Abstract
The study of eukaryotic gene transcription depends on methods to discover distal cis-acting control sequences. Comparative bioinformatics is one powerful strategy to reveal these domains, but still requires conventional wet-bench techniques to elucidate their specificity and function. The DNase I hypersensitivity assay (DHA) is also a method to identify regulatory domains, but can also suggest their function. Technically however, the classical DHA is constrained to mapping gene loci in small increments of ∼20 kb. This limitation hinders efficient and comprehensive analysis of distal gene regions. Here, we report an improved method termed mega-DHA that extends the range of existing DHAs to facilitate assaying intervals that approach 100 kb. We demonstrate its feasibility for efficient analysis of single-copy genes within a large and complex genome by assaying 230 kb of the human ADAMTS14-perforin-paladin gene cluster in four experiments. The results identify distinct networks of regulatory domains specific to expression of perforin and its two neighboring genes.
INTRODUCTION
The physiological transcription of eukaryotic genes hinges upon regulating chromatin structure. More often than not, this process depends on distal cis-acting control regions. Genome-wide studies suggest that the majority of these regulatory domains reside beyond 10 kb from sites of transcription initiation (1), some of which can be located hundreds of kilobases from their cognate promoters (2,3). Using currently available ‘wet-lab’ methodology, comprehensively identifying the entire suite of regulatory sites that control a given gene can be daunting.
Interspecies genome alignments to identify conserved non-coding sequences (CNS) are an efficient strategy to scour large regions for potential regulatory sites (4,5). These computational approaches are now quite powerful, owing to recent expansion of high-quality datasets from the various genome projects (6,7). As an exclusive tool however, they are not yet capable of predicting the specificity and potential regulatory function of CNS (7,8). Moreover, computational approaches that only rely on identifying candidate regions based on genomic sequences which align between species may nevertheless overlook conserved motifs with regulatory function, such as those conserved epigenetically (9,10). Therefore, traditional experiments that examine chromatin structure and epigenetic organization remain fundamental to both augment computational predictions as well as to independently identify functional regions.
For more than two decades, the DNase I hypersensitivity assay (DHA) has been instrumental for discovering regulatory domains (11). This method maps discrete sites in the genome where the conformation of chromatin renders its DNA hypersensitive to cleavage by the endonuclease DNase I (12). DNase I hypersensitive sites (DHSs) have been mapped to most categories of cis-acting regulatory domains including promoters, enhancers, silencers, insulators and locus control regions (13–16). The ubiquitous or, alternatively, lineage and activation specific formation of DHSs at these respective regulatory domains frequently belies their function (17,18). DHSs can indicate transient remodeling of chromatin as in the case of transcriptional induction, or stable, epigenetic inheritance of programmed changes to a locus during differentiation (19). Characterizing the formation of DHSs remains a basic, albeit robust strategy to determine the specificity and potential function of putative regulatory domains.
Technical limitations restrict classical DHA methodology to analysis of genes in ∼20 kb increments. A long-range approach to DHA has been reported for analysis of a gene-amplified locus (20), but a method capable of detecting DHSs in single-copy genes of mammalian genomes has not. Recently, PCR based methods and genome-wide strategies have been developed as an alternative to map DHSs, though most still require validation using classical approaches, and also require automation or high-throughput capabilities, making them inaccessible to most labs (1,21,22). In this report, we provide a straightforward approach using field inversion gel electrophoresis (FIGE) and demonstrate the ability to display authentic DHSs in single-copy genes of the human genome at ranges near 100 kb. This method is as good or better than the classical DHA and can be readily introduced into any conventional molecular biology laboratory. In essentially four experiments, we assayed 230 kb of the human perforin locus, a region that includes both of its nearest neighboring genes. This approach discovered three distinct networks of DHSs that are likely to coordinate the regulation of perforin and its neighbors.
METHODS
Cell culture
Jurkat (clone E6-1) is a cell-line derived from a T-cell leukemia and was obtained from ATCC. YT is a human NK-like lymphoma (23). Both YT and Jurkat cells were cultured in IMDM supplemented with 10% heat inactivated fetal bovine serum (FBS). HA(10)A is a microcell-hybrid, mouse fibroblast line that carries one copy of a neor tagged human chromosome 10 and was obtained from Coriell Cell Repository (item GM11688) (24). HA(10)A is cultured in DMEM containing 10% FBS, and retains the human chromosome 10 by selection with 500 µg/ml G418. All culture media was supplemented with Gentamicin to 50 µg/ml.
Mega-DNase I hypersensitivity analysis (MDHA)
Cell lysis and nuclei isolation
Exponentially growing lymphocyte cultures were harvested at room temperature, then washed and concentrated in ice-cold Ca2+/Mg2+ free phosphate-buffered saline (PBS) before adjusting to 2 × 107 cells/ml in PBS. Cells were lysed on ice for 10 min by adding 4 vol of lysis buffer [12.5 mM Tris, pH 7.4, 45 mM KCl, 6.25 mM MgCl2, 375 mM sucrose and 0.125% nonidet P-40, supplemented with one complete (EDTA free) protease inhibitor cocktail (Roche)/50 ml] to 1 vol of cells. For fibroblast cultures, cells were detached by trypsinization, harvested and washed in PBS, although after the final wash they were re-suspended as 8 × 106/ml in fibroblast lysis buffer [12.5 mM Tris, pH 7.4, 5 mM KCl, 0.1 mM spermine tetrahydrochloride, 0.25 mM spermidine, 175 mM sucrose, supplemented with one complete (EDTA free) protease inhibitor cocktail tablet (Roche)/50 ml lysis buffer] and equilibrated on ice for 10 min. These cells were then lysed by adding 0.02 vol of 10% nonidet P-40, gently vortexing and inverting tubes three times to mix followed by an additional 5 min on ice. Nuclei were pelleted at 500 × g for 7 min (lymphocytes), or 1000 × g for 5 min (fibroblasts), in a pre-cooled swinging bucket rotor at 4°C. Nuclei from both cell types were gently re-suspended in one-tenth of the original lysis volume using nuclei wash buffer [10 mM Tris, pH 7.4, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2 and 300 mM sucrose] before pooling pellets 2:1 and adjusting volumes to half the original lysis volume using nuclei wash buffer; nuclei were pelleted again as above. Supernatants were completely aspirated and the pellets were re-suspended in nuclear wash buffer and adjusted to 2.5 × 108 nuclei/ml on ice.
DNaseI treatement and encapsulation in agarose
A DNase I cocktail (2× relative to DNase I, CaCl2 and BSA) was prepared on ice by supplementing nuclei wash buffer to 2 mM CaCl2, 100 µg/ml BSA and DNase I (Roche) to 800 U/ml. The DNase I was titrated by 2-fold serial dilution in the same buffer without DNase I. Nuclei were treated on a timed schedule. One volume of nuclei (2.5 × 108/ml) in a 1.5 ml microfuge tube and one DNase I concentration from the titration were each placed at room temperature for 4 min to equilibrate. Then, 1 vol of the DNase I cocktail was added to 1 vol of nuclei and digestions proceeded for 4 min at room temperature before being stopped by addition of 0.5 vol of ice-cold 5× stop buffer (nuclei wash buffer supplemented to 50 mM EDTA) and placing on ice. Nuclei were embedded in 1% agarose (final concentration) by addition of 2.5 vol of 2% low-melting (BioRad) point agarose dissolved in nuclei wash buffer supplemented with 10 mM EDTA and maintained at 50°C. Nuclei in molten agarose were gently mixed and immediately dispensed into 75 µl disposable plug molds (BioRad). The embedded nuclei were cooled at 4°C 15–20 min before being solubilized to purify genomic DNA.
