Abstract
Caenorhabditis elegans has been an excellent model for studying many aspects of biology including host responses to bacterial pathogens1,2, but it is not known to support replication of any virus. It is also not known if small RNA-guided RNAi provides protection against viruses in single-Dicer C. elegans and mammals as documented in plants3 and insects4, which encode multiple Dicers that recognize distinct precursors of small RNAs and act cooperatively5-9. Here we demonstrate complete replication of the Flock house virus (FHV) bipartite, plus-strand RNA genome in C. elegans. We show that FHV replication triggers potent antiviral silencing in C. elegans that requires RDE-1, a worm Argonaute protein10,11 essential for RNAi by small interfering RNA (siRNA) but not by microRNA. The worm immunity is capable of rapid virus clearance in absence of expression of FHV B2 protein, a broad-spectrum RNAi inhibitor4,12 that acts upstream of rde-1 by targeting the precursor of siRNA. This work establishes the first C. elegans model for genetic studies of animal virus-host interactions and indicates an antiviral potential for RNAi in mammals via the siRNA pathway.
We chose animal nodavirus FHV to determine if C. elegans supports virus replication because FHV replicates in yeast, plant, insect and mammalian cells13. The first genome segment of FHV, RNA1, encodes the entire viral contribution to the viral RNA-dependent RNA polymerase (RdRP) and replicates autonomously in absence of RNA2, which depends on RNA1 for replication13 (Fig. 1a). Worm strains were generated to carry either a chromosomally-integrated FHV RNA1 (FR1) or RNA2 (FR2) transgene, designed to yield transcripts FR1 and FR2 in the soma after heat induction which are identical in sequence to FHV genomic RNAs 1 and 2, respectively (Fig. 1b). Northern blot hybridizations detected high level accumulation of FHV RNA1 in worms carrying the FR1-3 transgene following transcription induction (Fig. 1c, lane 3). FHV RNA2 accumulated to high levels in strain FR1-3/FR2 carrying both FR1-3 and FR2 combined by genetic crosses (Fig. 1c, lane 5), but was undetectable in worms carrying FR2 alone (Fig. 1c, lane 6). Thus, the abundant viral RNAs detected in transgenic worm strains resulted from active RNA replication since in absence of RNA replication the initial heat-inducible transcripts were below the limit of detection in worms two days after induction.
Fig. 1.
Replication and silencing of FHV in C. elegans. a, Structure, replication and expression of FHV genome. b, Structure of FHV transgenes. HIP- heat inducible promoter. The junction region between HIP (lower case) and FHV cDNA (upper case) is shown. Transcriptional initiation sites from HIP were indicated. Rz — a self-cleaving ribozyme used to reduce non-viral extensions at the 3'-termini of heat-induced transcripts. Northern blot detection of FHV RNA accumulation in C. elegans (c) and fruit fly S2 cells (d). Four μg total RNA was loaded per lane in both c and d, except for lane 8 of c.
Both FR1-3 and FR1-3/FR2 worms also contained abundant RNA3 (Fig. 1c, lanes 3 and 5). RNA3 is a subgenomic RNA transcribed during RNA1 replication from an internal site of the complementary, replicative intermediate of RNA1, (-)-RNA1 (Fig. 1a), and is not required to initiate FHV infection unlike the genomic RNAs. RNA1 replication initiated from FR1-2 transcripts was inefficient and became detectable only in the presence of RNA2 replication (Fig. 1c, lanes 2 and 4), which directs expression of the pre-capsid protein (pre-CP) essential for virion package (Fig. 1a). Taken together, detection of self-replication of RNA1 and trans replication of RNA2 as well as production of a subgenomic RNA in these worm strains provide compelling evidence that C. elegans supports complete replication of the FHV RNA genome.
To investigate a possible induction of antiviral silencing by FHV RNA replication in worms, we first examined if FHV accumulation in C. elegans requires expression of the FHV-encoded B2 protein, an RNAi suppressor that is active across the animal and plant kingdoms4,12 and is not known to have a direct role in FHV RNA replication13. We created a derivative of FR1-3 transgene, FR1-3ΔB2 (Fig. 1b), containing a point mutation that abolished the B2 ORF but had no effect on the out-of-frame overlapping viral RdRP ORF (Fig. 1b). In contrast to high levels of RNA1 and RNA3 in FR1-3 worms (Fig. 1c, lane 3), we detected no accumulation of FHV RNA1 in worms carrying an integrated FR1-3ΔB2 transgene (Fig. 1c, lane 7). Thus, B2 is also essential for FHV accumulation in C. elegans as has been demonstrated in cultured insect cells4,12.
