiochem). The beads were washed six times with NT2 buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM MgCl2 and 0.05% NP-40]. Sample (a): for RNA analysis, the beads were incubated with 100 µl NT2 buffer containing 20 U RNase free DNase I for 15 min at 37°C. Proteins bound to the beads were then digested by adding 0.1% SDS and 0.5 mg/ml proteinase K to the reactions and the incubations were continued for 15 min at 55°C. RNA was extracted by ethanol precipitation and used to perform RT–PCR. The PCR products were visualized after electrophoresis in 1% agarose gels. Sample (b): for hnRNP A18 co-immunoprecipitation with eIF4G, an RNase cocktail (Ambion) was incubated with the cell lysates at 30°C for 30 min prior to the IP. Following the IP, the proteins were eluted with SDS loading buffer and loaded on an 8% SDS–PAGE gel for western analysis.
CAT assays
The CAT activity was measured essentially as described previously (6). Cellular extracts containing either 50 or 25 µg of protein were incubated at 37°C for 1–3 h and separated on TLC plates. The conversion of chloramphenicol was quantified on a PhosphorImager (Molecular Dynamics STORM) with the ImageQuant software.
RNase protection assay
RKO cells were transiently transfected with hnRNP A18 and either a CAT or a CAT-TRX 3′-UTR construct as described previously (6). Forty hours later the cells were treated with actinomycin D (10 µg/ml) for 8 h before harvesting. RNase protection was performed using the ribonuclease protection kit RPAIII (Ambion) and the procedures were performed as described in (6) with the exception that a new set of primers was used to prepare the antisense CAT RNA probe: 5′-TCAGCTGGTGCTGATGCCGCTG-3′ and 3′-GACGCTACTCACCGTGATCGG-ATATCACTCAGCATAAT-5′ (T7 polymerase binding site is underlined).
RESULTS
Specific interaction between hnRNP A18 and TRX 3′-UTR mRNA
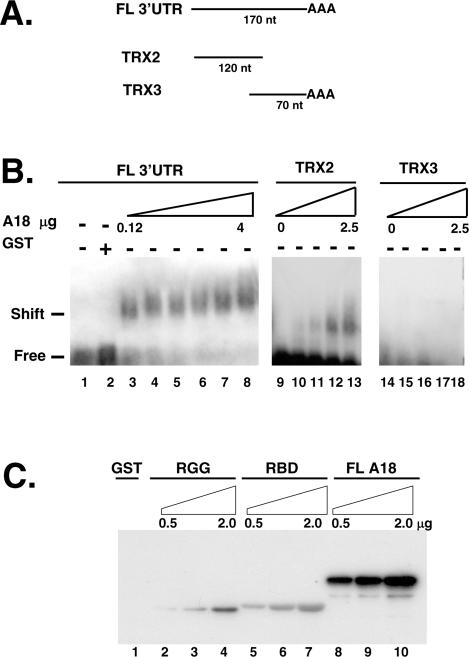
We have previously shown that hnRNP A18 specifically binds to several stress-activated transcripts including TRX (6). Binding of hnRNP A18 to TRX was specific to TRX 3′-UTR since no binding was detected with the ORF or the 5′-UTRs of TRX. In order to further characterize the interaction between hnRNP A18 and TRX mRNA in solution, mobility shift assays were performed with GST fusion hnRNP A18 and 32P-labeled TRX 3′-UTR RNA. Because of the size of the RNA probe (220 nt) we used 1% agrose gel. Data in Figure 1B show that hnRNP A18 formed a single complex with 32P-labeled TRX 3′-UTR RNA in a dose dependent manner (0.12 to 4 µg, lanes 3–8). No binding was observed with GST alone (lane 2). We also performed the RNA band shift with deletion mutants of the TRX 3′-UTR. Our data (lanes 9–13) indicate that the first 120 nt of the 3′-UTR are sufficient to confer binding. No binding was observed with the last 70 nt of the 3′-UTR (lanes 14–18). These data confirm the specific interaction between hnRNP A18 and TRX mRNA previously observed by Northwestern (6).
Figure 1.
hnRNP A18 binds to TRX 3′-UTR in solution. (A) Schematic representation of the different probes used. FL; full-length, nt; nucleotides. (B) RNA band shift was performed as described in the text. Increasing amounts (0.12 to 4 µg) of GST–hnRNP A18 was incubated with the indicated labeled TRX probe and run on a 1% agarose gel. (C) hnRNP A18 RBD and RGG domain are required for maximal RNA binding. Northwestern analysis was performed with increasing amounts (0.5 to 2 µg) of either full-length (FL) GST–hnRNP A18 (lanes 8–10), GST–hnRNP A18 RGG domain (RGG; lanes 2–4), GST–hnRNP A18 RBD (lanes 5–7) or GST alone (lane 1) and labeled TRX 3′-UTR as described in the text.
Most hnRNP proteins exert their RNA binding function through their RBD. However, it has been reported that RGG boxes may increase the general RNA binding affinity (20). To determine whether the hnRNP A18 domain containing the RGG boxes was involved in RNA binding, we engineered deletion mutants and analyzed them by Northwestern with TRX mRNA. The data shown in Figure 1C indicate that both the RBD (lanes 5–7) and the RGG domain (lanes 2–4) can bind RNA independently of each other but maximal binding is only observed when both domains are present (lanes 8–10).