Purification and restriction enzyme digestion of genomic DNA in agarose
Embedded nuclei were solubilized with stripping buffer [100 mM EDTA, 1% Sodium Laroyl Sarcosine, 0.2% Sodium Deoxycholate and 1 mg/ml protienase K (Roche)], by using a 10:1 (v/v) ratio of buffer to agarose block at 50°C, overnight, with gentle rocking and treating two times. Blocks were then washed five times, 45 min each wash, using a 10:1 ratio (v/v) wash buffer (50 mM EDTA, 20 mM Tris, pH 8.0) to agarose block, at 4°C with vigorous shaking. The second wash contained 1 mM phenylmethylsulfonyl fluoride. Agarose embedded DNA was stored under a residual volume of wash buffer at 4°C. For restriction enzyme digestion, agarose blocks were washed 2 × 30 min in TE (1 mM EDTA) to reduce EDTA from wash buffer, followed by two sequential 30 min equilibrations at 4°C using ≥10 vol (relative to agarose blocks) excess of the appropriate restriction enzyme buffers. The equilibration buffer was replaced with 4 vol of the appropriate restriction enzyme cocktail [Pac I (NEB) 90 U/∼4 × 106 genomes] and digestions proceeded for 16 h at 37°C. Reactions were quenched by removing cocktails and washing each plug with ice-cold TE, followed by equilibration in 0.5× TBE running buffer at 4°C without motion. One-half agarose block (∼37 µl of 5 × 107 nuclei/ml) was loaded per lane of 1% pulse field certified agarose (BioRad) gels prepared in 0.5× TBE and blocks were sealed with 1% low-melting point agarose also prepared in 0.5× TBE.
Field inversion gel electrophoresis, blotting and mapping DHSs
FIGE analysis was performed with a FIGE mapper (BioRad). The FIGE system is commercially available as part of the CHEF mapper XA system (BioRad). All FIGE mapper programs used linear shaped ramps for switch times, a forward voltage of 180 V, a reverse voltage of 120 V and were run in re-circulated 0.5× TBE maintained at 14°C.
| FIGE program | Separation (kb) | Switch times (s) forward (F) and reverse (R) | Run time (h) |
|---|---|---|---|
| P1 | 1–30 | 0.1–0.9 (F), 0.1–0.9 (R) | 15 |
| P2 | 1–60 | 0.2–1.2 (F), 0.2–1.2 (R) | 17.5 |
| P3 | 1 to >60 | 0.1–3.5 (F), 0.1–3.5 (R) | 18 |
Following electrophoresis, gels were stained for 40 min in 0.5 µg/ml ethidium bromide in autoclaved water at 4°C with constant agitation. The gels were photographed under preparative UV light and DNA was nicked using 70 mJ of UV irradiation before being blotted to nylon membranes (Hybond N+) in alkali (0.4 M NaOH, 1.5 M NaCl) using a capillary transfer (turboblotter Schleicher&Schuell) by transferring one liter of buffer through membranes. Membranes were then neutralized for 5 min in 0.5 M Tris, pH 7.0, and rinsed in 2× SSC before storing or hybridization.
To map the position of DHSs in the locus, best-fit curves were generated by manual measurements of the migration distances of DNA size standards during each FIGE analysis; gel-to-gel variation in resolution between independent runs of identical FIGE programs was extremely low (data not shown). The location of DHSs were determined by measuring the migration distance of each DNase I dependent sub-germline band after Southern analysis and computing an average from at least two independent experiments; bands separated by at least 1 mm were considered distinct. Based on the equation for the best-fit curve describing the resolution of each FIGE analysis, experimental fragment sizes were calculated using their respective average migration distance. For each region analyzed, the experimentally determined size of the intact germline fragment was normalized to its known length based on sequence information (25,26). The sizes of sub-germline DNase I dependent fragments were standardized accordingly and the locations of DHSs were expressed relative to perforin transcription initiation (set to +1) in the human genome (GenBank accession no. AL355344) (27). The difference in mapping the same DHS, in different nuclei preparations from the same cells, was calculated as a percentage relative to the size of the germline fragment in which it was mapped to quantify variability: ±%variability = [max difference in DHS fragments between independent experiments (bp)/germline fragment (bp)] × 50.
Northern and Southern blot analysis
Northern and Southern hybridizations were performed in glass bottles using a rotisserie oven. Blots were washed in 2× SSC followed by pre-hydridization using 0.5 ml hybridization solution (Clonetech ExpressHyb)/10 cm2 membrane, for 30–60 min at 68°C (Southern) to 70°C (Northern). An equal volume of hybridization solution containing 1 ng/ml of TCA precipitable probe was used to replace the pre-hybridization solution and blots were hybridized for 12 h at the same temperature as the pre-hybridizations. Blots were washed with 10 ml wash buffer/10 cm2 blot, four times at room temperature in 2× SSC–0.1%SDS for 10 min, one time in 0.5× SSC–0.1%SDS at 60°C for 20 min, and finally at 60°C for 20 min in 0.1× SSC–0.1%SDS. Blots were exposed to Kodack, BioMax MS film using HE intensity screens at −80°C, or to screens for phosphorimaging and analysis with the ImageQuant 2.0 software. All conventional probes were gel-purified restriction fragments labeled with [α-32P]dCTP by the random priming method, purified by size exclusion chromatography and quantified by TCA precipitation.
Probes for MDHA and Northern analysis
Probes for MDHA analysis were either cloned fragments, or synthetic oligonucleotides. Probes were located within ∼1 kb of each restriction site used in MDHA analysis and were from ∼600 to 1200 bp (cloned fragments) or 30–40 bp (oligos). Sequences for all probes were selected computationally for the absence of repetitive elements using RepeatMasker Open-3.0 (http://www.repeatmasker.org) and significant similarity to other sequences (BlastN NCBI) before testing empirically for single-copy specificity in genomic Southern analysis. Oligonucleotides were selected using OLIGO 2.0 software, comprised sequences of ∼50% GC content, predicted melting temperature (Tm) = 65–75°C and were predicted to not form dimers more stable than −5.0 kcal/mol, or secondary structures more stable than 2.0 kcal/mol (100 mM salt and 1.0 pM oligo). Oligonucleotide probes were typically hybridized at 12.5°C below their predicted Tm according the manufacturer's suggestions (3DNA—Genishpere; IDT Technologies) (28,29). Oligonucleotide probes used to analyze the four Pac I fragments (78, 39, 26 and 87 kb) were 3′ 78 kb-ACC-AGGTGCTAGTAATCATAGAGAGGGCTACAAAGACACA, 5′ 39 kb-CTGAAGCAAATGA-GGGGCAAGAAGTCAATCTGGTAAAAGG, 3′ 39 kb-ACAAGACCTGAGACAAACTGGG-AGAAAGAACTGACTGTAT, 5′ 26 kb-GGCGACCTCAAATGTTAAGGCAGGAATGGGGA-AATGGATA, 3′ 26 kb-ACCTGAGGGGAAGTCTCTCGTTCTGGAAGTATCGCAGTTA, 5′ 87 kb-GTCATAATAGGAGAGACGACAGGATCTTCCAGCCCCAACA.
