Abstract
Gene expression over time is, biologically, a continuous process and can thus be represented by a continuous function, i.e. a curve. Individual genes often share similar expression patterns (functional forms). However, the shape of each function, the number of such functions, and the genes that share similar functional forms are typically unknown. Here we introduce an approach that allows direct discovery of related patterns of gene expression and their underlying functions (curves) from data without a priori specification of either cluster number or functional form. Smoothing spline clustering (SSC) models natural properties of gene expression over time, taking into account natural differences in gene expression within a cluster of similarly expressed genes, the effects of experimental measurement error, and missing data. Furthermore, SSC provides a visual summary of each cluster's gene expression function and goodness-of-fit by way of a ‘mean curve’ construct and its associated confidence bands. We apply this method to gene expression data over the life-cycle of Drosophila melanogaster and Caenorhabditis elegans to discover 17 and 16 unique patterns of gene expression in each species, respectively. New and previously described expression patterns in both species are discovered, the majority of which are biologically meaningful and exhibit statistically significant gene function enrichment. Software and source code implementing the algorithm, SSClust, is freely available (http://genemerge.bioteam.net/SSClust.html).
INTRODUCTION
Time course microarray experiments, where thousands of genes are assayed repeatedly over many time-points, generate great amounts of high-dimensional data. Clustering algorithms have been crucial in reducing the dimensionality of such data to aid in biological inference. For the majority of time course gene expression experiments, however, the number and shape of expression patterns over time is not known. An ideal clustering method would provide a statistically significant set of clusters and curves derived from the data themselves without relying on a pre-specified number of clusters or set of known functional forms. Further, such a method should take into account the between time-point correlation inherent in time course data and be able to handle missing data. Some popular methods such as k-means clustering (1), self-organizing maps (SOM) (2) and hierarchical clustering (3) do not satisfy the above requirements. One promising approach is to use a general multivariate Gaussian model to account for the correlation structure (4); however, such a model ignores the time order of gene expression. As evidenced in our analysis of real data, the time factor is important in interpreting the clustering results of time course data. Another approach is to use an auto-regression model to describe the gene expression time series (cluster analysis of gene expression dynamics, ‘CAGED’) (5). Unfortunately, such models often require stationarity and the Markov property, which are unlikely to hold for most time course microarray data.
A curve-based clustering method called FCM was introduced in (6) to cluster sparsely sampled time course data. Similar approaches were proposed in (7–9) to analyze time course gene expression data. In these methods, the mean gene expression profiles are modeled as linear combinations of spline bases. However, with different choices of the number of bases and knots, one could get an array of quite different estimates of the underlying curves. Effective methods or guidance on how to select the number of bases and knots are still lacking, which hinders the effective use of these methods in real applications (10). Additionally, the spline bases for the ‘mean gene expression profiles’ are assumed to be the same in (6–8), which create difficulties when accommodating very dissimilar expression patterns in different clusters. Moreover, these methods utilize the basic Expectation-Maximization (EM) algorithm (11) and are computationally very expensive for the analysis of large-scale gene expression data as in our examples. More recently, a Bayesian curve-based hierarchical clustering algorithm has been proposed (12). However, in addition to utilizing a similar modeling strategy as in (6–9) (i.e. using spline basis functions), this algorithm favors highly unbalanced clusters a priori, which could lead to non-interpretable results. Furthermore, it is based on ad hoc combination of the EM and Markov chain Monte Carlo algorithms, which both lacks theoretical coherence and is very costly for large-scale gene expression data.
Here, we introduce a data-driven clustering method, called smoothing spline clustering (SSC), that overcomes the aforementioned obstacles using a mixed-effect smoothing spline model and the rejection-controlled EM algorithm (11). The SSC method not only provides gene-to-cluster assignment but a predicted mean curve for each cluster and associated confidence bands and R2 value for each cluster. A distinguishing feature of SSC is that it accurately estimates individual gene expression profiles and the mean gene expression profile within clusters simultaneously, making it extremely powerful for clustering time course data.
MATERIALS AND METHODS
Gene expression curves—smoothing spline model
Since gene expression values change over time in a smooth fashion, we wish to fit our data to a curved function (with respect to time ti). Thus, we assume that gene expression can be represented by the general model:
where the ɛj are the ‘errors’ modeled by a Gaussian distribution N (0, σ2). A standard practice for ‘curve fitting’ is to minimize the residual sum of squares (RSS):
| 1 |
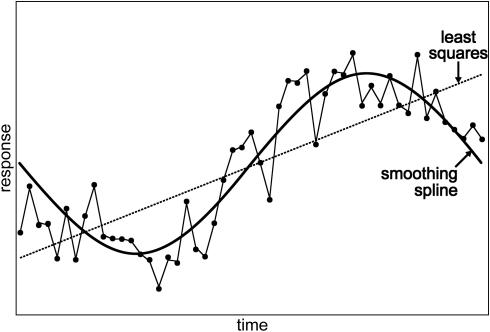
However, since we do not wish to impose a particular parametric form for the curve, there exist many functions that can pass through all the observed data points (the resulting RSS is thus zero, see Figure 1). A strategy employed in (6–9) to avoid such over-fitting is to parameterize f(t) by a set of pre-specified basis functions. A more flexible strategy is to impose a smoothness condition, which is also scientifically desirable here. Here, we adopt a standard constraint used in the statistics literature, i.e.