hnRNP A18 and TRX transcripts form a complex and co-localize on ribosomal fractions
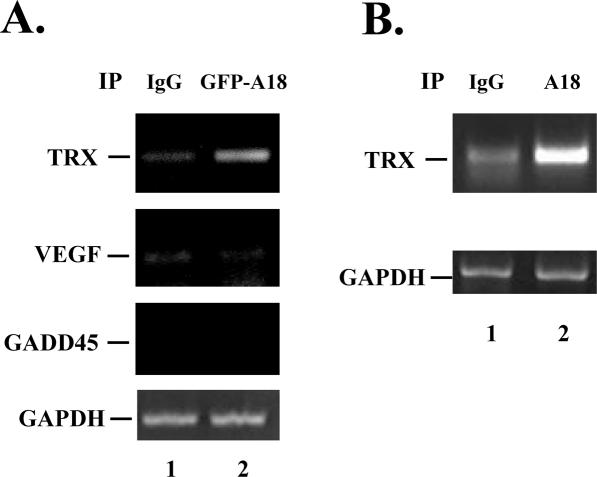
To verify if TRX is a target for hnRNP A18 in cells and to support a role for hnRNP A18 on TRX regulation, we performed RNA IP of cellular lysates extracted from RKO cells exposed to UV radiation. We initially used RKO cells stably transfected with GFP–hnRNP A18 to facilitate the detection of the immune complex. The cells were exposed to UV radiation to translocate hnRNP A18 to the cytosol (6) and the proteins were extracted 4 h later. Polysomes were then extracted under conditions that preserve the association of RBPs with target mRNA essentially as described before (19). To facilitate the detection of the bound mRNAs, the polysomes were disrupted by the addition of EDTA during the IP step. hnRNP A18-bound mRNAs were then eluted and used for RT–PCR to amplify the products. The data shown in Figure 2A indicate that endogenous TRX can be amplified from the material immunoprecipitated with anti-GFP antibody to pull down GFP–hnRNP A18 (lane 2) but not when nonspecific antibodies (IgG) (lane 1) were used. To verify the specificity of the interaction, we also amplified two oxidative stress-responsive transcripts (21,22) not targeted by hnRNP A18. Our data indicate that neither vascular endothelial growth factor (VEGF) nor GADD45 were preferentially amplified from the mRNAs pulled down by hnRNP A18. To determine whether the same amount of input material was used in both samples we also amplified trace amounts of the non-target GAPDH mRNA. Our data indicate that GAPDH could be amplified to the same extent in both samples but rather inefficiently since three times as much material as TRX had to be loaded to detect its presence. To verify whether endogenous hnRNP A18 could also associate with TRX transcripts in the cells, we repeated the experiment described above but used purified hnRNP A18 antibodies to immunoprecipate the complex. The data shown in Figure 2B indicate that endogenous hnRNP A18 also specifically associates with TRX transcripts (lane 2). These data confirm the in vitro binding of TRX transcript to hnRNP A18 (Figure 1) and indicate that TRX mRNA is a bona fide target for hnRNP A18 in cells.
Figure 2.
hnRNP A18 interacts with TRX mRNA in the cells. (A) Polysomes were extracted from RKO cells exposed to 20 J m−2 UV radiation. IP with either anti IgG or anti-GFP to pull down GFP-A18 hnRNP (A18) was performed in the presence of 20 mM EDTA to disrupt the polysomes but preserve the association of RBP with target mRNAs. IP was followed by RT–PCR to detect endogenous TRX. As a negative control, VEGF, GADD45 and GAPDH were amplified from the same fractions. To facilitate detection of GAPDH, three times as much material as the TRX products was loaded. (B) Same as (A) except that anti hnRNP A18 was used to pull down mRNAs bound to endogenous hnRNP A18.
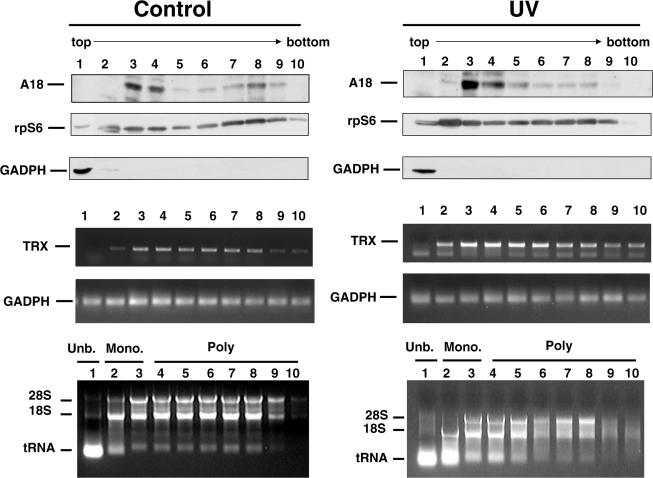
To determine whether hnRNP A18 interaction with TRX transcripts could influence translation of TRX, we first performed sucrose gradient analysis (Figure 3) to evaluate whether hnRNP A18 and TRX transcripts co-localize on ribosomal fractions. The RKO cells were exposed or not to UV radiation and the cells extracts were processed as described above in the absence of EDTA to preserve the polysomes integrity. The extracts were layered on 10–50% sucrose gradients and centrifuged. Our data (Figure 3) indicate that in the absence of UV radiation (Control), hnRNP A18 is mainly localized in fractions 3 and 4. According to the conventional distribution of the polysomal fractions and the localization of the ribosomal S6 protein and the ribosomal 18 and 28S RNA, these fractions correspond to the mono-ribosomal and low molecular weight polysomal fractions. hnRNP A18 also localized to some extent to fraction 8 which corresponds to a high molecular weight polysomal fraction. In contrast, the non-ribosomal protein GAPDH is exclusively found in fraction 1, which corresponds to the unbound, non-ribosomal fraction. RT–PCR of TRX transcript indicates that TRX mRNA is detected in fraction 2 but is more abundant in fractions 3–8. Interestingly, the presence of hnRNP A18 in fraction 3 correlates with an increase in TRX mRNA levels. Levels of GAPDH transcript and ribosomal RNAs are relatively constant through out the gradient. The co-localization of hnRNP A18 and TRX transcript in the mono-ribosomal and low molecular weight polysomal fractions suggest a role for hnRNP A18 in ribosomal assembly and/or translation initiation. Exposing the cells to UV radiation did not change the distribution of either hnRNP A18 or TRX transcript. However, UV radiation increased the levels of both, hnRNP A18 protein and TRX transcript. Again, hnRNP A18 and TRX transcripts co-localized in fractions 3–4. The presence of TRX transcript in high molecular weight polysomal fractions, which have no or low levels of hnRNP A18 (fractions 5–8), would suggest that hnRNP A18 is not involved in translation elongation.