Cloned fragments for probes were excised directly from genomic clones, or were cloned after PCR of human genomic DNA with the following primers: 5′ end of the PacI 79 kb fragment (F-ACA-AACGCTCCAGCAAACACTCAT, R-CCTCCTGTGGTTCCAGAATGTC-AC), the 3′ end of the PacI 79 kb fragment is a PstI–PacI fragment; the 5′ end of the Pac I 39 kb fragment is a Pac I–Pst I fragment, the 5′ end of the Pac I 26 kb fragment (F-GCTTAGATGCC-GTCATTTAGTTCA, R-GACCCCTTTCCTATTGATTTGTTA), the 3′ end of the Pac I 26 kb fragment (F-CCTTGACTCAACCTTGCAGACACT, R-TGAGGGAAAGATGTTGGCTTCA-G), the 5′ end of the Pac I 87 kb fragment (F-GGGCTCCTCAGCAAACACGACAA, R-CCCT-ACCACCCCTTCCCTCTAATG). Probes for Northern analysis were an NcoI–BamHI human perforin cDNA fragment, a 2.2 kb BamHI ADAMTS-14 cDNA (transcript A) fragment (30), and a 1.4 kb PstI fragment from the paladin cDNA (GenBank BC040163).
RESULTS
Premise and development of MDHA
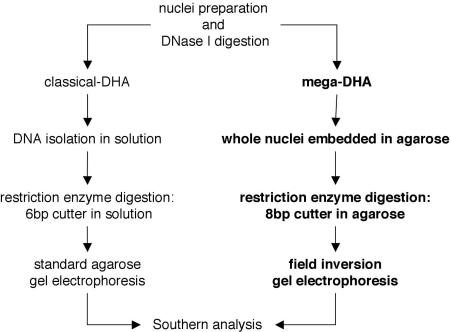
The manipulation of DNA in solution and standard agarose gel electrophoresis limits the range at which conventional DHA methodology can display DHSs. Mechanical forces during pipeting and the organic extractions or chromatography typically used to purify genomic DNA from DNase I treated nuclei causes random breaks in DNA. This ultimately limits the maximum size of fragments that can be analyzed and also increases background. Moreover, DNA molecules in excess of ∼20 kb cannot be systematically resolved using conventional gel electrophoresis conditions. We therefore incorporated several straightforward adaptations to overcome these limitations in order to establish the MDHA method (Figure 1). Nuclei for MDHA are isolated and treated with DNase I similar to existing assays. However, to prevent shearing forces from manipulation of DNA in solution, DNase I treated nuclei were directly embedded in low-melting point agarose. Both purification of genomic DNA and its subsequent digestion with restriction enzymes were performed in the context of the agarose blocks. To generate large restriction fragments for analysis, 8 bp cutting enzymes were used. These fragments were resolved using FIGE, a method of pulsed field gel electrophoresis that facilitates separation of DNA molecules in size from 1 to 103 kb (31). Based on previous reports (32,33), conditions for UV nicking of DNA and capillary blotting were empirically determined for MDHA (data not shown) to ensure efficient transfer of the large DNA from FIGE gels and its optimal hybridization during Southern analysis. Finally, two approaches for design and application of probes for Southern hybridization were examined. The first were oligonucleotides (30–40 bp) directly labeled by radioactive tailing with 32P (28), or those linked to multivalent scaffolds (3DNA dendrimer molecules) labeled with alkaline phosphotase (29). These strategies were compared with conventional DNA probes excised from genomic clones that were labeled with 32P by random priming (34).
Figure 1.
Comparison of conventional DHA versus MDHA. The classical DHA method was modified as follows: immediately following DNase I treatment, nuclei were embedded in low-melting point agarose. The DNA remains protected in the agarose for all subsequent manipulations, including digestion with rare-cleaving restriction enzymes. Finally, the DNA, containing substantially larger genomic fragments than previously existing DHA is fractionated by FIGE and analyzed Southern blot analysis.
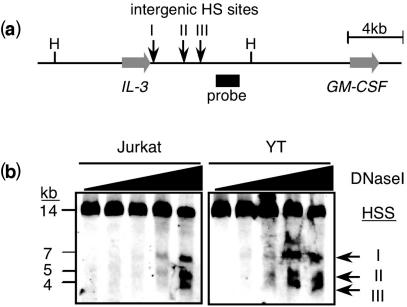
We initially validated the MDHA protocol by assaying known DHSs. To that end, the previously characterized intergenic region between the human IL-3 and GM-CSF genes was analyzed in two transformed cell-lines, the helper T-cell line Jurkat and the natural killer (NK)-like line, YT. The IL-3/GM-CSF intergenic region contains three constitutive DHSs that form in most cell types (35) (Figure 2a). Southern blots generated by the MDHA protocol were hybridized with 3DNA oligonucleotide probes and detected the appearance of three fragments in response to increasing concentrations of DNase I (Figure 2b). The DHSs formed at comparable DNase I concentrations in nuclei preparations from both the Jurkat and YT cells. The size of each DNase I dependent band corresponded to the location of the three previously reported DHSs between the IL-3 and GM-CSF genes. Thus, the MDHA approach detects DHSs in a manner similar to existing DHA. Of note, these experiments were accomplished with a ∼40 bp oligonucleotide probe under non-radioactive conditions, indicating it is possible to detect DHSs of single-copy genes without the need for cloning to generate conventional probes.
Figure 2.
MDHA of known DHSs in the IL-3/GM-CSF intergenic region. (a) Schematics of the IL-3/GM-CSF locus and the location of the three previously established intergenic DHSs. Horizontal arrows indicate the IL-3 and GM-CSF genes, and vertical arrows show the relative position of the known DHSs. A HindIII restriction fragment (H) was analyzed in (b) using the indicated probe. (b) MDHA analysis. Nuclei of Jurkat and YT cells were treated with increasing concentrations of DNase I (wedges above blots) and encapsulated into agarose blocks. The blocks were deproteinized, treated with HindIII and the DNA was fractionated by FIGE. The shown autorads represent hybridizations with a 40 bp 3DNA oligonucleotide probe that was conjugated to alkaline phosphotase. The germline HindIII fragment (H) and the three DNase I dependent fragments (I, II and III) correspond to the schematics shown in (a).
MDHA of 230 kb the perforin locus at human 10q21–22
Previously, we expended significant effort using transgenic methods to identify the essential regulatory regions that control perforin, a gene that is required for a form of cytolysis mediated by specialized immune cells known as cytotoxic lymphocytes. However, multiple transgenes that spanned 45 kb of the human perforin locus were not expressed physiologically in these cell types (36,37). Similarly, analysis of chromatin structure in proximal regions of perforin did not provide significant additional insight as to the essential regulatory elements governing the physiologically high expression of perforin in cytotoxic lymphocytes (38,39). We therefore presumed additional missing domains were located in far distal regions of the locus. MDHA was developed specifically to identify these potential domains. Encouraged by our preliminary analysis of the IL-3/GM-CSF locus, we turned our focus to the perforin gene located on human 10q21–22 (40).