| 2 |
for some specific η. According to the Lagrange multiplier method, minimizing Equation 1 under constraint 2 is equivalent to minimizing the combined function:
| 3 |
which results in a curve that is known as a cubic smoothing spline (13–15) [a spline is a smoothed, piecewise polynomial (13)], which has the following analytic form
where (.)+ denotes positive part of the number. This solution also has a likelihood (or Bayesian) interpretation. By writing f(t) = δ0 + δ1t + f1(t), one can show that the minimizer of Equation 3 is the posterior mean and mode of f(t) when a uniform prior distribution on (δ0, δ1) and an independent, zero-mean, Gaussian process prior on f1 are imposed (14,15).
Figure 1.
Curve fitting with an arbitrary function showing over-fitting, a least squares fit, and curve fitting with a smoothing spline.
Clustering expression curves using a mixed-effect model
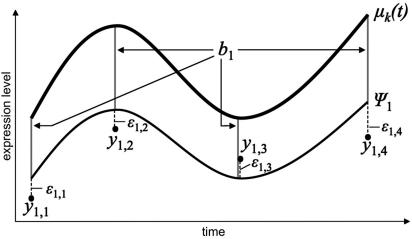
Now that we have a function-based framework for modeling gene expression data over time, we proceed to define gene expression clusters. Firstly, we model the ‘mean curve’ for each cluster of genes by a smoothing spline. The time course mRNA expression level of a gene in a given cluster is assumed to follow the shape of the mean curve, but with an additional gene-specific shift (b in Figure 2), which is called a ‘random effect’ in the statistics literature. The actual observations at pre-specified discrete time-points are subject to normally distributed measurement errors. More specifically, given that gene i is in cluster k, its observed mRNA expression at time tij (where the subscript j is a time-point label) can be written as:
| 4 |
where, μk is the mean curve, bi ∼ N (0, ) explains the gene-specific deviation from μk that is not due to measurement error, and ɛij ∼ N (0, σ2) is the Gaussian measurement error. An illustration of the mixed-effect model is shown in Figure 2. Model 4 has been extensively studied in statistical literature, e.g. (16,17). By expressing explicitly , where ET×T is a T × T matrix with all entries being 1, and IT×T is a T×T identity matrix, model 4 is equivalent to saying that yi ∼ N (μk, Σk), where yi and μk are the vector representations of the expression observations and the mean curve.
Figure 2.
Mixed-effect model representation of gene expression over time. The deviation of gene y1's expression from the mean curve μ(t) in cluster k, is a combination of the so-called random effect b and the measurement error ɛ. Ψ1 represents the ‘real’ expression curve of gene 1. Note that the b is constant over all time-points and captures between time-point dependence.
Since a gene's cluster membership is generally unknown, we can model the time course expression vector yi by a mixture Gaussian distribution, i.e.
| 5 |
where K is the total number of clusters, p1, p2, … , pK are relative sizes (proportions) of these clusters, and μk and Σk are as defined above. In words, we assume that gene i has probability pk to belong to cluster k a priori.
The maximum penalized likelihood approach to estimating parameters
Consider first only one cluster, cluster k, containing n genes. Based on the forgoing mixed-effect model formulation, we can write down the log-likelihood function (distribution) for y and b, together with penalty term for the curve's smoothness:
| 6 |
Intuitively, the first part of this expression describes the measurement error; the second part describes the gene-specific shift; and the third part, the smoothness penalty, which also forces the estimator of μk to be correlated between time-points. The maximization of Equation 6 results in an estimate of μk as a smoothing spline.
To incorporate the cluster assignment proportions described in Equation 5, we combine 5 and 6 to yield the complete data penalized log-likelihood:
| 7 |
Maximizing Equation 7 will provide us the most efficient estimates of the unknown parameters (including the curve μ), although it is analytically intractable. When multiple clusters are present, we also wish to simultaneously assign genes to appropriate clusters and to estimate the above parameters for each cluster. The following section presents our algorithmic approach to address these questions.
Model fitting using a variation of the EM algorithm
Since directly maximizing the penalized log-likelihood (Equation 7) is not analytically possible, we develop a variation of the EM algorithm (11) in conjunction with generalized cross-validation (GCV) (17,18) for the task. In our case, the expectation step of the EM algorithm is the computation of the probability that a particular gene belongs to each cluster given all the parameters in the model, which is simply,
| 8 |
The maximization step of the EM algorithm involves computing and maximizing the weighted version of the penalized log-likelihood (Equation 7) for each cluster:
| 9 |
The estimated gene expression in a particular cluster can be expressed as a linear combination of the observed gene expression, i.e. . The matrix obtained by arranging aijlm in proper entries is called the smoothing matrix.
