Abstract
Background
Haematological cancer is characterised by chromosomal translocation (e.g. MLL translocation in acute leukaemia) and two models have been proposed to explain the origins of recurrent reciprocal translocation. The first, established from pairs of translocated genes (such as BCR and ABL), considers the spatial proximity of loci in interphase nuclei (static "contact first" model). The second model is based on the dynamics of double strand break ends during repair processes (dynamic "breakage first" model). Since the MLL gene involved in 11q23 translocation has more than 40 partners, the study of the relative positions of the MLL gene with both the most frequent partner gene (AF4) and a less frequent partner gene (ENL), should elucidate the MLL translocation mechanism.
Methods
Using triple labeling 3D FISH experiments, we have determined the relative positions of MLL, AF4 and ENL genes, in two lymphoblastic and two myeloid human cell lines.
Results
In all cell lines, the ENL gene is significantly closer to the MLL gene than the AF4 gene (with P value < 0.0001). According to the static "contact first" model of the translocation mechanism, a minimal distance between loci would indicate a greater probability of the occurrence of t(11;19)(q23;p13.3) compared to t(4;11)(q21;q23). However this is in contradiction to the epidemiology of 11q23 translocation.
Conclusion
The simultaneous multi-probe hybridization in 3D-FISH is a new approach in addressing the correlation between spatial proximity and occurrence of translocation. Our observations are not consistent with the static "contact first" model of translocation. The recently proposed dynamic "breakage first" model offers an attractive alternative explanation.
Background
The Mixed Lineage Leukaemia (MLL) gene (also called HRX, HTRX or ALL-1) on chromosome band 11q23 is a recurrent target of reciprocal translocation in human acute leukaemias. The 11q23 translocation occurs in 5–6% of patients with acute myelogenous leukaemia (AML) and 7–10% of patients with acute lymphoblastic leukemia (ALL). More than 40 partners are involved in MLL translocation [2,3]; the most common partner being the AF4 gene, on band 4q21. Translocation t(4;11)(q21;q23) is involved in a third of all MLL translocation cases [4] and represents 95% of translocations involved in ALL, compared to only 3.3% in AML [5]. Another frequent partner of translocation is the ENL gene located on band 19p13.3, but whilst translocation t(11;19)(q23;p13.3) is found in both ALL and AML, it represents only 6% of all MLL translocations.
It has been demonstrated that pairs of genes (and chromosomal domains) which are frequently involved in reciprocal chromosomal translocation are close to each other in interphase nuclei. These observations have established a direct correlation between the spatial proximity of loci in interphase nuclei and the frequency of their translocation. This implies that the translocation takes place where and when the chromatin fiber containing the partner genes are co-localized (static "contact first" model). However, recent investigations, using alpha-particles for targeting double-strand breaks (DSBs), have demonstrated some displacements of DNA breaks into DNA repair foci where chromosomal exchange can occur. In other words, breaks formed at distal locations can subsequently be brought together to induce translocation (dynamic "breakage first" model).
In the present work, we propose a new direct approach to study the correlation between the relative position of genes in interphase nuclei and the corresponding frequencies of translocation. We have applied our approach to genes involved in 11q23 translocations (MLL, AF4 and ENL genes) in differentiated haematological cell lineages by 3D-FISH and in-depth image data analysis.