Figure 3.
hnRNP A18 is associated with ribosomes. Sucrose gradient analysis was performed as described in the text on RKO cells exposed or not (Control) to 14 J m−2 UV radiation. The fractions were collected from the bottom of the tubes and their proteins and RNA contents were analyzed. Western blot analysis was used to measure the levels of hnRNP A18 (A18), ribosomal protein S6 (rpS6) and GAPDH. The levels of TRX and GAPDH transcripts were evaluated by RT–PCR. Total amounts of RNA were evaluated by agarose gels. The positions of the tRNA, 18 and 28S ribosomal RNA are indicated. The absence of ribosomal RNA in the first fraction corresponds to the non-ribosomal fraction (Unb:unbound). The fractions 2 and 3 contain the Monoribosomes (Mono.) and the low and high molecular weight polysomes are in the fractions 4 to 10.
hnRNP A18 protein levels correlate with TRX protein levels
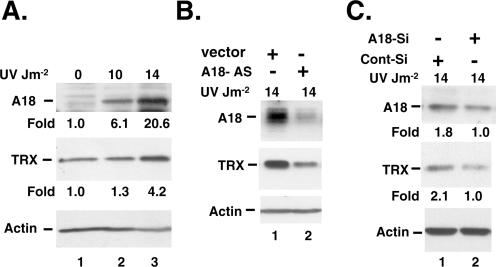
To verify whether hnRNP A18 binding and co-localization with TRX transcript could affect TRX protein levels, we then compared the levels of hnRNP A18 and TRX in response to UV radiation. Our data (Figure 4A) indicate that hnRNP A18 protein levels increase in a dose dependent manner in response to UV radiation. The basal levels of TRX are also increased in response to UV radiation and are the highest at 14 J m−2 where hnRNP A18 protein levels are also the highest. To determine whether hnRNP A18 contributed to TRX increased protein levels in response to UV radiation, we measured the levels of hnRNP A18 and TRX protein in RKO cells stably transfected with an hnRNP A18 antisense vector. Our data (Figure 4B) indicate that the levels of hnRNP A18 and TRX are markedly reduced in the antisense cell line. Because stable transfectants may sometimes lead to cellular aberrations, we also transiently transfected RKO cells with a 21 nt small interfering RNA (siRNA) matching only hnRNP A18 sequence in the GenBank database. The data shown in Figure 4C indicate that down regulation of hnRNP A18 (lane 2) reduced the accumulation of both hnRNPA18 and TRX in response to UV radiation (lane 3). This effect is specific since a scrambled RNA sequence of the same length had no effect on TRX (Figure 4C, lane 1). Interestingly, hnRNP A18 had no significant effect on TRX basal levels in absence of radiation (data not shown). This is in good agreement with our data showing that hnRNPA18 is mainly in the nucleus in absence of stress (6).
Figure 4.
hnRNP A18 protein levels correlate with TRX protein levels. (A) Western blots analysis performed on RKO protein extracts (50 µg) exposed or not to the indicated dose of UV radiation. The ‘Fold’ induction was measured by densitometry analysis and normalized to Actin. (B) Western blot analysis of RKO cells stably transfected with an hnRNP A18 antisense vector (A18-AS) or an empty vector (vector) and exposed to the indicated dose of UV radiation. (C) Western blots analysis of RKO cells transiently transfected (+) or not (−) with hnRNP A18 siRNA (A18-Si) or scrambled RNA (Cont-Si) and exposed to the indicated dose of UV radiation. Actin is used as a loading control. TRX: thioredoxin. Densitometry analysis is expressed as ‘Fold’ induction normalized to Actin and then to cells transfected with hnRNP A18 siRNA.
hnRNP A18 increases TRX translation in cells
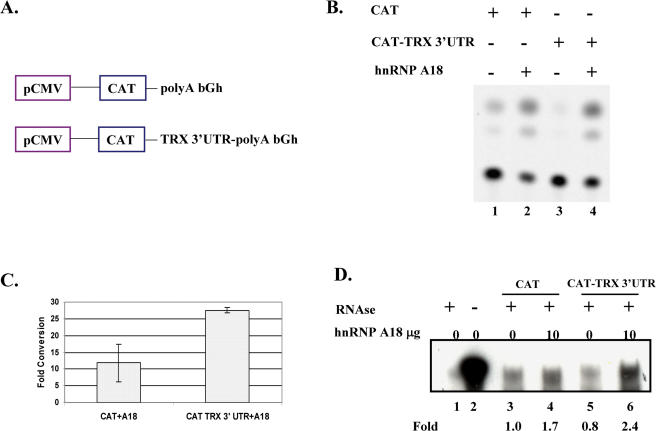
It is now becoming increasingly evident that proteins binding to 3′-UTR can play important roles in translational regulation (23). We have recently shown that hnRNP A18 binds to the replication protein A (RPA) 3′-UTR and increases translation by increasing its RNA stability (6). We thus performed functional assays to determine if the in vitro hnRNP A18 binding to TRX 3′-UTR RNA had any consequence on TRX in cells. We first performed a modified CAT assay to measure the in vivo effect of hnRNP A18 on translation of transcripts containing an RNA binding site of interest as described before (6). Briefly, we co-transfected hnRNP A18 with either a CAT construct or a CAT-TRX 3′-UTR construct (Figure 5A). The first construct (CAT) contains the CAT cDNA under the control of the pCMV protmoter and a poly(A) tail from the bovine growth hormone transcript (Figure 5A). The second construct (CAT-TRX 3′-UTR) is identical to the CAT construct except that we have inserted the TRX 3′-UTR without its poly(A) tail upstream of the bovine growth hormone poly(A)s. This system allows us to use the constitutive CAT activity as a measure of hnRNP A18 effect on translation. The data shown in Figure 5B indicate that co-transfection of hnRNP A18 with either construct (CAT or CAT-TRX 3′-UTR) produced an increase of the CAT activity (Figure 5B). Addition of the TRX 3′-UTR downstream of the CAT (lane 3) decreased the basal CAT activity suggesting that this sequence destabilized or repressed the translation of this transcript. Even though addition of hnRNP A18 increased similarly the translation of either construct (lanes 2 and 4), the overall effect of hnRNP A18 on the stimulation of the CAT activity was more pronounced when the TRX 3′-UTR was present. This is better illustrated by fold conversion of chloramphenicol measured with the two constructs (Figure 5C). Although addition of hnRNP A18 resulted in an 11-fold increase of the CAT activity with the CAT construct, a more than 27-fold increase was observed when hnRNP A18 was added to the CAT-TRX 3′-UTR construct. This indicate that hnRNP A18 preferentially stimulates translation by more than 2-fold when the TRX 3′-UTR is present. Similar data were also obtained previously with RPA 3′-UTR, another hnRNP A18 target (6).