We use a refined leave-one-out cross-validation procedure called GCV (17,18) to choose values for and λk. The use of GCV to choose values for and λk in this context has been shown to asymptotically minimize the discrepancy between the true and estimated expression profiles (17). The error variance σ2 can then be estimated by the RSS of the data.
The cluster proportion parameters pk are then updated by:
| 10 |
These steps are iterated until convergence. This process is illustrated in Figure 3.
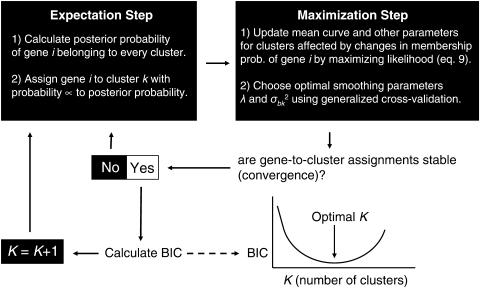
Figure 3.
Outline of the SSC algorithm.
Rejection-controlled EM (RCEM)
With thousands of genes under consideration, the exact EM algorithm is very costly to implement since the M-step involves maximizing a function that is the sum over all the genes in all clusters (with weights, Equation 9). The resulting algorithm is very unstable and error-prone. To alleviate the computational cost and to stabilize the algorithm, we propose the following RCEM algorithm [see ref. (19) for details of the rejection control method].
First, we set a ‘low’ threshold value c (e.g. c = 0.05) for gene-to-cluster membership probabilities. Genes with a cluster membership probability greater than this threshold are unaffected. However, genes with a gene-to-cluster membership probability less than c are reassigned either probability zero or probability c of belonging to the particular cluster at hand. The former assignment is made with probability 1 − P (genei ∈ k)/c and the latter with probability P (genei ∈ k)/c. By setting many ‘low-probability’ genes to zero, we greatly reduce the cost of the summation in Equation 9.
Note that when c = 0, the proposed algorithm is exactly the EM algorithm, whereas the proposed algorithm reduces to the Monte Carlo EM algorithm (20,21) when c = 1. In this way, it is possible to make very accurate approximations during the E-step while greatly reducing the cost of the M-step. Finally, in order to avoid local optima, the RCEM is run with multiple chains.
Choosing the final number of clusters
Without restriction, a model with a larger number of clusters will always be favored since it has more free parameters and will always have a better fit. But as the number of clusters increases, the model starts to over-fit the data, resulting in poor predictive power. In our likelihood framework, the Bayesian information criterion (BIC) (22) is a natural choice for penalizing model complexity. BIC in our context is defined as
where vk is the number of free parameters in the kth cluster, which is defined as the trace of its smoothing matrix obtained in the M-step (Equation 9) of RCEM [see Supplementary Data and (15,17) for details]. The BIC imposes a penalty on the total number of parameters, scaled by the logarithm of sample size, so as to strike a balance between the goodness-of-fit and the model complexity.
Starting with an initial set of two clusters derived from a simple clustering algorithm, for example k-means, we calculate the mean curve for each cluster and its associated parameters. Then, using the EM algorithm, we update both gene-to-cluster assignment and the estimated expression curve parameters iteratively for all genes until they converge. Based on the parameter estimates after convergence, we can calculate BIC. We then increase K by one and repeat the above steps. This process is repeated until the value of BIC begins to rise resulting in a roughly U-shaped BIC curve. The smallest value of BIC is then used to determine the final number of K clusters. Figure 3 outlines the main steps of the algorithm. Software and source code implementing the SSC algorithm, SSClust, is freely available (http://genemerge.bioteam.net/SSClust.html).
Measuring the strength of clusters
In our curve clustering procedure, an approximate sampling variance of each mean curve can be computed via the RCEM algorithm, which is used to construct 95% confidence intervals/bands (15,17). Another measure, R2, the analog of the R2 in a linear regression, estimates the fraction of variation within each cluster that can be explained by the mixed-effect model. The higher the value is, the tighter the cluster is.
RESULTS
Simulations
To assess the performance of SSC, we carried out extensive analyses on simulated datasets. First, 100 time series datasets consisting of 150 curves each across 10 time-points were generated from four different functions,
We randomly generated 30, 40, 50 and 30 curves from each function with Gaussian noise, respectively, using different variances and between time-point covariances,
30 observations (2% of all observations) were chosen randomly, removed and then treated as missing data. For k-means, CAGED and MCLUST, we impute the missing data by zeros.