Perforin is simply organized in three exons that span ∼6 kb (27) and is positioned between two apparently unrelated genes, ADAMTS-14 (GenBank accession no. NM_139155) and paladin (GenBank accession no. NM_014431). Transcription of ADAMTS-14 (30) initiates ∼70 kb upstream of perforin, while transcription of paladin initiates ∼124 kb downstream (Figure 3a) (GenBank NT_008583) (41). ADAMTS-14 codes for a metalloprotease that is expressed in fibroblasts and connective tissue (30). Paladin codes for a potential tyrosine phosphatase with unknown function and is broadly expressed (42). In contrast, perforin codes for an effector molecule of the immune system that is expressed in cytotoxic lymphocytes and deployed from these cells to lyse infected or malignant host cells (43,44). The perforin gene is constitutively expressed at high levels in NK cells (45) but typically is not expressed by non-cytotoxic, helper CD4+ T-cells (46). Therefore, we used MDHA to analyze the long-range chromatin structure of the perforin gene and its closest neighboring genes in YT cells (cytotoxic, NK-like cell-line) (23), Jurkat cells (non-cytotoxic helper T-cell line) (47) and HA(10)A cells (non-lymphoid fibroblast cell-line) (24).
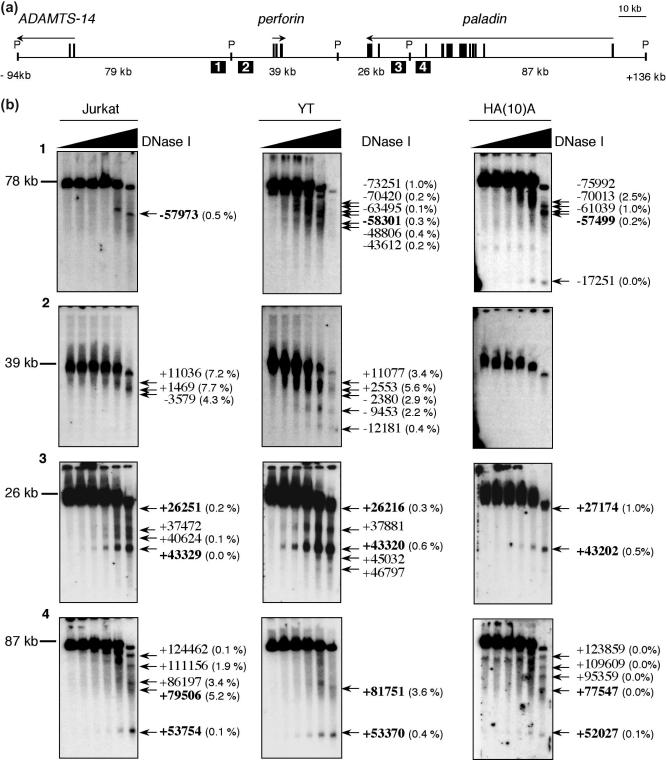
Figure 3.
MDHA of the ADAMTS14, perforin and paladin loci. (a) Map detailing the essential components used in the MDHA analysis. A total of 230 kb was analyzed in four experiments. The exons and transcriptional orientation of perforin and its two neighboring genes are indicated in relation to the PacI restriction fragments (P) that were analyzed. The size of PacI fragments are shown below the map in addition to the location of the hybridization probes (numbered squares) that were hybridized to the Southern blots in (b). (b) MDHA of perforin and its flanking genes in Jurkat and YT and HA(10)A cell-lines. The Southern blots are numbered according to the hybridization probes denoted in (a). DHSs are indicated with arrows and their computed map locations are reported relative to perforin transcription initiation (set as +1); the variability in mapping the same DHSs in independent experiments is indicated in parentheses (see Methods). Those shown in bold were interpreted to be the same DHSs in all three cell-lines, based on qualitative side-by-side comparisons following autoradiography. The autorads shown represent multiple experiments (at least two independent nuclei preparations) as well as hybridizations with different probes and labeling techniques (shown are images obtained with random primed labeled radioactive probes; images obtained from non-radioactive oligonucleotide probes and probes hybridizing to the reciprocal ends of the PacI restriction fragements are not shown).
A total of 230 kb was analyzed in four large restriction fragments (79, 39, 26 and 87 kb) (Figure 3a). The experiments assayed nuclei from at least two independent nuclei preparations for each cell-type and the entire region was mapped at least twice, using probes derived from restriction fragments labeled radioactively by random priming (Figure 3b), as well as, oligonucleotides linked to 3DNA molecules labeled with alkaline phosphatase (data not shown). Both approaches yielded analogous results, however, hybridizations with the radioactively labeled probes derived from cloned restriction fragments consistently resulted in less background and, consequently, improved sensitivity. The presence versus absence of each DHS was determined qualitatively by comparing autorads side-by-side and the locations of DHSs in the locus were mapped based on best-fit curves describing each FIGE separation (for details on mapping see Methods).
MDHA mapped 13 DHSs in Jurkat cells (Figure 3b, left panels). The majority of these sites were located across paladin and its intergenic region with perforin. Only two DHSs were detected in Jurkat cells across nearly 95 kb of perforin and its upstream flank. MDHA of YT cells mapped 18 DHSs (Figure 3b, middle panels). Unlike Jurkat cells, the majority of these DHSs were located over and upstream of perforin. Finally, in HA(10)A cells, 12 DHSs were detected. They mapped almost exclusively to regions overlapping the ADAMTS-14 and paladin genes (Figure 3b, right panels). The maximum variability in mapping any one DHS was ±7.7%, which translates to mapping the same DHSs within ±800 bp at a range of 10 kb (see Methods); precision comparable to conventional DHA. The largest variability correlated with DHSs that resolved in regions of gels where migration of DNA was the least linear; the migration rate of DNA during FIGE analysis is not linear across all molecule sizes when ramping between switch times is linear. However, this is usually confined to a small region of the gel, thus the majority of all DHSs were mapped to within ≤1.0% of the same position between independent experiments. We concluded MDHA reproducibly mapped genuine DHSs from single-copy genes in the human genome at ranges up to 87 kb.
DHSs detected by MDHA display known and unknown regulatory regions
Only one report has previously mapped DHSs of human perforin, and this centered on a region 1.4 kb upstream and 1.3 kb downstream of transcription initiation. Two areas of DHSs were identified, one near −0.2 kb and a second cluster spanning −0.8 to −1.0 kb; they are present in YT cells and cytotoxic T-cells as well as helper T-cells, but are absent in fibroblasts (39). Two DHSs are present in analogous regions of the mouse locus in perforin expressing cells (38). Similarly, MDHA mapped two broad DHSs in YT cells, one centered at −2.4 kb and a second area at + 2.6 kb. We presume the first collectively comprises the previously identified DHSs near the promoter and perhaps an additional DHS near conserved sequences spanning −1.7 to −1.9 kb (Table 1). The second DHS maps to perforin's second intron, a region wherein analysis has not been reported in the human locus, but is near a CNS and an analogous region in mouse perforin where two DHSs have been reported (38). Thus, MDHA results were consistent with the prior studies. Nevertheless, given the focused interval examined in the previous studies, an entirely systematic comparison with the long-range analysis by MDHA is difficult.
