Abstract
The gene SHOOT MERISTEMLESS (STM) is required for the initiation and the maintenance of the shoot apical meristem (SAM) in Arabidopsis and encodes a MEINOX/three amino acid loop extension (TALE)-HD-type transcription factor. Translational fusions with the green fluorescent protein showed that STM is not nuclear by default. In a yeast two-hybrid screen performed with a meristem-enriched cDNA library, three interacting BLH (Bel1-like homeodomain) transcription factors were identified. According to bimolecular fluorescence complementation, STM is targeted into the nuclear compartment through heterodimerization with BLH partner proteins, which are expressed in distinct SAM domains from the center to the periphery. On a functional level, overexpression experiments in transgenic Arabidopsis plants suggest that individual heterodimers provide distinct contributions. These results contribute to our understanding of the STM transcription factor function in the SAM and also shed new light on the evolution of the TALE-HD super gene family in animal and plant lineages.
INTRODUCTION
The aerial organs of angiosperm plants, leaves, stem and flowers arise through activity of the shoot apical meristem (SAM), which initiates new organs in a predictable and regular pattern. The SAM is a highly organized group of cells which can be divided into distinct domains with different functions (1–3). The central zone is involved in meristem maintenance and provides a permanent source of stem cells. Surrounding this is the peripheral zone, where new primordia are initiated. Although the SAM appears as an extremely stable structure, its individual cells divide and have to acquire new fates before they finally differentiate.
The gene SHOOT MERISTEMLESS (STM) is required for the initiation and maintenance of the SAM in Arabidopsis. Embryos homozygous for strong loss-of-function mutations in the STM gene form cotyledons and other embryonic structures but fail to establish a population of self-renewing stem cells (4,5). Characteristic cell divisions that generate the typical tunica/corpus organization are also missing in stm mutant embryos. Plants homozygous for weak stm alleles form SAMs but these are prone to premature termination (6,7). STM encodes a Knotted-like homeodomain-containing protein and is likely to act as a transcriptional regulator in promoting SAM development and maintenance [(7) and recently reviewed in (8,9)]. This assumption is supported by the transcription pattern, as STM activity is restricted to the SAM but down-regulated in founder cells (P0) of newly arising lateral organ primordia (5).
The STM homeodomain (HD) belongs to the three amino acid loop extension (TALE) class of homeodomains. A second conserved protein motif is the KNOX domain, more generally known as the MEINOX domain (10). Although initially identified in plant Knotted-related homeobox genes, the KNOX domain was subsequently also found in animal myeloid ecotropic viral integration site (MEIS) proteins. This combination of MEINOX and TALE-HD domains in animal and plant proteins is suggestive of a common ancestry (11). Whereas the TALE-HD serves to interact with DNA-target sites, the MEINOX domain in animal proteins has been shown to mediate interactions with a second group of TALE-HD proteins—the pre-B cell (PBC) class (12–16). Also in plants there is accumulating evidence that the MEINOX/TALE-HD proteins interact with a second class of TALE-HD gene products, the BEL1-like homeodomain (BLH) proteins (17–19). In total, the Arabidopsis genome contains 12 BLH genes, besides BEL1 (20) and the founding member ATH1 (21) systematically numbered BLH1 to BLH10. A functional interaction between plant MEINOX-HD and BLH genes is substantiated by the synergism observed when pennywise a null-allele of BLH9 (18), allelic to bellringer, replumless, vaamana (19,22,23) is combined with the brevipedicellus1 (bp1) mutant (24). Guided by an evolutionarily conserved structure and heterodimerization of two TALE-HD super gene family members in plants and animals, it has been proposed that KNOX/BLH as MEIS/PBC genes share a common ancestry (25). However, amino acid sequence conservation outside the homeodomain gene is low, although members of the plant BLH or animal PBC gene families each share a typical and lineage-specific domain, the BELL domain and the bipartite PBC-A/B domain, respectively (11,17,25,26).
In contrast to divergence of the BELL and the PBC-A/B domains, the MEINOX domain is highly conserved between plant KNOX and animal MEIS proteins. The MEINOXSTM domain is essential for STM function, as overexpression of the isolated MEINOXSTM domain strongly phenocopies stm loss-of function alleles (27). This dominant-negative function upon expression of the MEINOXSTM domain is strengthened in combination with the Drosophila engrailed repressor domain and best explained by squelching: the abundant MEINOXSTM domain depleting the native STM protein from essential interaction partners. Interestingly, although BREVIPEDICELLUS1 (BP1) is thought to act redundantly with STM in the SAM (24) the same experiment performed with the MEINOXBP1 domain results in bp1 mutant phenocopies (27). Putative protein interactions mediated by the MEINOXSTM domains therefore exhibit selectivity, consistent with domain-swap experiments performed with NTH1 and NTH15 in transgenic tobacco (28).