We applied SSC to each of the 100 simulated datasets to determine how well the algorithm was able to recover: (i) the true number of clusters, (ii) the mean curve for each function and (iii) the true classification of expression profiles (curves). These were assessed by determining the misclassification rate: the number of misclassified curves/total number of curves, and, when applicable, the overall success rate: the fraction of times an algorithm recovered the correct number of clusters × (1 − the misclassification rate). We compared the SSC algorithm with the k-means algorithm (1), MCLUST (4), CAGED (5) as well as FCM (6). Since a cluster number must be specified a priori with k-means and the partially implemented FCM software, we gave a significant starting advantage to the k-means and FCM algorithms by letting the number of clusters k be the true number of clusters (four). Since the k-means algorithm is easily stuck in local optima, we ran it five times with random initial cluster configurations and reported the lowest misclassification rate. For MCLUST, eight models with different covariance structures were fitted to the data. The clustering result from the model with optimal BIC was reported.
In this study, CAGED and MCLUST chose the correct number of clusters 14 out of 100 times (14%) and 77 out of 100 times (77%), respectively, whereas SSC chose the correct number of clusters 100 out of 100 times (100%). The estimated mean curve using SSC for each cluster fit the true mean curves (functions) exceptionally well (Figure 4). We found that across 100 replicates the average misclassification rates of the k-means algorithm using Euclidean distance and Pearson's r, and using FCM was 9.73, 2.64 and 0.75%, respectively, whereas that of SSC was only 0.13% with an overall success rate of 98.7%. For 14 replicates where CAGED chose the correct number of clusters, the average misclassification rate was 11.07% yielding an overall success rate of 2.93%. For the 77 replicates where MCLUST chose the correct number of clusters, the average misclassification rate was 0.38% yielding an overall success rate of 69.5%. The analysis above was repeated for all algorithms using complete data (no missing data). We found SSC still had the lowest misclassification rate (0.13%) and highest overall success rate (98.7%). These results suggest that SSC outperforms k-means and FCM (even under the ideal scenario where the correct number of k clusters is provided to k-means and FCM a priori) as well MCLUST and CAGED. The computational complexity of one iteration of SSC is approximately O (n2 × t2).
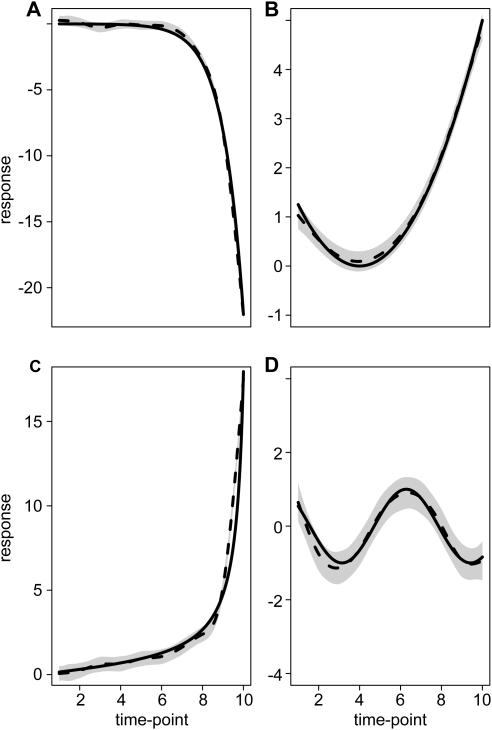
Figure 4.
Estimated mean curves and 95% confidence bands for one of each the 4 functions of the 100 simulated time series datasets. The true mean functions are shown as solid lines, the dashed lines are the estimated mean curves, and grey bands are 95% confidence bands for mean curves for each cluster.
Drosophila expression time course data
In a previous study, the mRNA levels of 4028 genes in wild-type flies (Drosophila melanogaster) were obtained using cDNA microarrays during ∼70 time-points beginning at fertilization and spanning embryonic, larval, pupal stages and the first 30 days of adulthood (23). mRNA was extracted from mixed male and female populations until adulthood where males and females were sampled separately. Each experimental sample was hybridized to a common reference sample from pooled mRNA from all stages of the life-cycle.
Starting with an initial cluster of k = 2 based on k-means clustering, the 3873 non-redundant genes on the array were clustered by SSC into a final optimal set of 17 clusters after examining up to k = 28 clusters. For each gene expression cluster we obtained its mean expression curve and the associated 95% point-wise confidence interval and R2 value. A complete list of genes in each cluster, raw and mean expression curves for all 17 clusters can be found in the Supplementary Data. To accommodate expression differences between the sexes in adult flies, we used two branches after metamorphosis to model the expression of adult male and female flies separately using branching splines (24) (Supplementary Data).