Figure 5.
hnRNP A18 increases TRX translation. (A) Schematic representation of the constructs used in the CAT assay. The poly A tract of the bovine growth hormone (bGh) was placed downstream of the CAT gene under the control of a constitutive pCMV promoter. (B) Representative CAT assay performed by transient co-transfections in RKO cells of, either the CAT construct or the CAT construct containing the TRX 3′-UTR, with hnRNP A18. (C) Average of two CAT assays performed with 25 µg of cellular extracts. The conversion of the chloramphenicol was quantified with a Phosphorimager and expressed as a relative fold conversion of total counts of [14C]chloramphenicol compared to basal levels (0 µg hnRNP A18) of % conversion. (D) RNase protection assay was performed as described in the text. The controls yeast RNA (lanes 1 and 2) were digested (+) or not (−) with RNase. Assays were performed in the absence (0) or presence (10 µg) of hnRNP A18 expression vector with the indicated CAT construct. Levels of transcripts were quantified with a Phosphorimager and expressed as relative fold induction compared to basal levels (0 µg hnRNP A18, lane 3).
To determine whether the increase in translation was due to stabilization of the transcripts, we performed ribonuclease protection assay on cell extracts treated with actinomycin D. Our data (Figure 5D) indicate that indeed over-expression of hnRNP A18 stabilized both transcripts (lanes 4 and 6). This stabilization is even more pronounced (about 3-fold) when the TRX 3′-UTR sequence is present (lane 6). These data indicate that hnRNP A18 is more efficient at stabilizing transcripts that contain targeted sequences. However, we cannot rule out the possibility that other mechanisms are also involved in the overall effect of hnRNP A18 on translation since it stimulates translation at a higher level than it stabilizes mRNAs.
hnRNP A18 interacts with the translation initiation factor eIF4G
Recent evidence indicate that 3′-UTR binding proteins can regulate the rate of translation initiation by interacting with 5′ cap binding proteins (24). Here, we used co-immunoprecipitation assays to evaluate the capacity of hnRNP A18 to interact with components of the basal translational machinery. More specifically we evaluated the capacity of hnRNP A18 to interact with the eukaryotic initiation factors eIF4G, one component of the cap binding complex eIF-4. The cells were exposed to UV radiation and lysed under conditions that preserve the integrity of polysomes (Figure 5). The lysates were then incubated with RNAse in the presence of EDTA to dissociate the ribosome. The presence of eIF4G in the cell extracts (InPut) was verified on a small fraction (Figure 6, lane 3). GFP–hnRNP A18 was then immunoprecipitated with GFP antibodies (Santa Cruz) and a negative control was performed with IgG antibodies. The data (Figure 6) indicate that indeed eIF4G form a complex with hnRNP A18 on cells exposed to UV radiation (lane 2). The interaction is specific since eIF4G was not detected in the fraction immunoprecipitated with the negative control IgG (lane 1). Interaction of hnRNP A18 with eIF4G could thus also contribute to the increased translation observed in Figure 5.
Figure 6.
hnRNP A18 forms a complex with eIF4G. The protein fractions of the polysomes extracts (2 mg) were treated with RNase and immunoprecipitated in the presence of EDTA (20 mM) with either control antibodies (IgG) or GFP antibodies to immunoprecipiate GFP-hnRNP A18. The protein complexes formed on dissociated ribosomes in the absence of RNA were resolved on SDS–PAGE and the blot was reacted with eIF4G antibodies (western blot). In lane 3, a fraction of the protein extract (50 µg, 2.5%) was used to assess the amount of eIF4G present in the InPut. The position of the eIF4G is indicated.
Phosphorylation of hnRNP A18
Post-translational modification such as phosphorylation has been reported to affect the translocation and RNA binding activity of a number of RBPs. For example, phosphorylation of hnRNP A1 is associated with its translocation to the cytosol (25) and phosphorylation of the RBP nucleolin increased its RNA binding activity (26) while dephosphorylation of the hnRNP C is required for its binding to RNA (27). Thus, it is important to determine whether hnRNP A18 could be phosphorylated and, if so, whether it affects its RNA binding activity. We first used several purified protein kinases and recombinant hnRNP A18 to investigate which protein kinase may target hnRNP A18 in vitro. The analysis of hnRNP A18 primary sequence reveals several consensus phosphorylation sites for stress-activated kinases. The hnRNP A18 primary sequence indicates two possible Casein Kinase 2 (CK2) sites, several glycogen synthase kinase 3 (GSK3) and potential ATM and Rad3-related kinase ATR. CK2 targets several stress-activated proteins such as p53 (28), the oxidative stress-inducible protein A170 (29) and the murine STI1 protein (30). GSK3 was originally described as a constitutively active regulator of glycogen synthase but has recently been described as being activated during development and under prolonged hypoxia (17). ATR phosphorylates several substrates in response to UV radiation (31) including the RBP nucleophosmin (NPM) (32). Hypoxia can activate ATR as well (33). Recently another member of the ATM/ATR protein kinase has been isolated. This kinase was originally named ATX and subsequently renamed SMG1. Like ATM and ATR, SMG1ATX is activated by UV and X-ray radiation and share substrates specificity with these kinases (34).