If lack of FHV RNA accumulation in FR1-3ΔB2 worms was indeed due to loss of B2 suppression of RNAi, we would expect to find FR1-3ΔB2 accumulation rescued in worm mutants defective in support of RNAi. To test this hypothesis, worm strains carrying FR1-3 and FR1-3ΔB2 were each crossed into an rde-1 (ne219) mutant background10. Unlike many RNAi mutants such as dcr-1/Dicer that exhibit multiple developmental defects due to disruption of miRNA function8, rde-1 mutants are otherwise very healthy10. Northern blot analysis revealed that, whereas FHV RNA1 was undetectable in wild type N2 worms carrying FR1-3ΔB2 (Fig. 1c, lane 7), the same FR1-3ΔB2 transgene directed abundant accumulation of FHV RNAs 1 and 3 in the rde-1 mutant background (Fig. 1c, lane 8). In fact, the accumulation level of FHV RNAs in FR1-3ΔB2/rde-1 worms was similar to that in FR1-3 worms (Fig. 1c, compare lanes 3 and 8: note that amount of total RNAs loaded in lane 8 was about 1/3 loaded in lane 3; see also Fig. 2 below). Thus, the mutant RNA1 from FR1-3ΔB2 transgene is not defective in self-directed replication, indicating that the reduced virus RNA accumulation in FR1-3ΔB2/N2 worms (Fig. 1c, lane 7) was due to induction and clearance of virus RNAs by antiviral silencing in an rde-1-dependent siRNA pathway.
Fig. 2.
FHV RNAi suppressor is active in rde-1 worms. Total RNA was extracted from worms of either wild type or rde-1 genotype carrying an integrated FR1-3 or FR1-3ΔB2 transgene two days after transcriptional induction. Northern blot hybridizations were carried out as in upper panels of Fig. 1c/d.
Both FHV RNAs 1 and 2 produced in worms were biological active following transfection into cultured Drosophila S2 cells (Fig. 1d, lanes 3-5), known to initiate potent RNAi-mediated antiviral silencing upon FHV challenge4. In contrast, no viral RNA was detected in S2 cells transfected with total RNA extracted from either FR1-3ΔB2/N2 or FR1-3ΔB2/rde-1 worms (Fig. 1d, lanes 7-8), in spite of the fact that the total RNA sample extracted from FR1-3ΔB2/rde-1 worms contained abundant FHV RNA1 (Fig. 1c, lane 8). This finding shows that progeny RNA1 of FR1-3ΔB2 produced in FR1-3ΔB2/rde-1 worms remained defective in B2 expression and incapable of preventing virus clearance by RNAi-mediated antiviral silencing in S2 cells4. We therefore conclude that abundant accumulation of FHV RNAs detected in FR1-3ΔB2/rde-1 worms does not result from a genetic reversion in the progeny of FR1-3ΔB2, but is due to genetic suppression of the viral B2 deletion by the loss-of-function mutation of rde-1 in the worm genome. This genetic complementation of loss-of-function mutations in a viral RNAi suppressor gene and a host RNAi pathway gene provides the first direct evidence that viral RNAi suppressors enhance virus accumulation and facilitate viral infection by suppressing the host antiviral silencing mechanism.