We initiated a search for potential STM interaction partner genes starting from a yeast two-hybrid screen performed with the MEINOXSTM domain as bait for a meristem-enriched cDNA library. However, in planta analysis of protein–protein interactions demonstrated that the transcription factor STM is not an obligatory nuclear protein but that nuclear uptake depends on partner proteins. Described here are experiments with three different BLH partner proteins, which target STM into the nuclear compartment and which are expressed in distinct SAM subdomains. Combinatorial gain-of-function experiments in transgenic Arabidopsis plants provide evidence that individual STM/BLH heterodimers may provide different functions.
MATERIALS AND METHODS
Yeast two-hybrid screen and immature inflorescence library
The Matchmaker GAL4 system (Clontech) was used to perform the yeast two-hybrid screen. As bait the sequences encoding the MEINOXSTM domain (amino acids 116–220 of the STM protein) or the slightly longer MEINOX-ELKSTM (amino acids 116–287) domain were cloned into the vector pGBKT7 (Clontech) and expressed in fusion with the GAL4 DNA-binding domain. Directional cloning of PCR fragments was performed by the addition of in-frame NcoI or BamH1 restriction sites to gene-specific PCR primers.
The meristem-enriched cDNA library in the vector pACT2 (Clonetech) was prepared from immature inflorescence shoots (Lumbrineris erecta, 4–5 mm size) containing inflorescence and multiple floral meristems (FMs). cDNA synthesis exactly followed the manual for the cDNA synthesis kit (Stratagene). Directional cloning into pACT2 was achieved via an XhoI site addition to the oligo-dT primer (3′) and an EcoRI adapter at the cDNA 5′-terminus. The cDNA library (1.25 × 106 clones) was colony amplified in Escherichia coli, plasmid DNA purified by CsCl2-gradient centrifugation and used for yeast transformation (strain AH109). Screening of the library followed the Matchmaker manual applying quadruple selection (HIS3, ADE2, LacZ and MEL1). In total 2.5 × 106 colonies were screened for encoded polypeptides which interact with the MEINOX-ELKSTM domain. Positive clones (223) were isolated and cDNA inserts in pACT2 were subjected to N-terminal sequence analysis. The specificity of each protein interaction was confirmed by retransformation of sequence-verified cDNA clones (ATH1, BHL3 or BHL9) into yeast strains providing the MEINOXSTM or MEINOXBP1 bait constructs in pGDKT7. No interaction was observed with the empty pACT2 prey vector.
Full-length protein coding regions and RNA in situ hybridizations
Complete coding regions of ATH1, BHL3 and BHL9 were isolated by reverse transcriptase PCR, with primers (20–23mers) positioned at the translation start codon. Where possible, directional cloning was facilitated by embedding the ATG start codon in a NcoI restriction site, and the addition of a BamH1 site (alternatively BglII site) after the stop codon. Total RNA was extracted from 3-week old seedlings according to Chomczynski and Sacchi (29) and used for first strand cDNA synthesis following the cDNA synthesis kit protocol (Stratagene). Resulting PCR products were generally cloned into the vector pCRII-TOPO (Invitrogen) and subjected to complete DNA sequence analysis to exclude PCR-based amino acid alterations. Verified cDNA inserts were either directly transferred into appropriate secondary vectors for bimolecular fluorescence complementation (BiFC), co-immunoprecipitation or transgenic experiments and served as templates in second-round PCR-amplification to remove the stop codon or to adjust open reading frames (ORFs) for translational fusions.
Non-radioactive in situ hybridization experiments were performed essentially as described in Bradley et al. (30). Paraffin wax-embedded tissue was sectioned by the use of a rotary microtome and 7 µm sections were used for hybridization. Digoxigenin-labeled probes were transcribed from PCR fragments after addition of the T7 promoter at the 5′ end of the reverse-primer.