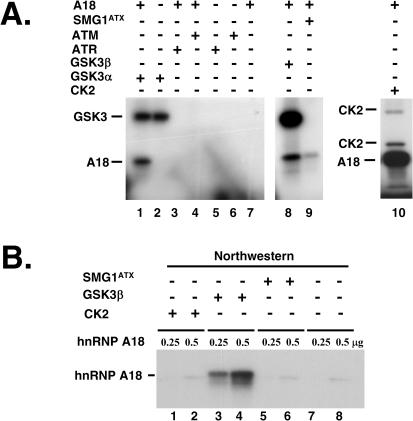
Therefore, we investigated whether hnRNP A18 could be phosphorylated by CK2, ATM, ATR, SMG1ATX and the two GSK3 isoforms, α and β. The in vitro kinase assay was performed as described before (32). The data in Figure 7A indicate that hnRNP A18 is a good in vitro substrate for GSK3 α and β (lanes 1and 8), CK2 (lane 10) and to some extent for SMG1ATX (lane 9) but not for ATM or ATR.
Figure 7.
Phosphorylation of hnRNP A18. (A) In vitro phosphorylation with recombinant hnRNP A18 and recombinant CK2 (lane 10), GSK3α (lane 1), GSK3β (lane 8) and FLAG immunoprecipitated SMG1ATX (lane 9), ATM (lane 4), ATR (lane 3) as described in the text. The positions of phosphorylated hnRNP A18 and autophosphorylated GSK3 and CK2 are indicated. (B) Northwestern analysis was performed with increasing amounts (0.25 to 0.5 µg) of recombinant hnRNP A18 phosphorylated in vitro with cold ATP and the indicated kinases (SMG1ATX; lanes 5 and 6, GSK3β; lanes 3–4, CK2; lanes 1–2). The blot was reacted with labeled TRX 3′-UTR.
To determine if phosphorylation of hnRNP A18 could affect its RNA binding activity, we phosphorylated hnRNP A18 in vitro with different kinases and cold phosphate and performed Northwestern assay with TRX 3′-UTR. We used only 0.25 and 0.5 µg of recombinant protein in order to prevent saturation and facilitate the detection of the effect of phosphorylation. The data shown in Figure 7B indicate that even though hnRNP A18 is a good in vitro substrate for CK2 and SMG1ATX (Figure 7A), these kinases have no effect on hnRNP A18 RNA binding activity. However, phosphorylation by GSK3β (Figure 7B lanes 3 and 4) increases considerably hnRNP A18 binding to TRX 3′-UTR. Interestingly, five consensus GSK3 phosphorylation sites (S144, S148, S152, S155 and S159) are located in hnRNP A18 RGG domain. This domain is required for maximal RNA binding activity (Figure 1C).
The UV-activated hnRNP A18 RNA binding activity is GSK3β dependent
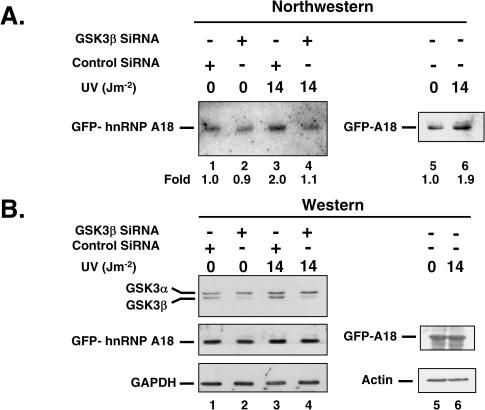
The hnRNP A18 was originally identified based upon its rapid induction by UV radiation (7). We subsequently showed (6) that the nuclear hnRNP A18 is also translocated to the cytoplasm in response to UV radiation. Accumulation of hnRNP A18 in the cytoplasm does not require de novo protein synthesis since pre-treatment of the cells with cycloheximide (100 µM for 4 h) does not prevent the UV-induced translocation (data not shown). To determine whether hnRNP A18 RNA binding activity is affected by UV radiation, we used a GFP antibody to immunoprecipitate GFP–hnRNP A18 from cells exposed or not to UV radiation and measured the RNA binding activity of the immunoprecipitated proteins by Northwestern. The data shown in Figure 8A indicate that the binding activity of hnRNP A18 is increased 2-fold upon exposure to UV radiation (lane 3). The level of induction of the RNA binding activity is in the same range as what has been reported for other RBPs (26) when exposed to cellular stress. Western blot analysis of the same extracts (Figure 8B) indicates that the overall levels of GFP–hnRNP A18 protein levels are not affected by UV radiation (lanes 1 and 3). This suggests that post-translational modifications could contribute to enhance hnRNP A18 RNA binding activity. Our in vitro data (Figure 7B) indicate that GSK3β can increase hnRNP A18 RNA binding activity. To determine whether the UV-induced activation of hnRNP A18 was GSK3β dependent, we down regulated GSK3β with specific siRNA and repeated the Northwestern. Our data indicate that the siRNA were indeed specific for GSK3β since they decreased the levels of GSK3β considerably but did not affect the levels of GSK3α (Figure 8B, lane 2). Moreover, down regulation of GSK3β prevented the activation of hnRNP A18 RNA binding activity upon UV exposure (Figure 8A, lane 4). This effect is specific since scrambled siRNA did not prevent the activation of hnRNP A18 RNA binding activity (lane 3) that is normally observed following UV exposure (lane 6). These data suggest that phosphorylation of hnRNP A18 by GSK3β following UV radiation exposure is necessary to increase hnRNP A18 RNA binding activity. The presence of GSK3β phosphorylation sites in hnRNP A18 RGG domain and the fact that it co-immunoprecipitates with GSK3β, but not GSK3α (data not shown), also support that hnRNP A18 is a bona fide oxidative stress-responsive GSK3β substrate.