Table 1.
DHSs detected by MDHA align with CNS and sequences identified in genome-wide DHSs analysis
| Probes | MDHA | Ref. (48) human/mouse | Ref. (22) CD4+ T | ||
|---|---|---|---|---|---|
| Jurkat | YT | HA(10)A | CNS | DHS cluster | |
| DHS | DHS | DHS | |||
| * | −73251 (1.0%) | −75992 (2.5%) | −76927, −77026 | ||
| −69460, −69677 | |||||
| * | −70420 (0.2%) | −70013 (2.5%) | −69113, −69211 | ||
| −68875, −68994 | |||||
| −61970, −62368 | −63080, −63149 | ||||
| * | −63495 (0.1%) | −61039 (1.0%) | −60964, −61063 | ||
| 1 | −57973 (0.5%) | −58301 (0.3%) | −57499 (0.2%) | −54438, −54585 | |
| −50416, −50631 | |||||
| * | |||||
| * | |||||
| * | |||||
| * | −9453 (2.2%) | * | * | ||
| 2 | −3579 (4.3%) | −2380 (2.9%) | * | −1753, −1855 | |
| −6, −169 | |||||
| +1469 (7.7%) | +2553 (5.6%) | * | +2761, +2646 | ||
| +11036 (7.2%) | +11077 (3.4%) | * | +12873, +12707 | ||
| +26251 (0.2%) | +26216 (0.3%) | +27174 (1.0%) | * | +23078, +22471 | |
| +37472 | +37881 | * | * | +38380, +37210 | |
| 3 | +40624 (0.1%) | * | * | +40263, +40158 | |
| +41789, +41660 | |||||
| +43329 (0.0%) | +43320 (0.6%) | +43202 (0.5%) | +43233, +43117 | ||
| * | +45032 | * | * | ||
| * | +46797 | * | +47211, +47056 | ||
| +47442, +47303 | |||||
| +53754 (0.1%) | +53370 (0.4%) | +52027 (0.1%) | * | +57944, +57216 | |
| +79506 (5.2%) | +81751 (3.6%) | +77547 (0.0%) | * | ||
| 4 | +86197 (3.4%) | * | +95359 (0.0%) | +94170, +94011 | |
| +111156 (1.9%) | * | +109609 (0.0%) | +111370, +111212 | +109129, +108916 | |
| +124462 (0.1%) | * | +123859 (0.0%) | +124292, +124162 | +125115, +124860 | |
| +125575, +125447 | |||||
The table reports the predicted map location of DHSs identified by MDHA, nearby sequences conserved between humans and mice (CNS), and sequence tag clusters identified by genome-wide recovery of DHSs, relative to the transcription initiation site of perforin (+1). DHSs identified by MDHA are shown with the difference (±percentage of germline band) in predicted map locations between independent experiments within the same cell-lines. Those shown in bold were qualitatively interpreted to be the same DHSs in side-by-side comparison following Southern analysis in all three cell-lines analyzed. Non-coding sequences conserved between humans and mice (70% sequence identity across 100 bp) are indicated and were retrieved via the VISTA browser (http://pipeline.lbl.gov/cgi-bin/gateway2), using the human May 2004 genome, sequences from chr10: 71 900 000–72 130 000, aligned with the mouse May 2004 genome (48). Sequence tags are those clustered within 500 bp (bold), or 1000 bp (http://research.nhgri.nih.gov/DnaseHS) (22).
Recent genome scale analysis of DHSs in human helper T-cells based on sequencing cloned fragments resulting from DNase I cleavage of nuclear DNA has been reported (22). We compared DNase I cutting events in the perforin locus that were identified by this strategy (http://research.nhgri.nih.gov/DNaseHS), to the location of those we mapped by MDHA (Table 1 and Figure 4b). In the genome-wide assay, separate validation indicates confidence in predicting a genuine DHS depends on identifying multiple tags within 500 bp of each other (22). Four sites in the perforin locus were identified with at least two tags clustered within 500 bp. The core size of a nuclease sensitive domain in a DHS is on the order of 150–200 bp, though the extent of nuclease sensitivity may extend beyond this core (21). Therefore, we also considered sequenced tags in the perforin locus that clustered within 1000 bp, which revealed two additional sites, that were also proximal to the predicted map location of DHSs identified by MDHA. Thus results from the MDHA analysis validate those from a genome-wide approach, but also identified additional genuine DHSs not detected by the whole genome strategy—at least in context of the number of tags sequenced, and cells assayed, in that analysis.
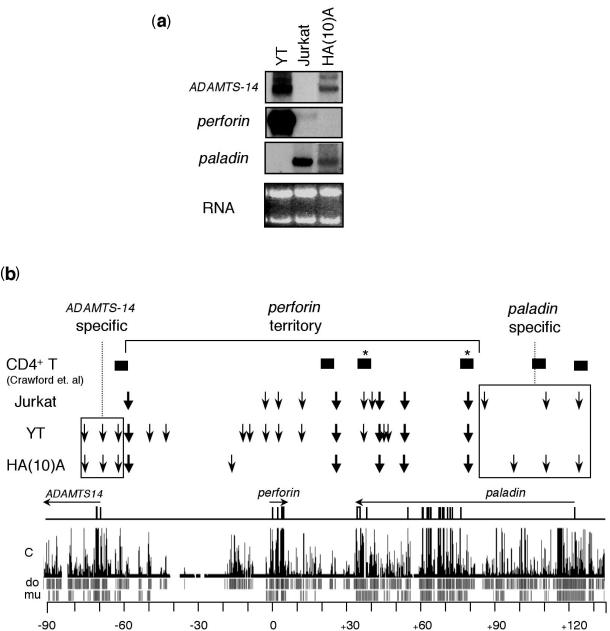
Figure 4.
Gene expression and DHS formation at 10q21-22. (a) Northern blot analysis of mRNA expression from perforin and its neighboring genes in Jurkat, YT and HA(10)A cells. For Northern analysis, total RNA was extracted from cells using TRIZOL (Invitrogen) and 20 µg of total RNA was electrophoresed through formaldehyde agarose gels and blotted to nylon membranes. The blots were sequentially hybridized with cDNA probes from each of the indicated genes. Ethidium bromide staining of the membranes following transfer (RNA) is shown for loading consistency. (b) Schematic summary of DHSs identified by MDHA in Jurkat, YT and HA(10)A cells. The DHSs identified by MDHA (vertical arrows) and sequence tags clustered within 500 bp (closed boxes; asterisks denote those clustered within 1 kb) from genome-wide recovery of DHSs were placed on a map aligned with the locus and a conservation plot from the UCSC genome browser (chr10: 71 900 000–72 130 000; http://genome.ucsc.edu/, May 2004 release) (22,41). The likelihood of conservation in the human genome and the individual pair-wise alignments from the mouse and dog genomes are shown below the conservation plot. The locations of genes, their exons (vertical bars) and their transcriptional orientation (horizontal arrows) are indicated.