Comparative analysis of the viral RNA accumulation among FR1-3/rde-1, FR1-3/N2 and FR1-3ΔB2/rde-1 worms reveals two interesting results. First, accumulation of FHV RNA1 derived from the same FR1-3 transgene array was much lower in N2 worms than that in rde-1 worms (Fig. 2, compare lanes 3/4 and 5/6). This difference could be due to an incomplete B2 suppression of the worm rde-1-dependent antiviral silencing in FR1-3 worms, or alternatively to the recently discovered, rde-1-dependent transcriptional transgene silencing in the soma14. Second, FHV RNA1 accumulated to much higher levels in FR1-3/rde-1 worms than that in FR1-3ΔB2/rde-1 worms (Fig. 2, compare lanes 5/6 and 9/10), indicating that B2 expression also enhances viral accumulation in absence of rde-1. Since B2 is a broad-spectrum RNAi suppressor active in both the animal and plant kingdoms4,12 but it does not influence the rate of FHV RNA replication13, our data suggests the presence of active antiviral RNAi in rde-1 worms, which may be mediated by one or more of the remaining 26 worm AGO genes, among which differential requirements for alg-1, alg-2, ppw-1 and ppw-2 in various RNAi processes have been documented15.
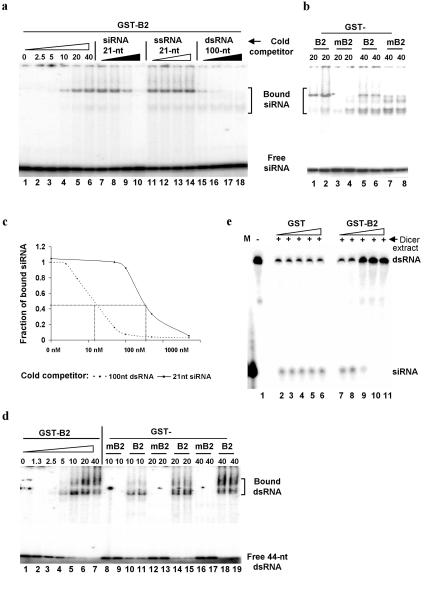
Our genetic analysis indicates that the viral RNAi suppressor is active in both wild type and rde-1 worms, suggesting that B2 act upstream of AGO. AGO plays a key role in the RNA-induced silencing complex by binding to siRNA for recognition and cleavage of the mRNA target8,11. No known protein domain could be recognized in the 106-residue FHV B2. We found that B2 synthesized as a GST fusion, bound in vitro to 21-nt siRNA duplex (Fig. 3a, lanes 1-6) independent of its overhang nucleotides (data not shown); siRNA binding has been reported for p19, a silencing suppressor encoded by the plant tombusvirus16. However, unlike p19, which binds much weaker to dsRNA longer than 23 nt, B2 also bound 25-nt siRNA and dsRNA of 44-nt (Fig. 3d) and 100-nt (data not shown) in length. Furthermore, competition experiments indicated a much higher affinity of B2 for long dsRNA than siRNA duplexes since the 100-nt dsRNA was approximately 30 times more effective than 21-nt siRNA in inhibiting the formation of the siRNA/B2 complex (Fig. 3a/c).
Fig. 3.
FHV B2 is a dsRNA-binding protein and inhibits siRNA production in vitro. a — d. GST-tagged B2 protein (GST-B2) binds both 21-nt siRNA duplex (a and d) and 44-nt dsRNA (d) in vitro. Volumes of the top siRNA/B2 band in lanes 7-10 and 15-18 were quantified by phosphoimager and plotted against the concentrations of the cold competitor RNAs (c). Note the presence of non-specific signals in lanes 1, 8, 13 and 16 of d. e. B2 inhibits in vitro processing of a labeled long dsRNA by the Dicer extracts from fruit fly S2 cells. Concentration gradients of GST or GST-B2 used were 50, 200, 800, 3,200 or 10,000 nM. A labeled 21-nt siRNA was used as a marker (M) at left.
A mutational analysis identified the substitution of Arg to Gln at position 54 of B2, which, when introduced into FHV RNA1, led to at least 20-fold reduction in the accumulation of FHV RNAs as compared to wild type FHV RNA1 (data not shown). Notably, the same R54Q mutation completely abolished the B2 activity to bind long dsRNA (Fig. 3d, lanes 8-19). The effect of the R54Q mutation on siRNA binding was less dramatic: Whereas the upper migrating siRNA/B2 complexes disappeared, the faster migrating siRNA/B2 complexes remained visible (Fig. 3b, compare lanes 1, 2, 5, 6 with lanes 3, 4, 7, 8). Furthermore, we found that Dicer processing of a labeled 500-nt dsRNA into siRNAs was inhibited by FHV B2 fused with GST beginning at 800 nM (Fig. 3e, lane 9), but not by GST up to 10,000 nM (Fig. 3e, lane 6), and that the inhibitory effect was essentially eliminated by the R54Q mutation (data not shown). These findings indicate a new mechanism of viral suppression of anitiviral silencing by targeting the dsRNA precursor of siRNAs although they do not rule out a possible role of siRNA binding16-18. This model is consistent with the genetic analysis placing B2 upstream of AGO and explains why B2 is active in both the animal and plant kingdoms.