Figure 8.
The UV-induced hnRNP A18 RNA binding activity is GSK3β dependent. (A) Northwestern was performed with TRX 3′-UTR RNA probe and GFP–hnRNP A18 immunoprecipitated from RKO cells. The cells were transiently transfected with GSK3β siRNA (lanes 2 and 4), scrambled RNA (Control SiRNA, lanes 1 and 3) or untransfected (lanes 5 and 6) and exposed to the indicated dose of UV radiation. Levels of RNA binding were quantified with a Phosphorimager and expressed as relative fold induction compared to levels in the absence of UV radiation (lane 1 for samples 1–4 and lane 5 for sample 5–6). (B) Western blot analysis was performed on the same extracts with the indicated antibodies. GFP antibody was used to detect GFP–hnRNP A18 (GFP-A18). GAPDH or Actin were used as loading controls.
DISCUSSION
It is now becoming increasingly evident that oxidative stress is implicated in several human diseases. The molecular understanding of the oxidative stress response is particularly relevant to solid tumor development and neurodegenerative diseases. The small (12 kDa) oxidoreductase TRX is a ubiquitous multifunctional protein that plays important parts in the oxidative stress response. TRX can regulate cellular signaling and gene transcription in addition to protecting the cells through the quenching of ROS (1). The redox activity of TRX increases the DNA binding and transactivating activity of transcription factors. A recent report (35) indicates that TRX increases the expression and activity of the hypoxia inducible factor (HIF1) and its downstream effector gene the VEGF. This underscores the pivotal role of TRX in the oxidative stress response. Because of its role in stimulating cancer cell growth and inhibiting apoptosis, TRX offers a target for the development of drugs to treat and prevent cancer. Although, studies have focused on the TRX gene expression at the transcriptional level, little is known on the post-transcriptional or translational regulation of TRX in response to stress.
Since UV and hypoxia generate ROS and both induce hnRNP A18 expression (10), we believe that hnRNP A18 is an oxidative stress-inducible protein that can participate in the cellular response to oxidative stress. Our immuno-fluorescence data indicate that UV (6) and hypoxia (CoCl2 200 µM for 6 h, data not shown) can induce translocation of the nuclear hnRNP A18 to the cytoplasm. One function of the hnRNP A18 in the cytoplasm could be to interact with and regulate the expression of targeted mRNAs transcripts. Using a modified version of the SELEX technique 46 potential hnRNP A18 mRNA targets were identified (6). Among these transcripts, more than 20, including TRX, were identified as stress-responsive mRNA targets (6). In this work, IP of mRNA–rotein complexes (Figure 2) verified that TRX is a bona fide target of hnRNP A18 in vivo. In addition, our in vitro binding data indicate that hnRNP A18 bind to TRX 3′-UTR in a dose dependent manner (Figure 1). Interestingly, both the RBD and the RGG domain can bind TRX 3′-UTR independently but maximal binding is only observed when both domains are present (Figure 1C). The hnRNP A18 RGG domain (residues 84–172) contains the RGG boxes (residues 93–119) and several phosphorylation sites. Our in vitro data (Figure 7) indicate that phosphosylation by the hypoxia inducible GSK3β (17) increases hnRNP A18 RNA binding activity considerably. The GSK3β phosphorylation sites are all located within the RGG domain between residues 144 and 159. Because the phosphorylation of hnRNP A18 is likely to increase its net negative charge and should be more repulsive of the negatively charged RNAs targets we believe that phosphorylation of the RGG domain triggers a conformational change that facilitates RNA binding. This however remains to be investigated. Our data underscore the uniqueness of hnRNP A18 in that the RGG domain does not merely increase the general RNA binding activity but is actively involved in the binding. So far, only hnRNP U had been reported to use an RGG box in specific RNA binding (36). In fact, the RGG box constitutes the entire RBD in this protein (36).
The significance of hnRNP A18 binding to TRX 3′-UTR is highlighted by our data on translation. Our modified CAT assay (Figure 5) indicates that over-expression of hnRNP A18 increases general translation and more specifically TRX translation. Addition of the TRX 3′-UTR downstream of the CAT gene reduced the basal translation of this transcript (Figure 5B) suggesting that the sequence destabilizes or represses the translation of the transcript. However, addition of hnRNP A18 increased translation to similar or even higher levels than the ones observed on construct lacking the TRX 3′-UTR (Figure 5B). This suggest that hnRNP A18 could bind to the TRX 3′-UTR, as shown in Figure 1, and stabilize the transcript, as shown in Figure 5D, to increase translation. Another possibility is that hnRNP A18 could compete out repressive proteins bound to TRX 3′-UTR and thus de-repress translation. This possibility is currently under investigation. The capacity to destabilize or repress translation seems to be particular to transcripts targeted by hnRNP A18 since adding the 3′-UTR sequence of another hnRNP A18 target, RPA also reduced translation of this gene (6) but the sequence of an unrelated transcript (GADD45) had no effect on this gene basal translation (Chakravarty, D. and Carrier, F., unpublished data). We have recently shown (6) and also observed here (Figure 5) that hnRNP A18 could stimulate translation at higher levels than it stabilizes mRNA transcripts. This led us to suggest that hnRNP A18 could also interact with components of the basal translational machinery to stimulate translation. The data presented here indicate that indeed, hnRNP A18 interacts with the translation initiation factor eIF4G (Figure 6). The effect of hnRNP A18 on translation is thus reminiscent of the effect of certain transcription factor such as p53 on transcription. p53 is a specific transcription factor that can also interact with components of the basal transcription machinery such as TBP and TAFs to regulate transcription (37).