Methods
Four hematological cancer cell lines were chosen ; [NALM-6, IM-9, AML-193 and PLB-985] for their human near-diploid karyotype and because the chromosomes of interest (4, 11 and 19) were unmodified. NALM-6 is a bona fide lymphoid leukemia cell line, derived from the peripheral blood of a 19-year-old male with B-precursor acute lymphoblastic leukemia. IM-9 is a non-malignant lymphoid cell line derived by Epstein-Barr virus (EBV) tranformation of residual normal B-cells from a patient with multiple myeloma. AML-193 is a bona fide myeloid leukemia cell line, derived from a 13-year-old female with acute myeloid leukemia, subtype M5. PLB-985 is a false cell line, the result of a cross-contamination with the myeloid leukemia cell line HL-60. Hence cultures labeled as PLB-985 contain in reality HL-60 cells. (All cells were purchased from DSMZ – Braunschweig, Germany – except the NALM-6 cell line which was kindly provided by L. Lagneaux – Belgium). Cells were not synchronized and, in order to preserve their natural nuclear structure, no drugs were added. 3D-FISH was performed according to the standard protocol [16]. Bacterial Artificial Chromosomes (BAC) and Phage Artificial Chromosomes (PAC) were obtained from the Resources for Molecular Cytogenetics database (Italy) [17] : CTD-217A21 for the MLL gene and RP11-476C8 for the AF4 gene. RP11-2344B19 for the ENL gene was purchased from Invitrogen (Carlsbad, California). dUTP-Alexa 488, dUTP-Cy3 and dUTP-Cy5 were used to label BAC/PAC DNA (using random priming protocols) corresponding to MLL, AF4 and ENL genes respectively. Simultaneous hybridization was performed in order to compare the distance between the MLL gene and each potential translocation partner inside a single nucleus (figure 1). Confocal microscopy was carried out using the TCS confocal imaging system, equipped with a 63× objective with a numerical aperture set to 1.4 nm. The confocal pinhole was adjusted to allow a minimum field depth. The focus step between sections was 0.35 μm and the XY pixel was set to 100 nm. Gene to gene distances were calculated using Smart 3D-FISH, an ImageJ plugin, recently developed in our laboratory [19]. More than 1000 nuclei were analysed. Only cells (415) displaying two loci and adequate morphological preservation of chromatin – as assessed by 4'-,6-diamino-2-phenylindole (DAPI) staining – were retained for analysis. Inter-loci distances were normalized to nuclear diameter: first, the nuclear volume was calculated from the total volume of DAPI stained DNA; a spherical approximation was then used to calculate the diameter of the nucleus from this volume. The minimum value of measured distances was expressed in microns (figure 2) according to gene proximity criterion in which all loci separated by less than 2 μm, could be in contact by Brownian motion prior to the translocation event (according to the "contact first" model). Statistical analysis was performed using Student's t test with a level of significance : α = 5%.
Figure 1.
Example of 4-color images from simultaneous 3D-FISH experiments in NALM-6 cell nucleus. Labeled genes were MLL, AF4 and ENL represented by green, white and red spots respectively. Nuclear DNA stained with DAPI is displayed in blue.
Figure 2.
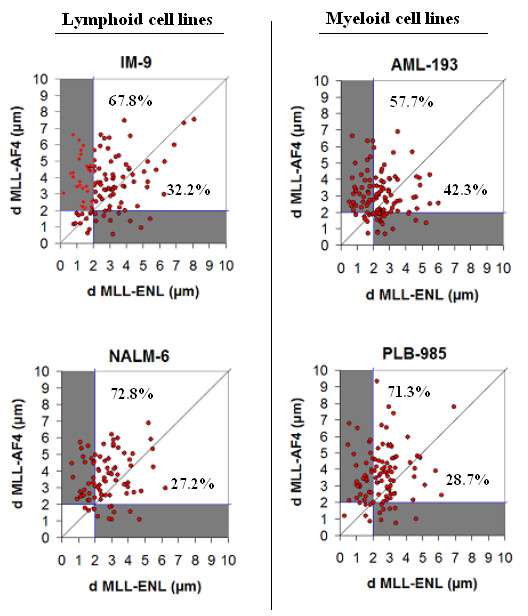
2D – Plot of minimum value of measured distances between MLL to AF4 and MLL to ENL genes for all cells. Distances are expressed in μm. Vertical and horizontal blue lines symbolize the gene proximity criterion (2 μm) below which genes could be potentially translocated. Values express the percentage of cells in which the distances reported in one axis is smaller than the distance on the other axis. Grey areas correspond to nuclei where only one of gene could be translocated.
Results and discussion
The mean inter-loci distance measurements between MLL, ENL and AF4 genes in the four cell lines are reported in table 1 (expressed as a percentage of nuclear diameter). No significant overall difference was observed in inter-loci distances between myeloïd and lymphoïd cell lines, despite cell line specific genome organization[21].
Table 1.
| Cell Lineage [No of loci] | ||||
| NALM-6 [n = 324] | IM-9 [n = 448] | PLB-985 [n = 432] | AML-193 [n = 444] | |
| MLL-ENL | 45.0 ± 1.1% | 45.3 ± 1.0% | 45.7 ± 0.9% | 39.4 ± 0.8% |
| MLL-AF4 | 54.8 ± 1.1% | 55.4 ± 1.0% | 57.8 ± 1.0% | 46.6 ± 0.9% |
| AF4-ENL | 48.5 ± 1.1% | 50.6 ± 0.9% | 43.7 ± 0.8% | 49.5 ± 1.0% |
Mean inter-loci distances between the MLL, AF4 and ENL genes in NALM-6, IM-9, PLB-985, AML-193 cell lines (expressed as a percentage of nuclear diameter).