Endogenous mammalian gene silencing by miRNAs initiates in the nucleus with Drosha cleavage of primary miRNA transcripts into precursor miRNAs, which are then exported to, and further processed into mature miRNAs by Dicer in the cytoplasm8. Much less is known about the initiation of antiviral RNA silencing in any organism. For RNA viruses, current models consider both dsRNA produced during RNA replication and highly structured elements in single-stranded viral RNAs as potential initiators via the siRNA and miRNA pathway, respectively16,18,19. Our genetic analyses (ref. 4 and this study) suggest a more prominent role for siRNA pathway in the recognition of the viral silencing initiators since neither Drosophila AGO2 nor C. elegans RDE-1 is essential for miRNA function20, in spite of their requirement in antiviral silencing. Furthermore, with a few exceptions such as influenza viruses12 and unlike DNA viruses21, most RNA viruses replicate exclusively in the cytoplasm and may escape detection by the miRNA pathway initiating in the nucleus. Nevertheless, potent antiviral silencing detected in a single Dicer organism rules out an essential role for multiple Dicers found in plants and insects5-9, indicating an antiviral potential for the mammalian RNAi machinery via the siRNA pathway in addition to targeting viral mRNAs by cellular miRNAs22. The C. elegans model established in this work will facilitate genetic studies of animal virus-host interactions, many aspects of which cannot be addressed in the alternative model in. Saccharomyces cerevisiae23, a unicellular organism that does not appear to encode an RNAi pathway8. Although most nodaviruses are pathogens of insect and fish hosts, Nodamura virus infects and kills sucking mice and sucking hamsters13, indicating a potential of the worm model for studying pathogenesis of mammalian viral diseases.
Methods
Transgene constructs and transgenic worms
Constructs were generated by standard methods using the vector pPD49.83 (a gift from A. Fire) which contains the promoter of the hsp16-41 gene24. Full-length cDNAs to FHV RNA1 and RNA2 together with a self-cleaving ribozyme from the tobacco ringspot virus satellite RNA were obtained from the infectious FHV cDNA clones previously constructed for infection of Drosophila S2 cells4
Nematodes were propagated and maintained by standard protocols (Brenner, 1974). The following strains were used: N2 (wild-type), unc-119(ed4) III, and rde-1(ne219) V. Animals were made transgenic by gonadal microinjection25. FHV plasmids were mixed with either the rol-6D plasmid pRF4 for injection into wild-type animals25, or the unc-119(+) plasmid pDP#MM016B, for injection into unc-119 mutants26. Integrated lines were generated with treatment of ∼30 transgenic hermaphrodites with 3500 rad of gamma rays from a 137Cs source, followed by screening for integrated animals in the F2 generation.
rol-6D and unc-119(+) transgenes were combined as described26. To generate rde-1; rol-6D strains, rde-1(ne219) males were crossed with rol-6D transgene hermaphrodites. F2 animals were grown individually on E. coli HT115 expressing unc-22 dsRNA (ref27), and plates generating no Unc-22 animals were assumed to be homozygous for rde-1(ne219).
Assay for FHV replication in C. elegans
Expression of FHV transgenes was achieved for each strain as follows. First, ten young adult hermaphrodites were placed on a seeded 10-cm plate and allowed to grow to the next generation for 5 days at 20°C. Plates were incubated for 2 hr at 33°C and returned to 23°C for two days, after which they were washed off plates into water and stored at -80°C. Total RNA was obtained by homogenizing thawed worm pellets using a Tissue Tearor (BioSpec Products, Inc.) followed by extraction with TRIzol (Invitrogen), then a column-based RNA purification using the RNeasy kit (Qiagen) according to the manufacturer's instructions. RNA concentrations were normalized and used for Northern blot analysis according to standard protocols4 using a 32P-labeled cDNA probe corresponded either to FHV RNA2, or to the last 387 nt of FHV RNA1, thus hybridizing to both RNAs 1 and 3.