Regulatory elements are found in CNSs, and consequently they often map in or near DHSs (7). We therefore compared the location of non-coding sequences conserved between humans and mice with the locations of DHSs that were identified by MDHA (Table 1). Most DHSs mapped within ∼1000 bp of CNSs identified by VISTA analysis (48). In addition, several DHSs that were not co-localized with regions of human–mouse conservation were nevertheless mapped near to sequences that were conserved between humans and other species, such as the dog (Figure 4b) (49). The predicted map location of DHSs identified by MDHA in proximity to CNSs did not precisely overlap all CNS, but was not surprising. There is not necessarily a strict correlation between the peaks in sequence conservation and the exact position of an associated DHS. DHS has also been identified in regions that are not considered CNS (7,21). Therefore, although the resolution of each FIGE program does not map DHSs to the base pair, MDHA mapped DHSs with sufficient precision to enumerate particular CNSs that are likely to be functionally relevant among all conserved regions. It also clearly identified additional regions that do not share significant nucleotide identity between species but that nevertheless may be functional.
DHSs detected by MDHA correlate with gene expression
To determine the specificity and infer the potential role of the distinct patterns of DHSs in Jurkat, YT and HA(10)A cells, the mRNA expression of ADAMTS14, perforin and paladin was determined by Northern blot analysis (Figure 4a). ADAMTS-14 transcripts were expressed in both YT cells and in HA(10)A cells, but were undetectable in Jurkat cells (Figure 4a). This correlated with the DHSs pattern over ADAMTS-14 in these three cell types. Three of the same DHSs, one which maps to the ADAMTS-14 promoter region, were present in both HA(10)A and YT cells but were not formed in Jurkat cells (Figures 3 and 4b), suggesting these DHSs may be positive regulatory domains specific to ADAMTS-14 expression.
Perforin mRNA was undetectable in HA(10)A cells but was expressed in YT cells and Jurkat cells (Figure 4a). Quantitatively, the YT cells expressed >100-fold more perforin mRNA than what was detected in Jurkat cells. In fact, the abundance of perforin mRNA in Jurkat cells corresponds to only 5% of what is physiological, as compared to primary human cytotoxic lymphocytes (M. E. Pipkin and M. G. Lichtenheld, unpublished data). However, Jurkat and YT cells shared a similar pattern of DHSs in regions proximal to the perforin gene, and in the intergenic region between it and paladin (Figure 4b). This included DHSs near the perforin promoter and its immediate upstream flank and intronic regions. This was similar to the DHSs identified by traditional DHA in primary human helper T-cells and YT cells (39). However, perforin's extended upstream region was virtually devoid of DHSs in Jurkat cells, while YT cells demonstrated additional DHSs in the extended 5′ flank. Several of these DHSs were closer to the ADAMTS-14 gene, yet these DHSs were not present in HA(10)A cells which transcribe ADAMTS-14, but not perforin. Therefore, these distal DHSs may be positive regulatory domains specific to perforin.
Finally, paladin mRNA was expressed in HA(10)A and Jurkat cells, but was not expressed in YT cells (Figure 4a). HA(10)A and Jurkat cells displayed a nearly identical DHS pattern across the paladin gene, including a DHS that mapped near its putative transcription initiation site (Figure 4b). In contrast, YT cells did not form this DHS and also lacked the remaining DHSs over the 5′ portion of the paladin gene that were present in both the HA(10)A and Jurkat cells (Figure 4b). This is consistent with the conclusion that DHSs downstream of the DHS located near +80 kb, as being positive regulatory domains specific to paladin.
DISCUSSION
In this report, we adapted the classical DHA into an approach we termed MDHA in order to facilitate the long-range analysis of single-copy genes in large and complex genomes (Figure 1). MDHA reproducibly displayed DHSs from multiple loci in both conventionally sized regions (Figure 2) as well as in fragments up to 87 kb (Figure 3b). The MDHA displayed previously established DHSs from the IL-3/GM-CSF locus and it discovered new DHSs in the perforin locus. DHSs from the perforin locus mapped in proximity to CNS regions and clustered sequence tags identified in genome-wide DHSs assays (Table 1). Distinct networks of DHSs identified by MDHA correlated with the cell-type restricted expression of perforin and its neighboring genes (Figure 4). This report emphasizes that it is practical to detect genuine DHSs in single-copy genes from large genomes at ranges on the order of 100 kb. Thus, comprehensive analysis of all regions of a particular gene or gene cluster, including distal locales, can be systematically and efficiently assayed in a single study.
Advantages and limitations of MDHA
MDHA can analyze 100 kb using similar resources and in a similar time frame to what is required by traditional approaches to analyze a 20 kb region; at least a 5-fold improvement in throughput over classical DHA. An earlier long-range approach developed to analyze a gene-amplified locus in cell-lines, suggests assaying even larger regions may be feasible (20). All of our experiments were performed on 15 cm gels, which limits the maximum size region that can be adequately resolved to discriminate distinct DHSs. The resolution of DHSs depends on the length of the DNA investigated, the actual location of DNase I cleavage sites, and the FIGE analysis program used to separate the DNA. Consequently, the resolution of MDHA might be low in certain cases. In particular, distinguishing neighboring DHSs that result from narrowly spaced DNase I cutting events located distal to the probe used in the analysis can be difficult. However, such DHS clusters can be delineated by displaying the DHSs from the reciprocal direction using a different probe, or mapping at higher resolution in the context of smaller restriction fragments. In any case, MDHA is at least informative of the approximate location of DHSs in large regions and establishes the basis of more focused studies. Using longer gels we anticipate to extend the distance that MDHA can analyze in a single experiment, with improved resolution.
At present, the resolution of MDHA is similar to classical DHA methods, but does not compare with recently developed high-throughput methods founded on the DHA concept. Both a quantitative PCR strategy and genome-wide methods have been reported for localizing DHSs (1,21,22,50). Both class of strategies provide at, or near, base pair resolution of DNase I cutting events. However, the PCR approach would require at least 1000 optimized primer sets to analyze the same region of perforin that was assayed by MDHA using only four hybridization probes. Moreover, reliable prediction of genuine DHSs by genome-wide approaches relates exponentially upon the frequency of sequenced tags clustering to a small genomic interval. Simulations in silico based on the most robust method estimates that, at a false positive rate of <10%, 4 × 106 sequenced tags will only predict 20% of all DHSs in the human genome (21). The need to compare multiple cell types, or stimulation conditions, renders these scales unfeasible for application by the typical bench scientist. Although MDHA is not sufficient for mapping DHSs to the base pair, DHSs in the perforin locus that had been predicted by one genome-wide approach were mapped by MDHA with considerable accuracy, even at very long ranges (Table 1). Therefore, we conclude that MDHA is sufficiently precise to accurately localize DHSs for further functional studies, although some cases may warrant additional fine mapping. Importantly, dissecting the regulation of a particular gene does not initially depend upon knowing the location of DHSs to the base pair. Rather, it is essential to map candidate regulatory sites, within some quantifiable degree of error, around which functional analyses are designed to ultimately delineate an active module to its genetic elements. Methods to efficiently manipulate genomic clones propagated in bacterial artificial chromosome (BAC) vectors, for transgenes and targeting constructs, are now well established (51). Thus, to guide those designs, MDHA will be a practical tool for the molecular biologist as it can efficiently map hundreds of kilobases for authentic DHSs but does not require high-throughput or automated systems.