As a check on the biological validity of the clusters obtained, we used GeneMerge (25) to identify the annotated functions of all genes in each of the 17 clusters to determine if any particular functional categories were statistically over-represented in each dataset. Gene functions in the ‘biological process’ category were obtained from the Gene Ontology Consortium (26) and were used as input for GeneMerge. Note that the entire hierarchy of gene ontology terms for each gene was used when assessing functional over-representation, therefore nested categories are reported that often contain the same sets of genes. Bonferroni corrected P-values are given unless otherwise noted. These annotated functions were also used in conjunction with the estimated cluster mean gene expression profiles over time to check the biological validity of the results.
We found good agreement between the gene expression clusters discovered using SSC and known or expected biological functions and discovered several new, biologically meaningful patterns of gene expression not previously reported. Of the 17 clusters discovered, 12 clusters (70%) exhibited significant functional over-representation of genes with known biological processes in the fly (P < 0.05, Supplementary Data).
Mean expression curves obtained by SSC for each cluster were also used in the biological interpretation of each cluster. The mean gene expression curves for four of these clusters and their 95% point-wise confidence bands are given in Figure 5. All clusters with mean curves and raw expression profiles can be found in the Supplementary Data.
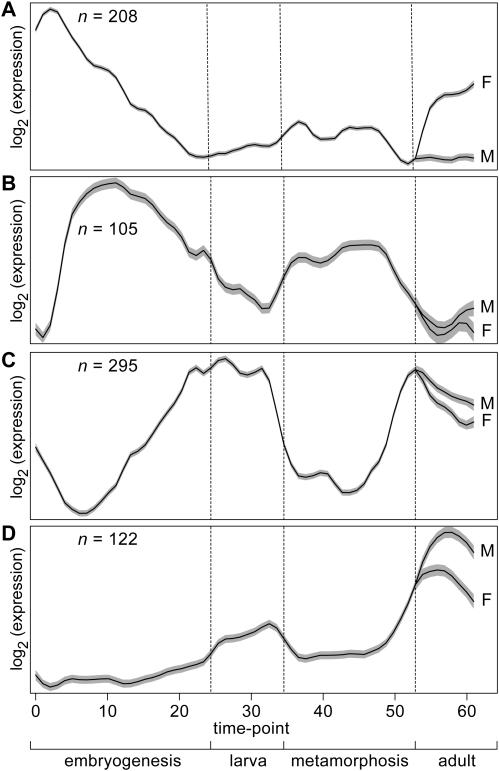
Figure 5.
Estimated mean expression curves (solid lines) and 95% confidence bands (grey bands) for four of 17 clusters discovered by SSC in the D.melanogaster (fly) time course microarray data (23). Adult male and female mean expression curves are labeled M and F, respectively.
The mean expression curve of cluster 5A (R2 = 0.560) which contains 208 genes shows a peak in gene expression in older females and the early embryo (Figure 5). This pattern, which has been described previously (23), is thought to represent female production of eggs and cell proliferation in the developing embryo. Consistent with this view, an over-representation of genes involved in DNA replication (P < 10−18), mitosis (P < 10−9) and cell proliferation (P < 10−12) was present in this cluster (Table 1) among other related functions (Supplementary Data).
Table 1.
Functional over-representation of genes in 4 of 16 clusters discovered by SSC in D.melanogaster (fly) time course micoarray data (19) corresponding to Figure 5
| Cluster | Gene ontology description | Fraction | Corrected P |
|---|---|---|---|
| 5A | DNA replication and chromosome cycle | 34/208 | 1.40E-19 |
| Cell proliferation | 53/208 | 7.36E-13 | |
| DNA replication | 17/208 | 1.86E-09 | |
| Nuclear organization and biogenesis | 18/208 | 1.22E-08 | |
| Mitosis | 24/208 | 1.80E-08 | |
| DNA packaging | 15/208 | 2.03E-06 | |
| 5B | Morphogenesis | 47/109 | 4.17E-19 |
| Organogenesis | 46/109 | 4.69E-19 | |
| Development | 52/109 | 1.68E-15 | |
| Neurogenesis | 27/109 | 1.52E-11 | |
| Embryonic development | 23/109 | 2.01E-10 | |
| Cell-fate determination | 11/109 | 1.68E-06 | |
| 5C | Oxidative phosphorylation | 20/295 | 6.26E-15 |
| Carboxylic acid metabolism | 28/295 | 6.22E-05 | |
| Lipid metabolism | 30/295 | 0.00022 | |
| Amino acid metabolism | 19/295 | 0.001624 | |
| Carbohydrate metabolism | 24/295 | 0.005684 | |
| Energy derived by oxidation of organic compounds | 13/295 | 0.008079 | |
| 5D | Phototransduction | 8/122 | 4.84E-06 |
| Detection of external stimulus | 11/122 | 2.64E-05 | |
| Response to abiotic stimulus | 14/122 | 0.000101 | |
| Sensory perception | 8/122 | 0.005561 | |
| Phototransduction, UV | 3/122 | 0.009644 | |
| Visual perception | 7/122 | 0.011772 |
From the estimated mean curve of cluster 5B (Figure 5), it can be observed that many genes that are up-regulated during embryogenesis are also up-regulated during metamorphosis, suggesting that many genes used for pattern formation during embryogenesis (the transition from egg to larva) are re-deployed during metamorphosis (the transition from larva to fly). Not surprisingly, this cluster (R2 = 0.557) was enriched for genes primarily involved in development (P < 10−14) (Table 1). A similar two-peak expression pattern was found using a peak-finding algorithm by (23) without suggesting statistical significance. Of the 109 genes in this cluster, 47 genes are known to be involved in morphogenesis (P < 10−18), 27 in neurogenesis (P < 10−10) and 8 involved in cell-fate specification (P < 10−5), as well and several other aspects of fly pattern formation (Supplementary Data). This pattern of gene expression is both biologically meaningful and statistically robust.