In all the studied cell lines, the average separation between MLL and ENL loci was significantly smaller than that observed for MLL and AF4 (P < 0.0001). The mean smallest intergenic distance (corresponding to the minimum value of each of the four inter-loci distances) follows the same tendancy (26% of nuclear diameter for MLL-ENL distance vs. 34% for MLL-AF4 distance in NALM-6 and IM-9 cell lines, 26% vs. 36% for PLB-985 cell lines and 24% vs. 29% for AML-193 cell lines).
For each nucleus, the two smallest intergenic distance – dMLL-AF4 and dMLL-ENL – are displayed in figure 2. Firstly, our results show that, for the majority of nuclei, the ENL gene is closer to the MLL gene than to the AF4 gene. Secondly, according to the gene proximity criterion, we have calculated the number of nuclei where only the AF4 gene should be translocated with MLL gene (i.e. dMLL-AF4 < 2 μm and dMLL-ENL > 2 μm, greyed area in figure 2). In the same way, we computed the number of cells in which only the ENL gene should be translocated with the MLL gene (i.e. dMLL-ENL < 2 μm and dMLL-AF4 > 2 μm, greyed area in figure 2). The ratio of these two values, respectively, indicates a greater probability of a translocation event between MLL and ENL. (0.33, 0.35, 0.30 and 0.68 for NALM-6, IM-9, PLB-985 and AML-193 cell lines respectively).
Published MLL translocation frequencies report a high level of translocation t(4;11) compared to translocation t(11;19). According to the static "contact first" model, AF4 loci should be closer to MLL than ENL loci. Clearly, this is not in accordance with our data. In the case of the 11q23 translocation, the dynamic of chromatin must be taken into account and our results could be the first observation of the dynamic breakage first model proposed by Kanaar and co-workers[12].
It should be noted that published translocation frequencies were established after manifestation of cancer. Taking into account that a second genetic event seems to be required to induce cancer [22], we cannot exclude that the chromatin architecture context after MLL-AF4 translocation may be more favourable to the induction of a second event compared to MLL-ENL translocation. This could explain the high level of translocation t(4;11) detected in acute leukaemia. This could be illustrated by the relatively small distances between the ENL gene and the AF4 gene which could be favorable to translocation while the corresponding translocation is never reported in literature[3].
Conclusion
This work, based on simultaneous multi-probe hybridization in 3D-FISH experiments describes a new approach to study translocation mechanisms in a chromatin context. This method offers the advantage of a direct comparison of several translocation frequencies with localization of corresponding loci in each nucleus. Furthermore, it allows the simultaneous measurement of multiple intergenic distances in a cell by cell manner in a single experiment, without the chromatin distortion common to existing procedures.
Our results are not in accordance with the static model of translocation mechanism and may represent the first observation of the dynamic "breakage first" model. We are currently extending our approach to study MLL and up to six partner genes to confirm these intriguing results.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
MG carried out the cytogenetics studies and data analyses
TB participated in the data analyses and coordination of the study
JSS participated in the design of the study
All authors read and approved the final manuscript
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
The authors would like to thank Maïté Coppey and Christophe Chamot at Institut Jacques Monod (Paris, France) for providing access and assistance with confocal microcopy, Marriono Rocchi (University of Bari, Italy) for providing BAC/PAC FISH clones, and Laurence Lagneaux (Belgium) for NALM-6 cell line. Thanks to Jason Martin for reading the manuscript.
Contributor Information
Michaël Gué, Email: michael.gue@curie.u-psud.fr.
Jian-Sheng Sun, Email: sun@mnhn.fr.
Thomas Boudier, Email: thomas.boudier@snv.jussieu.fr.