Recently, biomedical evidences have been provided to indicate that many 3′-UTR binding proteins can regulate translation initiation at the 5′ end. This phenomenon led to a model of circularization of mRNA (38) in which 3′ and 5′ ends of RNA form a 'kissing' loop through the interaction between 3′ RBPs and the cap binding protein eIF4F. Such end-to-end complex is thought to increase translation efficiency and permits 3′-UTR mediated translation control (23). There are three important elements involved in this mRNA circularization model: first, the binding of an RBP to the 3′-UTR; second, interaction with the 5′ end through bridge protein(s); and third, the effect of this end-to-end complex on translation. The data presented here indicate that all three elements are present in the case of hnRNP A18; (i) binding to 3′-UTR (Figure 1), (ii) interaction with 5′ end through bridge protein, eIF4G (Figure 6) and (iii) effect on translation (Figures 4 and 5). hnRNP A18 might thus fit into the ‘kissing’ loop model to increase translation in general and in particular stress related transcripts such as TRX. The specificity of hnRNP A18 translational control on TRX transcript is further supported by the correlation of the down regulation of hnRNP A18 with the down regulation of TRX protein levels (Figure 4).
Regulation of RBPs activity can be mediated through post-translational modifications (25). The data presented here indicate that hnRNP A18 is an in vitro substrate for several stress related kinases including CK II, SMG1 and GSK3 but not ATR and ATM. Most interestingly, only GSK3 phosphorylation increased hnRNP A18 RNA binding activity in vitro (Figure 7B). GSK3 was originally thought of as a constitutively expressed cytosolic protein involved in glycogen metabolism. The phosphorylation of several transcription factors by GSK3β implied a nuclear localization as well (17). Moreover, certain cellular stresses can lead to GSK3β accumulation in the nucleus (39). Actually phosphorylation of the transcription factor NFATc by GSK3β induces its transport out of the nucleus (40). It thus seems likely that GSK3β could affect hnRNP A18 similarly in response to oxidative stress. Because GSK3β, hnRNP A18 and TRX are all activated by hypoxia, our data suggest an interesting regulatory pathway where in response to hypoxia or oxidative stress generated by UV radiation hnRNP A18 would be phosphorylated by GSK3β (Figure 7), translocate to the cytosol (CoCl2 100 µM, 4 h data not shown), bind to TRX transcript (Figures 2 and 3) and increase its translation (Figures 4 and 5) by interacting with the general translational machinery (Figure 6). Even though the basic principles of this model have been presented here the molecular details and the kinetics of this model remain to be elucidated.
Acknowledgments
This work was supported by a Departmental Research Initiative Fund (F.C.) and Public Health Service grant GM58888 from National Institute of General Medicine (D.J.W.). The authors would like to thank Dr Ashish Lal for technical advice on RNAs immunoprecipitation and sucrose gradient analysis and Dr Devulapalli Chakravarty for technical advice on Northwestern blots. Funding to pay the Open Access publication charges for this article was provided in part by the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tanaka T., Kondo S., Iwasa Y., Hiai H., Toyokuni S. Expression of stress-response and cell proliferation genes in renal cell carcinoma induced by oxidative stress. Am. J. Pathol. 2000;156:2149–2157. doi: 10.1016/S0002-9440(10)65085-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokomizo A., Ono M., Nanri H., Makino Y., Ohga T., Wada M., Okamoto T., Yodoi J., Kuwano M., Kohno K. Cellular levels of thioredoxin associated with drug sensitivity to cisplatin, mitomycin C, doxorubicin, and etoposide. Cancer Res. 1995;55:4293–4296. [PubMed] [Google Scholar]
- 3.Powis G., Mustacich D., Coon A. The role of the redox protein thioredoxin in cell growth and cancer. Free Radic. Biol. Med. 2000;29:312–322. doi: 10.1016/s0891-5849(00)00313-0. [DOI] [PubMed] [Google Scholar]
- 4.Berggren M., Gallegos A., Gasdaska J.R., Gasdaska P.Y., Warneke J., Powis G. Thioredoxin and thioredoxin reductase gene expression in human tumors and cell lines, and the effects of serum stimulation and hypoxia. Anticancer Res. 1996;16:3459–3466. [PubMed] [Google Scholar]
- 5.Funasaka Y., Ichihashi M. The effect of ultraviolet B induced adult T cell leukemia-derived factor/thioredoxin (ADF/TRX) on survival and growth of human melanocytes. Pigment Cell Res. 1997;10:68–73. doi: 10.1111/j.1600-0749.1997.tb00469.x. [DOI] [PubMed] [Google Scholar]
- 6.Yang C., Carrier F. The UV-inducible RNA-binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response. J. Biol. Chem. 2001;276:47277–47284. doi: 10.1074/jbc.M105396200. [DOI] [PubMed] [Google Scholar]
- 7.Fornace A.J., Jr, Alamo I., Jr, Hollander M.C. DNA damage-inducible transcripts in mammalian cells. Proc. Natl Acad. Sci. USA. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krecic A.M., Swanson M.S. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 9.Dreyfuss G., Matunis M.J., Pinol-Roma S., Burd C.G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 10.Wellmann S., Buhrer C., Moderegger E., Zelmer A., Kirschner R., Koehne P., Fujita J., Seeger K. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J. Cell. Sci. 2004;117:1785–1794. doi: 10.1242/jcs.01026. [DOI] [PubMed] [Google Scholar]
- 11.Gray N.K., Wickens M. Control of translation initiation in animals. Annu. Rev. Cell. Dev. Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 12.Sheikh M.S., Fornace A.J., Jr Regulation of translation initiation following stress. Oncogene. 1999;18:6121–6128. doi: 10.1038/sj.onc.1203131. [DOI] [PubMed] [Google Scholar]