In cluster 5C, an almost opposite pattern of gene expression was found using SSC where gene expression peaks during larval and adult life and is at a minimum during developmental periods (Figure 5C). The 295 genes in this cluster (R2 = 0.593) were significantly enriched for oxidative phosphorylation (P < 10−14), a process by which energy is generated in the form of ATP. Additionally, cluster 5C contains an over-representation of genes involved in the metabolism of carbohydrates, lipids and amino acids (P < 0.01, for all) (Table 1). This up-regulation of energy production and metabolism genes during larval and adult life is expected since these are the stages during which nutrients are obtained from the environment, stored for morphogenesis and reproduction, and utilized for locomotion. This exciting pattern has not been described before and was not detected using either SOM or hierarchical clustering methods used in (23).
Finally, we observed a cluster of genes (n = 122) that peak in expression primarily in adult males (Figure 5D). Interestingly, this cluster (R2 = 0.553) is enriched with genes involved in phototransduction (P < 5 × 10−6) (Table 1). A total of 8 of 17 genes on the array known to be involved in this process were found in cluster 5D. This novel pattern, not detected using either SOM or hierarchical clustering methods used by Arbeitman et al. (23), was independently confirmed by separate microarray analyses conducted on adult male and female flies using different cDNA microarrays (27). Analysis of these experiments showed that genes in the phototransduction network are more highly expressed in males than females (data not shown).
Caenorhabditis elegans expression time course data
The time course of fly development in (23) was particularly densely sampled so we were interested in knowing whether SSC would be equally powerful when applied to more sparsely sampled time course data. cDNA microarray expression data for 17 871 genes were collected over the life-cycle of the nematode C.elegans by Jiang et al. (28) and contained 6 time-points, including eggs, larval stages: L1, L2, L3 and L4 and young adults. Because smoothing spline methods are not recommended for fewer than 5 time-points, this dataset challenges the lower limit of the SSC algorithm. Since the data of Jiang et al. (28) are sparse and contained no controls (28) we filtered them to include only those genes that were significantly modulated over the time course by a χ2 criterion (P < 0.01) (29). After filtering, a total of 3118 genes were found to be significantly modulated and were clustered using SSC. Starting with an initial cluster of k = 2 as above, the 3118 genes were clustered into a final optimal set of 16 clusters. The mean expression curves and associated 95% point-wise Bayesian confidence bands for three of these clusters are given in Figure 6. A complete list of genes in each cluster and raw and mean expression curves for all 16 clusters can be found in the Supplementary Data.
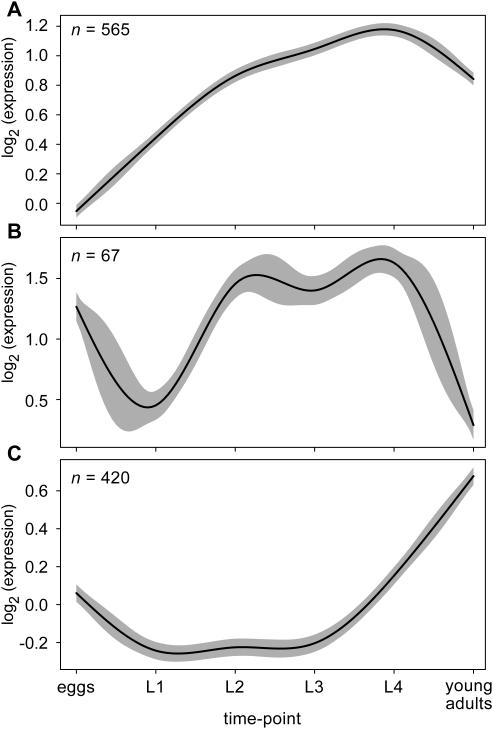
Figure 6.
Estimated mean expression curves (solid lines) and 95% confidence bands (grey bands) for three of 16 clusters discovered by SSC in the C.elegans (worm) time course microarray data (28).
Once again, we found good agreement between the SSC-based gene expression clusters and known or expected biological functions, and discovered several new and meaningful patterns of gene expression not previously reported. Of the 16 clusters discovered by SSC, 8 (50%) exhibited significant functional over-representation of known biological processes (P < 0.05, Supplementary Data).