References
- Rowley JD. Molecular genetics in acute leukemia. Leukemia. 2000;14:513–517. doi: 10.1038/sj.leu.2401600. [DOI] [PubMed] [Google Scholar]
- Huret JL, Dessen P, Bernheim A. An atlas of chromosomes in hematological malignancies. Example: 11q23 and MLL partners. Leukemia. 2001;15:987–989. doi: 10.1038/sj.leu.2402135. [DOI] [PubMed] [Google Scholar]
- Huret JL, Senon S, Bernheim A, Dessen P. An Atlas on genes and chromosomes in oncology and haematology. Cell Mol Biol (Noisy-le-grand) 2004;50:805–807. [PubMed] [Google Scholar]
- Secker-Walker LM, Moorman AV, Bain BJ, Mehta AB. Secondary acute leukemia and myelodysplastic syndrome with 11q23 abnormalities. EU Concerted Action 11q23 Workshop. Leukemia. 1998;12:840–844. doi: 10.1038/sj.leu.2401021. [DOI] [PubMed] [Google Scholar]
- Johansson B, Moorman AV, Haas OA, Watmore AE, Cheung KL, Swanton S, Secker-Walker LM. Hematologic malignancies with t(4;11)(q21;q23)--a cytogenetic, morphologic, immunophenotypic and clinical study of 183 cases. European 11q23 Workshop participants. Leukemia. 1998;12:779–787. doi: 10.1038/sj.leu.2401012. [DOI] [PubMed] [Google Scholar]
- Moorman AV, Hagemeijer A, Charrin C, Rieder H, Secker-Walker LM. The translocations, t(11;19)(q23;p13.1) and t(11;19)(q23;p13.3): a cytogenetic and clinical profile of 53 patients. European 11q23 Workshop participants. Leukemia. 1998;12:805–810. doi: 10.1038/sj.leu.2401016. [DOI] [PubMed] [Google Scholar]
- Neves H, Ramos C, da Silva MG, Parreira A, Parreira L. The nuclear topography of ABL, BCR, PML, and RARalpha genes: evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood. 1999;93:1197–1207. [PubMed] [Google Scholar]
- Kozubek S, Lukasova E, Mareckova A, Skalnikova M, Kozubek M, Bartova E, Kroha V, Krahulcova E, Slotova J. The topological organization of chromosomes 9 and 22 in cell nuclei has a determinative role in the induction of t(9,22) translocations and in the pathogenesis of t(9,22) leukemias. Chromosoma. 1999;108:426–435. doi: 10.1007/s004120050394. [DOI] [PubMed] [Google Scholar]
- Nikiforova MN, Stringer JR, Blough R, Medvedovic M, Fagin JA, Nikiforov YE. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science. 2000;290:138–141. doi: 10.1126/science.290.5489.138. [DOI] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- Parada L, Misteli T. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 2002;12:425–432. doi: 10.1016/S0962-8924(02)02351-6. [DOI] [PubMed] [Google Scholar]
- Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- Drexler HG. The Leukemia-Lymphoma Cell Line FactsBook. Academic Press. 2001.
- Drexler HG, Dirks WG, Matsuo Y, MacLeod RA. False leukemia-lymphoma cell lines: an update on over 500 cell lines. Leukemia. 2003;17:416–426. doi: 10.1038/sj.leu.2402799. [DOI] [PubMed] [Google Scholar]
- Urbani L, Sherwood SW, Schimke RT. Dissociation of nuclear and cytoplasmic cell cycle progression by drugs employed in cell synchronization. Exp Cell Res. 1995;219:159–168. doi: 10.1006/excr.1995.1216. [DOI] [PubMed] [Google Scholar]
- Spector LD, Goldman DR. A laboratory manual. 1998. p. 111.19 111.36.
- Ressources for Molecular Cytogenetics : [http://www.biologia.uniba.it/rmc]
- von Bergh A, Emanuel B, van Zelderen-Bhola S, Smetsers T, van Soest R, Stul M, Vranckx H, Schuuring E, Hagemeijer A, Kluin P. A DNA probe combination for improved detection of MLL/11q23 breakpoints by double-color interphase-FISH in acute leukemias. Genes Chromosomes Cancer. 2000;28:14–22. doi: 10.1002/(SICI)1098-2264(200005)28:1<14::AID-GCC2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Gue M, Messaoudi C, Sun JS, Boudier T. Smart 3D-fish: Automation of distance analysis in nuclei of interphase cells by image processing. Cytometry A. 2005;67:18–26. doi: 10.1002/cyto.a.20170. [DOI] [PubMed] [Google Scholar]
- Alcobia I, Dilao R, Parreira L. Spatial associations of centromeres in the nuclei of hematopoietic cells: evidence for cell-type-specific organizational patterns. Blood. 2000;95:1608–1615. [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;5:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukemia. Nature reviews. 2003;3 doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]