MDHA and a DHSs blueprint of the human perforin locus
Using MDHA, DHSs that formed in the ADAMTS-14, perforin and paladin loci have been associated with the differential expression of each gene in three model cell lines. The cell-type restricted pattern of expression suggests that expression from each of the three genes is not co-regulated. Therefore, it is possible that each locus is organized in a large territory or chromatin domain subdivided by insulators (52,53). The organization of DHSs identified in this study by MDHA is consistent with that hypothesis. MDHA mapped five DHSs at, or near, the same location in all three cell-lines (−58, +26, +43, +53 and +80 kb), regardless of ADAMTS-14, perforin and paladin expression (Figure 4b), which suggests they form independent of the expression from the three neighboring genes. This is characteristic of many previously characterized insulators, which often form more or less ubiquitously among tissues and do not correlate with expression of nearby genes (17,54). In contrast, DHSs located upstream of −58 kb were only present in cells expressing ADAMTS-14. Therefore, it is conceivable that the DHSs near −58 kb indicate one boundary between territories that control expression of ADAMTS-14 and perforin. Similarly, DHSs downstream of +80 kb were only present in cells expressing paladin, potentially suggesting it as a boundary between the paladin and perforin territory.
It is likely that most if not all regulatory domains required for physiological perforin expression are located between the DHSs mapped at −58 and +80 kb. Between these DHSs, MDHA mapped 10 additional DHSs in cells that express large amounts of perforin (YT cells), and these were absent from cells that do not express perforin [HA(10)A]. This suggests they comprise positive regulatory regions (i.e. enhancers and a locus control region) important for perforin expression. Of these DHSs, five were also present in the perforin-low cells (Jurkat) (Figure 4b), implying that these sites are some, but perhaps not all regulatory domains necessary for the physiologically high levels of perforin expression in cytotoxic lymphocytes. This result is consistent with prior studies that demonstrated sequences of perforin spanning −21 to + 24 kb collectively mediate tissue specific and inducible perforin expression, but do not confer copy number dependent or physiological amounts of expression to perforin transgenes in cytotoxic lymphocytes from transgenic mice (37). Since results here show that several DHSs found only in YT cells [i.e. not in Jurkat or HA(10)A cells] were located outside the regions assayed by the transgenes, several potentially key regulatory domains were not included. Two of these DHSs near −49 to −44 kb migrated in a portion of the FIGE gel where small differences in migration corresponded to large differences in fragment sizes, and thus may represent multiple closely spaced DHSs. An additional experiment to improve the resolution of this region is underway. Nevertheless, we anticipate that the long-range capability of MDHA has discovered the potentially instrumental regulatory regions that had eluded several earlier transgenic studies of the perforin locus. Therefore, in general, we propose creating a DHS blueprint of a locus by mapping with MDHA is the foundation for more rational design of transgenic experiments using large genomic clones from the widely available BAC and PAC resources.
Acknowledgments
The authors express their gratitude for stimulating discussions with Dr Marcela Nouzova and her critique of the manuscript. We also thank Dr Betty Nusgens for kindly providing the ADAMTS-14 cDNA. Funding to pay the Open Access publication charges for this article was provided by a Grant from the Sylvester Comprehensive Cancer Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sabo P.J., Humbert R., Hawrylycz M., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A. Genome-wide identification of DNaseI hypersensitive sites using active chromatin sequence libraries. Proc. Natl Acad. Sci. USA. 2004;101:4537–4542. doi: 10.1073/pnas.0400678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felsenfeld G., Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 3.Spitz F., Gonzalez F., Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 4.Gottgens B., Barton L.M., Gilbert J.G., Bench A.J., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., et al. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 2000;18:181–186. doi: 10.1038/72635. [DOI] [PubMed] [Google Scholar]
- 5.Loots G.G., Locksley R.M., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 6.Pennacchio L.A., Rubin E.M. Genomic strategies to identify mammalian regulatory sequences. Nature Rev. Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- 7.Nardone J., Lee D.U., Ansel K.M., Rao A. Bioinformatics for the ‘bench biologist’: how to find regulatory regions in genomic DNA. Nature Immunol. 2004;5:768–774. doi: 10.1038/ni0804-768. [DOI] [PubMed] [Google Scholar]
- 8.Hardison R., Krane D., Vandenbergh D., Cheng J.F., Mansberger J., Taddie J., Schwartz S., Huang X.Q., Miller W. Sequence and comparative analysis of the rabbit alpha-like globin gene cluster reveals a rapid mode of evolution in a G + C-rich region of mammalian genomes. J. Mol. Biol. 1991;222:233–249. doi: 10.1016/0022-2836(91)90209-o. [DOI] [PubMed] [Google Scholar]
- 9.Siggia E.D. Computational methods for transcriptional regulation. Curr. Opin. Genet Dev. 2005;15:214–221. doi: 10.1016/j.gde.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Roh T.Y., Cuddapah S., Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elgin S.C. DNAase I-hypersensitive sites of chromatin. Cell. 1981;27:413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- 12.Gross D.S., Garrard W.T. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 13.Sawada S., Littman D.R. Identification and characterization of a T-cell-specific enhancer adjacent to the murine CD4 gene. Mol. Cell. Biol. 1991;11:5506–5515. doi: 10.1128/mcb.11.11.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sands J.F., Nikolic-Zugic J. T cell-specific protein–DNA interactions occurring at the CD4 locus: identification of possible transcriptional control elements of the murine CD4 gene. Int. Immunol. 1992;4:1183–1194. doi: 10.1093/intimm/4.10.1183. [DOI] [PubMed] [Google Scholar]
- 15.Burgess-Beusse B., Farrell C., Gaszner M., Litt M., Mutskov V., Recillas-Targa F., Simpson M., West A., Felsenfeld G. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl Acad. Sci. USA. 2002;99:16433–16437. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talbot D., Collis P., Antoniou M., Vidal M., Grosveld F., Greaves D.R. A dominant control region from the human beta-globin locus conferring integration site-independent gene expression. Nature. 1989;338:352–355. doi: 10.1038/338352a0. [DOI] [PubMed] [Google Scholar]
- 17.Saitoh N., Bell A.C., Recillas-Targa F., West A.G., Simpson M., Pikaart M., Felsenfeld G. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agarwal S., Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 19.Agarwal S., Avni O., Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity. 2000;12:643–652. doi: 10.1016/s1074-7613(00)80215-0. [DOI] [PubMed] [Google Scholar]