For example, of 596 genes in cluster 6A (Figure 6) (R2 =0.505), 165 were involved in the process of worm growth according the Gene Ontology annotations, representing a highly significant enrichment of gene function in this cluster (P < 10−49) (Supplementary Data). Also enriched in this expression cluster were genes involved in development, metabolism and reproduction (P << 10−20 for all) (Supplementary Data). The steady rise in expression of genes belonging to cluster 6A, beginning from eggs to the last larval stage (L4) with a slight drop in young adults, is consistent with the normal growth, development and concomitant metabolic expenditure of maturing worms over the course of their life-cycle. A complete list of significantly enriched gene functions for this and all other clusters can be found in the Supplementary Data.
In cluster 6B (R2 = 0.760), which consists of only 67 genes, gene expression is at a maximum during L2 through L4 (Figure 6B). Here we find an over-representation of genes involved in locomotory behavior (P < 10−7), the regulation of growth (P < 10−3) and cuticle biosynthesis (P < 0.007) (Supplementary Data). A similar pattern of gene expression was found in one of the SOM clusters by (28). When this SOM cluster was examined for functional over-representation as done here, a similar pattern of functional enrichment was found (data not shown).
Cluster 6C (R2 = 0.323) is particularly interesting in that it contains genes with an expression pattern similar to that of cluster 5A in the fruit fly (Figure 5). Just as in the fly, cluster 6C is significantly enriched for genes involved in cell proliferation, the cell-cycle, and DNA replication (P < 0.01, P < 0.01 and P < 0.0003, uncorrected, respectively) (Supplementary Data). This pattern of functional enrichment was not found among any of the SOM clusters reported by (28) (data not shown). Although the young adults sampled are reported to have been collected ‘without eggs’ at the time of mRNA extraction (28), spermatogenesis begins during L3 and oogenesis during L4 in C.elegans (30) which is hermaphroditic. Thus, gametogenesis is likely to have been underway at time of worm collection resulting in the observed pattern of cluster 6C.
Indeed, 20 genes known to be involved in gametogenesis were found in this cluster (P < 0.0078, uncorrected) and 2 of the 5 genes in the C.elegans genome known to be involved in female gamete generation were also found in this cluster (P < 0.0053, uncorrected). Thus, it seems likely that gametogenesis in hermaphroditic worms, as in female flies, is responsible for the peak in gene expression of genes involved in cell proliferation in adults.
Overall, in the sparsely sampled nematode developmental time course data of Jiang et al. (28), SSC facilitated the automatic detection of previously described and novel patterns of gene expression. In total 50% of the clusters discovered by SSC exhibited statistically significant functional enrichment, a result that compares favorably with the 70% of clusters with significant functional enrichment in the more densely sampled expression data in D.melanogaster (23). This suggests that SSC is effective in detecting data-driven patterns of gene expression in both densely and sparsely sampled time course data.
DISCUSSION
Existing clustering methods such as k-means, SOM and others often require a priori specification of either the number of expected patterns in the data, a set of expected functional curves, or, in the case of hierarchical clustering, an ad hoc pruning procedure to generate clusters. SSC overcomes these limitations by modeling the natural properties of gene expression over time, taking into account differences in gene expression within a cluster of similarly expressed genes, and the effects of experimental measurement error. Furthermore, SSC provides a visual summary of each cluster's gene expression function and goodness-of-fit by way of the mean curve construct and associated confidence bands. The algorithm handles missing data automatically and is able to incorporate prior biological knowledge, if available. Finally, SSC provides a data-driven and statistically grounded criterion for determining the number of clusters in the data.