- 13.Baden H.P., Pearlman C. The effect of ultraviolet light on protein and nucleic acid synthesis in the epidermis. J. Invest. Dermatol. 1964;42:71–75. [PubMed] [Google Scholar]
- 14.Hinnebusch A.G. The eIF-2 alpha kinases: regulators of protein synthesis in starvation and stress. Semin. Cell. Biol. 1994;5:417–426. doi: 10.1006/scel.1994.1049. [DOI] [PubMed] [Google Scholar]
- 15.Bertram J., Palfner K., Hiddemann W., Kneba M. Overexpression of ribosomal proteins L4 and L5 and the putative alternative elongation factor PTI-1 in the doxorubicin resistant human colon cancer cell line LoVoDxR. Eur. J. Cancer. 1998;34:731–736. doi: 10.1016/s0959-8049(97)10081-8. [DOI] [PubMed] [Google Scholar]
- 16.Duncan R.F., Hershey J.W. Translational repression by chemical inducers of the stress response occurs by different pathways. Arch. Biochem. Biophys. 1987;256:651–661. doi: 10.1016/0003-9861(87)90622-9. [DOI] [PubMed] [Google Scholar]
- 17.Kim L., Kimmel A.R. GSK3, a master switch regulating cell-fate specification and tumorigenesis. Curr. Opin. Genet. Dev. 2000;10:508–514. doi: 10.1016/s0959-437x(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 18.Kastan M.B., Zhan Q., el-Deiry W.S., Carrier F., Jacks T., Walsh W.V., Plunkett B.S., Vogelstein B., Fornace A.J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 19.Tenenbaum S.A., Lager P.J., Carson C.C., Keene J.D. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 20.Burd C.G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 21.Sasaki H., Ray P.S., Zhu L., Galang N., Maulik N. Oxidative stress due to hypoxia/reoxygenation induces angiogenic factor VEGF in adult rat myocardium: possible role of NFkappaB. Toxicology. 2000;155:27–35. doi: 10.1016/s0300-483x(00)00274-2. [DOI] [PubMed] [Google Scholar]
- 22.Patton G.W., Paciga J.E., Shelley S.A. NR8383 alveolar macrophage toxic growth arrest by hydrogen peroxide is associated with induction of growth-arrest and DNA damage-inducible genes GADD45 and GADD153. Toxicol. Appl. Pharmacol. 1997;147:126–134. doi: 10.1006/taap.1997.8227. [DOI] [PubMed] [Google Scholar]
- 23.Mazumder B., Seshadri V., Fox P.L. Translational control by the 3′-UTR: the ends specify the means. Trends. Biochem. Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 24.Gray N.K., Coller J.M., Dickson K.S., Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO. J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Houven van Oordt W., Diaz-Meco M.T., Lozano J., Krainer A.R., Moscat J., Caceres J.F. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell. Biol. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C., Maiguel D.A., Carrier F. Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins. Nucleic Acids Res. 2002;30:2251–2260. doi: 10.1093/nar/30.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayrand S.H., Dwen P., Pederson T. Serine/threonine phosphorylation regulates binding of C hnRNP proteins to pre-mRNA. Proc. Natl Acad. Sci. USA. 1993;90:7764–7768. doi: 10.1073/pnas.90.16.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meek D.W., Simon S., Kikkawa U., Eckhart W. The p53 tumour suppressor protein is phosphorylated at serine 389 by casein kinase II. EMBO. J. 1990;9:3253–3260. doi: 10.1002/j.1460-2075.1990.tb07524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanagawa T., Yuki K., Yoshida H., Bannai S., Ishii T. Phosphorylation of A170 stress protein by casein kinase II-like activity in macrophages. Biochem. Biophys. Res. Commun. 1997;241:157–163. doi: 10.1006/bbrc.1997.7783. [DOI] [PubMed] [Google Scholar]
- 30.Lassle M., Blatch G.L., Kundra V., Takatori T., Zetter B.R. Stress-inducible, murine protein mSTI1. Characterization of binding domains for heat shock proteins and in vitro phosphorylation by different kinases. J. Biol. Chem. 1997;272:1876–1884. doi: 10.1074/jbc.272.3.1876. [DOI] [PubMed] [Google Scholar]
- 31.Tibbetts R.S., Brumbaugh K.M., Williams J.M., Sarkaria J.N., Cliby W.A., Shieh S.Y., Taya Y., Prives C., Abraham R.T. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes. Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maiguel D.A., Jones L., Chakravarty D., Yang C., Carrier F. Nucleophosmin sets a threshold for p53 response to UV radiation. Mol. Cell. Biol. 2004;24:3703–3711. doi: 10.1128/MCB.24.9.3703-3711.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammond E.M., Dorie M.J., Giaccia A.J. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J. Biol. Chem. 2003;278:12207–12213. doi: 10.1074/jbc.M212360200. [DOI] [PubMed] [Google Scholar]
- 34.Brumbaugh K.M., Otterness D.M., Geisen C., Oliveira V., Brognard J., Li X., Lejeune F., Tibbetts R.S., Maquat L.E., Abraham R.T. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol. Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Welsh S.J., Bellamy W.T., Briehl M.M., Powis G. The redox protein thioredoxin-1 (Trx-1) increases hypoxia-inducible factor 1alpha protein expression: Trx-1 overexpression results in increased vascular endothelial growth factor production and enhanced tumor angiogenesis. Cancer Res. 2002;62:5089–5095. [PubMed] [Google Scholar]
- 36.Siomi H., Matunis M.J., Michael W.M., Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farmer G., Colgan J., Nakatani Y., Manley J.L., Prives C. Functional interaction between p53, the TATA-binding protein (TBP), andTBP-associated factors in vivo. Mol. Cell. Biol. 1996;16:4295–4304. doi: 10.1128/mcb.16.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Craig A.W., Haghighat A., Yu A.T., Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 39.Bijur G.N., Jope R.S. Proapoptotic stimuli induce nuclear accumulation of glycogen synthase kinase-3 beta. J. Biol. Chem. 2001;276:37436–37442. doi: 10.1074/jbc.M105725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beals C.R., Sheridan C.M., Turck C.W., Gardner P., Crabtree G.R. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]