To verify the biological activities of FHV RNAs produced in C. elegans, cultured Drosophila S2 cells were transfected with 2μg total RNA extracted from individual worm strains two days after heat induction. Three days after transfection total RNA was extracted from S2 cells for Northern blot detection of FHV RNA accumulation. pMT-FR1 was used as a control and contained the full-length cDNA of FHV RNA1 under transcriptional control of the CuSO4-inducible metallothionein promoter as described4.
Fusion protein expression, dsRNA binding and Dicer activity assays
Expression of GST-tagged B2 proteins (GST-B2) in E. coli and RNA binding assays were carried out as described previously12,28. Both single-stranded and duplex siRNAs were chemically synthesized and end-labeled by exchange reaction. Long dsRNA (44-nt and 100-nt) was in vitro transcribed and annealed, either with or without kinase end-labeling after dephosphorylation. The final concentration for all labeled RNAs was 50nM whereas 4 different concentrations (50, 100, 500 and 5000nM) were used for each cold competitor RNA. mB2 contained an Arg to Gln substitution at position 54 of FHV B2. The concentrations of GST-B2 or GST-mB2 used in RNA binding ranged from 0 to 40ng/μl as indicated. Binding reactions were resolved in native 6% polyacrylamide gel electrophoresis (PAGE) for binding to the 44-nt dsRNA or 8% for siRNA binding. Gels were dried before autoradiography.
Preparation of Dicer extracts from Drosophila S2 cells, labeling of long dsRNA, and Dicer activity assay were as described9. A 32P-labeled 500-nt dsRNA at 7.5 nM was incubated with the Dicer extracts in presence of either GST or GST-B2 in five different concentrations, 50, 200, 800, 3,200 and 10,000 nM. After incubation RNAs were fractionated by 15% PAGE along with a chemically synthesized 21-nt siRNA labeled with γ-32P-ATP as a marker.
Acknowledgements
The authors thank Dr. Xun Huang for recommending the use of pPD49.83 and Caenorhabditis Genetics Center funded by the National Center for Research Resources of the National Institutes of Health for some of the strains used in this work. This project was supported by NIH grant R01 AI52447 and USDA National Research Initiative Competitive Grants Program Award 2001-02678 and 2004-03258 (to S.W.D.).
Footnotes
Competing interests statement. The authors declare that they have no competing financial interests.
References
- 1.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 4.Li HW, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 8.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–29. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in Celegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 11.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 12.Li WX, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A. 2004;101:1350–5. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ball LA, Johnson KL. Reverse genetics of nodaviruses. Adv Virus Res. 1999;53:229–44. doi: 10.1016/s0065-3527(08)60350-4. [DOI] [PubMed] [Google Scholar]
- 14.Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19:683–96. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vastenhouw NL, Plasterk RH. RNAi protects the Caenorhabditis elegans germline against transposition. Trends Genet. 2004;20:314–9. doi: 10.1016/j.tig.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Silhavy D, Burgyan J. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 2004;9:76–83. doi: 10.1016/j.tplants.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Li WX, Ding SW. Viral suppressors of RNA silencing. Current Opinion in Biotechnology. 2001;12:150–154. doi: 10.1016/s0958-1669(00)00190-7. [DOI] [PubMed] [Google Scholar]
- 18.Voinnet O. Nat Rev Genet. 2005. Induction and suppression of RNA silencing: insights from viral infections. [DOI] [PubMed] [Google Scholar]
- 19.Ding SW, Li H, Lu R, Li F, Li WX. RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res. 2004;102:109–15. doi: 10.1016/j.virusres.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 20.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–66. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–6. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 22.Lecellier CH, et al. A cellular microRNA directs antiviral immunity in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 23.Lee WM, Ahlquist P. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J Virol. 2003;77:12819–28. doi: 10.1128/JVI.77.23.12819-12828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stringham EG, Dixon DK, Jones D, Candido EP. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol Biol Cell. 1992;3:221–33. doi: 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–70. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol Cell. 2001;7:475–85. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- 27.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 28.Silhavy D, et al. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]