Application of SSC to time course microarray data from D.melanogaster and C.elegans life-cycles with 69 and 6 time-points, respectively, yielded significant clusters of gene expression that recapitulated results of the original analyses and facilitated the discovery of several new and biological meaningful patterns of gene expression not found with SOM, hierarchical clustering or peak-finding algorithms used in the original analyses. Since SSC is a general method, it may be applied to other types of time course data where between time-point dependence occurs, missing data may be present, and where the discovery of different functional forms and their number is desired.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work is supported in part by the National Science Foundation grant DMS-0244638 and DMS-0204674 and the China National Science Foundation grant CNSF 10228102. Special thanks to Chong Gu, Christian Landry and Yu Zhu for suggestions and comments and Renate Hellmiss-Peralta for help with figure design. The authors also thank two anonymous referees for their constructive suggestions. Funding to pay the Open Access publication charges for this article was provided by National Science Foundation, USA and China National Science Foundation, China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hartigan J.A., Wong M.A. A K-means clustering algorithm. App. Statist. 1978;28:100–108. [Google Scholar]
- 2.Kohonen T. Self-Organizing Maps. NY: Springer; 1997. [Google Scholar]
- 3.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraley C., Raftery A.E. Model-based clustering, discriminant analysis, and density estimation. J. Amer. Statist. Assoc. 2002;97:611–631. [Google Scholar]
- 5.Ramoni M., Sebastiani P., Kohane P.R. Cluster analysis of gene expression dynamics. Proc. Nat. Acad. Sci. USA. 2002;99:9121–9126. doi: 10.1073/pnas.132656399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James G., Sugar C. Clustering for sparsely sampled functional data. J. Amer. Statist. Assoc. 2003;98:397–408. [Google Scholar]
- 7.Luan Y., Li H. Clustering of time-course gene expression data using a mixed-effects model with B-spline. Bioinformatics. 2003;19:474–482. doi: 10.1093/bioinformatics/btg014. [DOI] [PubMed] [Google Scholar]
- 8.Luan Y., Li H. Model-based methods for identifying periodically regulated genes based on the time course microarray gene expression data. Bioinformatics. 2004;20:332–339. doi: 10.1093/bioinformatics/btg413. [DOI] [PubMed] [Google Scholar]
- 9.Storey J.D., Xiao W., Leek J.T., Tompkins R.G., Davis R.W. Significance of time course microarray experiments. Proc. Natl Acad. Sci. USA. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruppert D., Wand M.P., Carroll R.J. Semiparametric Regression. Cambridge, UK: Cambridge University Press; 2003. [Google Scholar]
- 11.Dempster A., Laird N., Rubin D. Maximum likelihood from incomplete data via the EM algorithm. J. Royal Statist. Soc. B. 1977;39:1–38. [Google Scholar]
- 12.Heard N.A., Holmes C.C., Stephens D.A., Dimopoulos G., Hand D.J. Bayesian coclustering of Anopheles gene expression time series: study of immune defense response to multiple experimental challenges. Proc. Natl Acad. Sci. USA. 2005;102:16939–16944. doi: 10.1073/pnas.0408393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoenberg I.J. Spline functions and the problem of graduation. Proc. Natl Acad. Sci. USA. 1964;52:947–950. doi: 10.1073/pnas.52.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahba G. Spline Models for Observational Data.; CBMS-NSF Regional Conference Series in Applied Mathematics. SIAM; Philadephia. 1991. [Google Scholar]
- 15.Gu C. Smoothing Spline ANOVA Models. NY: Springer; 2002. [Google Scholar]
- 16.Wang Y. Mixed-effects smoothing spline ANOVA. J. Royal Statist. Soc. B. 1998;60:159–174. [Google Scholar]
- 17.Gu C., Ma P. Optimal smoothing in nonparametric mixed-effect models. Ann. Statist. 2005;33:1357–1379. [Google Scholar]
- 18.Craven P., Wahba G. Smoothing noisy data with spline functions: estimating the correct degree of smoothing by the method of generalized cross-validation. Numer. Math. 1979;31:377–403. [Google Scholar]
- 19.Liu J.S., Chen R., Wong W.H. Rejection control for sequential importance sampling. J. Amer. Statist. Assoc. 1998;93:1022–1031. [Google Scholar]
- 20.Celeux G., Diebolt J. A probabilistic teacher algorithm for iterative maximum lilkelihood estimation. In: Bock H.H., editor. Classification and Related Methods of Data Analysis. Vol. I. Amsterdam: North Holland/Elsevier; 1988. pp. 617–623. [Google Scholar]
- 21.Wei G.C., Tanner M.A. A Monte carlo implementation of the EM algorithm and the poor man's data augmentation algorithms. J. Amer. Statist. Assoc. 1990;85:699–704. [Google Scholar]
- 22.Schwarz G. Estimating the dimension of a model. Ann. Statist. 1978;6:461–464. [Google Scholar]
- 23.Arbeitman M., Furlong E., Imam F., Johnson E., Null B.H., Baker B.S., Krasnow M.A., Scott M.P., Davis R.W., White K.P. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- 24.Silverman B.W., Wood T.J. The nonparametric estimation of branching curves. J. Amer. Statist. Assoc. 1987;82:551–558. [Google Scholar]
- 25.Castillo-Davis C.I., Hartl D.L. Genemerge: post-genomic analysis, data-mining and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- 26.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ranz J.M., Castillo-Davis C.I., Meiklejohn C.D., Hartl D.L. Sex-dependent gene expression and the evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 28.Jiang M., Ryu J., Kiraly M., Duke K., Reinke V., Kim S.K. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2001;98:218–223. doi: 10.1073/pnas.011520898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson R.A., Wichern D.W. Applied Multivariate Statistical Analysis. 5th edn. NJ: Prentice Hall; 2002. p. 234. [Google Scholar]
- 30.Hirsh D., Oppenheim D., Klass M. Development of the reproductive system of Caenorhabditis elegans. Dev. Biol. 1976;49:200–219. doi: 10.1016/0012-1606(76)90267-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.