- 20.Mautner J., Bornkamm G.W., Polack A. Long-range chromatin analysis of the human MYC locus by pulsed-field gel electrophoresis. Genes Chromosomes Cancer. 1996;16:247–253. doi: 10.1002/(SICI)1098-2264(199608)16:4<247::AID-GCC4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 21.Sabo P.J., Hawrylycz M., Wallace J.C., Humbert R., Yu M., Shafer A., Kawamoto J., Hall R., Mack J., Dorschner M.O., et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc. Natl Acad. Sci. USA. 2004;101:16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crawford G.E., Holt I.E., Mullikin J.C., Tai D., Blakesley R., Bouffard G., Young A., Masiello C., Green E.D., Wolfsberg T.G., et al. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl Acad. Sci. USA. 2004;101:992–997. doi: 10.1073/pnas.0307540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yodoi J., Teshigawara K., Nikaido T., Fukui K., Noma T., Honjo T., Takigawa M., Sasaki M., Minato N., Tsudo M., et al. TCGF (IL 2)-receptor inducing factor(s). I. Regulation of IL 2 receptor on a natural killer-like cell line (YT cells) J. Immunol. 1985;134:1623–1630. [PubMed] [Google Scholar]
- 24.Pershouse M.A., Stubblefield E., Hadi A., Killary A.M., Yung W.K., Steck P.A. Analysis of the functional role of chromosome 10 loss in human glioblastomas. Cancer Res. 1993;53:5043–5050. [PubMed] [Google Scholar]
- 25.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 26.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 27.Lichtenheld M.G., Podack E.R. Structure of the human perforin gene. A simple gene organization with interesting potential regulatory sequences. J. Immunol. 1989;143:4267–4274. [PubMed] [Google Scholar]
- 28.Behlke M.A., Dames S.A., McDonald W.H., Gould K.L., Devor E.J., Walder J.A. Use of high specific activity StarFire oligonucleotide probes to visualize low-abundance pre-mRNA splicing intermediates in S. pombe. Biotechniques. 2000;29:892–897. doi: 10.2144/00294pf01. [DOI] [PubMed] [Google Scholar]
- 29.Orentas R.J., Rospkopf S.J., Casper J.T., Getts R.C., Nilsen T.W. Detection of Epstein-Barr virus EBER sequence in post-transplant lymphoma patients with DNA dendrimers. J. Virol. Methods. 1999;77:153–163. doi: 10.1016/s0166-0934(98)00149-9. [DOI] [PubMed] [Google Scholar]
- 30.Colige A., Vandenberghe I., Thiry M., Lambert C.A., Van Beeumen J., Li S.W., Prockop D.J., Lapiere C.M., Nusgens B.V. Cloning and characterization of ADAMTS-14, a novel ADAMTS displaying high homology with ADAMTS-2 and ADAMTS-3. J. Biol. Chem. 2002;277:5756–5766. doi: 10.1074/jbc.M105601200. [DOI] [PubMed] [Google Scholar]
- 31.Lalande M., Noolandi J., Turmel C., Rousseau J., Slater G.W. Pulsed-field electrophoresis: application of a computer model to the separation of large DNA molecules. Proc. Natl Acad. Sci. USA. 1987;84:8011–8015. doi: 10.1073/pnas.84.22.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Devanter D.R., Von Hoff D.D. Acid depurination after field inversion agarose gel electrophoresis reduces transfer of large DNA molecules. Appl. Theor. Electrophor. 1990;1:189–192. [PubMed] [Google Scholar]
- 33.Lee H., Birren B., Lai E. Ultraviolet nicking of large DNA molecules from pulsed-field gels for southern transfer and hybridization. Anal. Biochem. 1991;199:29–34. doi: 10.1016/0003-2697(91)90265-u. [DOI] [PubMed] [Google Scholar]
- 34.Feinberg A.P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 35.Cockerill P.N., Shannon M.F., Bert A.G., Ryan G.R., Vadas M.A. The granulocyte-macrophage colony-stimulating factor/interleukin 3 locus is regulated by an inducible cyclosporin A-sensitive enhancer. Proc. Natl Acad. Sci. USA. 1993;90:2466–2470. doi: 10.1073/pnas.90.6.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lichtenheld M.G., Podack E.R., Levy R.B. Transgenic control of perforin gene expression. Functional evidence for two separate control regions. J. Immunol. 1995;154:2153–2163. [PubMed] [Google Scholar]
- 37.Zhang J., Scordi I., Smyth M.J., Lichtenheld M.G. Interleukin 2 receptor signaling regulates the perforin gene through signal transducer and activator of transcription (Stat)5 activation of two enhancers. J. Exp. Med. 1999;190:1297–1308. doi: 10.1084/jem.190.9.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Youn B.S., Kim K.K., Kwon B.S. A critical role of Sp1- and Ets-related transcription factors in maintaining CTL-specific expression of the mouse perforin gene. J. Immunol. 1996;157:3499–3509. [PubMed] [Google Scholar]
- 39.Lu Q., Wu A., Ray D., Deng C., Attwood J., Hanash S., Pipkin M., Lichtenheld M., Richardson B. DNA methylation and chromatin structure regulate T cell perforin gene expression. J. Immunol. 2003;170:5124–5132. doi: 10.4049/jimmunol.170.10.5124. [DOI] [PubMed] [Google Scholar]
- 40.Fink T.M., Zimmer M., Weitz S., Tschopp J., Jenne D.E., Lichter P. Human perforin (PRF1) maps to 10q22, a region that is syntenic with mouse chromosome 10. Genomics. 1992;13:1300–1302. doi: 10.1016/0888-7543(92)90050-3. [DOI] [PubMed] [Google Scholar]
- 41.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Rapp B.A., Wheeler D.L. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Podack E.R., Hengartner H., Lichtenheld M.G. A central role of perforin in cytolysis? Annu. Rev. Immunol. 1991;9:129–157. doi: 10.1146/annurev.iy.09.040191.001021. [DOI] [PubMed] [Google Scholar]
- 44.Lichtenheld M.G. Control of perforin expression: A paradigm for understanding cytotoxic lymphocytes? In: Sitkovsky M.V., Henkart P.A., editors. Cytotoxic cells: Basic mechanisms and medical applications. Philadephia: Lippincott Williams & Wilkins; 2000. pp. 123–145. [Google Scholar]
- 45.Salcedo T.W., Azzoni L., Wolf S.F., Perussia B. Modulation of perforin and granzyme messenger RNA expression in human natural killer cells. J. Immunol. 1993;151:2511–2520. [PubMed] [Google Scholar]
- 46.Lu P., Garcia-Sanz J.A., Lichtenheld M.G., Podack E.R. Perforin expression in human peripheral blood mononuclear cells. Definition of an IL-2-independent pathway of perforin induction in CD8 T cells. J. Immunol. 1992;148:3354–3360. [PubMed] [Google Scholar]
- 47.Weiss A., Wiskocil R.L., Stobo J.D. The role of T3 surface molecules in the activation of human T cells: a two-stimulus requirement for IL 2 production reflects events occurring at a pre-translational level. J. Immunol. 1984;133:123–128. [PubMed] [Google Scholar]
- 48.Couronne O., Poliakov A., Bray N., Ishkhanov T., Ryaboy D., Rubin E., Pachter L., Dubchak I. Strategies and tools for whole-genome alignments. Genome Res. 2003;13:73–80. doi: 10.1101/gr.762503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dorschner M.O., Hawrylycz M., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., et al. High-throughput localization of functional elements by quantitative chromatin profiling. Nature Methods. 2004;1:219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- 51.Warming S., Costantino N., Court D.L., Jenkins N.A., Copeland N.G. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Labrador M., Corces V.G. Setting the boundaries of chromatin domains and nuclear organization. Cell. 2002;111:151–154. doi: 10.1016/s0092-8674(02)01004-8. [DOI] [PubMed] [Google Scholar]
- 53.Patrinos G.P., de Krom M., de Boer E., Langeveld A., Imam A.M., Strouboulis J., de Laat W., Grosveld F.G. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 2004;18:1495–1509. doi: 10.1101/gad.289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bulger M., Schubeler D., Bender M.A., Hamilton J., Farrell C.M., Hardison R.C., Groudine M. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse beta-globin locus. Mol. Cell. Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]