In planta bimolecular fluorescence complementation
The ORFs of genes encoding candidate partner proteins (ATH1, BHL3 or BHL9) and STM were cloned in-frame into the transient expression vectors pUC-SPYCE and pUC-SPYNE, respectively, through the BamHI site (31), containing either the N- or C-terminal parts of the coding regions of the yellow fluorescent protein (YFP). As controls, green fluorescent protein (GFP) fusions with STM and BLH partner proteins were created in the vector pRTΩNotI/AscI (32). To create NLS-GFP-STM, a synthetic oligonucleotide providing the virD2 nuclear localization signal (NLS) (33) was inserted into a NcoI site at the translation start of the GFP ORF. For transient expression in leek epidermal cells, 50 µl of tungsten particles (1.1 µm diameter, BioRad) were mixed with 10–15 µg of plasmid DNA, 60 µl of 2.2 M CaCl2 and 20 µl of 0.1 M spermidine in a total volume of 150 µl. The DNA was precipitated on the tungsten particles at room temperature by adding 200 µl ethanol, for 3 min with continuous shaking. After brief centrifugation, the tungsten pellet was washed three times in 100% (v/v) ethanol before being resuspended in 30 µl of 100% (v/v) ethanol. Aliquots (10 µl) of DNA-coated tungsten were spotted on macro carriers and used to transform leek epidermal cells at 1100 psi rupture disc bursting pressure using a Biolistic PDS-1000-He apparatus (BioRad) The bombarded tissue was kept in Petri dishes on damp filter paper for 12–16 h in the dark and YFP or GFP fluorescence was visualized using a LEICA MZFLIII stereomicroscope. All images were processed using the Adobe Photoshop software (version 7.0) package.
Co-immunoprecipitations and western blotting
Epitope-tagged proteins were prepared in the EasyXpress protein system (Qiagen), via an in vitro transcription/translation system based on the T7 promoter. T7 promoter transcription templates of STM partner proteins were obtained via nested PCRs on the ATH1, BHL3 or BHL9 constructs in pUC-SPYCE which encodes the HA epitope prior the YFP C-terminal domain. Both the T7 promoter and the 6× His-tag were added to the STM coding region via a nested PCR strategy. STM and partner proteins were co-transcribed/translated by mixing size- and quality-checked PCR-amplicons of STM with those of ATH1, BHL3 and BLH9. Control reactions were performed with single STM or BLH amplicons.
The MACS epitope tagged protein isolation kit (Miltenyi Biotec) was used to precipitate candidate partner proteins via the HA epitope. Eluted protein samples were split (1/3 and 2/3), size-fractionated in parallel on two polyacrylamide SDS-gels (concentration adjusted to protein size), electroblotted onto Immobilon-P membrane (Millipore) and subjected to antibody detection. HA-tagged (1/3 aliquot) partner proteins were identified using an anti-HA horseradish peroxidase (HRP)-conjugated antibody (clone 3F10; Roche 2012819). The co-precipitated 6× his-tagged STM protein was visualized by a primary penta-His mouse antibody [α-(H)5; Qiagen 34660] and secondary HRP-coupled goat anti-mouse IGG (Dianova 115-035-062). Peroxidase activity was detected non-radioactively via chemiluminescence (ECL PLUS kit; Amersham Biosciences) and documented on Kodak-X-omat AR films. Epidermal proteins after bombardement of leek epidermal cells were isolated in 50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% Triton and size-fractionated by SDS–PAGE. GFP fusion proteins were detected after transfer to Immobilon-P membrane (Millipore) by a mouse monoclonal anti-GFP HRP-conjugated antibody (IgG1; Milteny Biotec 130-091-833).
Transgenic plants
For overexpression, the protein coding regions of ATH1, BHL3 or BHL9 and modified STM versions were cloned behind the CaMV 35S promoter in a modified pRTΩNotI/AscI vector and the flanking AscI sites were used to transfer the expression cassette into the binary vector pGPTV-BAR/AscI (32). To create NLS-STM, a synthetic oligonucleotide providing the virD2 NLS (33) was inserted into an NcoI site created at the translation start of the STM ORF. Transgenic plants were generated by vacuum infiltration of Arabidopsis thaliana ecotype Columbia recipient plants by use of Agrobacterium tumefaciens strain GV3101 (34) and primary transformants (T1) were selected using resistance against the herbicide BASTA. A minimum of 65 transgenic plants was obtained for each construct. Representative lines (5–10) were selected for the analysis of transgene and phenotype transmission in the T2 generation. Based on segregation (3:1 for BASTAR), putative single transgene insertion lines were chosen for crosses with the heterozygous STM-GR effector line (35) and detailed phenotypic characterization (Figure 5E).
Double-transgenic progeny of crosses were selected for transmission of ATH1, BHL3 or BHL9 transgenes by BASTAR. Dexamethasone induction was carried out by spraying plants with a solution containing 1 µm Dex (Sigma D8893). The subsequent development of single- and double-transgenic plants as untreated controls was documented by photography on a daily basis. The co-expression of both transgenes in Dex-responsive/BASTAR plants was verified on the transcriptional level by RNA gel blot analysis. Total RNA was isolated from pooled explant material of double-transgenic progeny from each cross and transfered Nytran-SPC filters (Schleicher & Schuell) after electrophoresis. Filters were successively hybridized with firstly ATH1, BHL3 or BHL9 probes and secondly with the STM probe labelled radioactively by incorporation of [α32P]dATP.
RESULTS
The STM protein is not nuclear by default
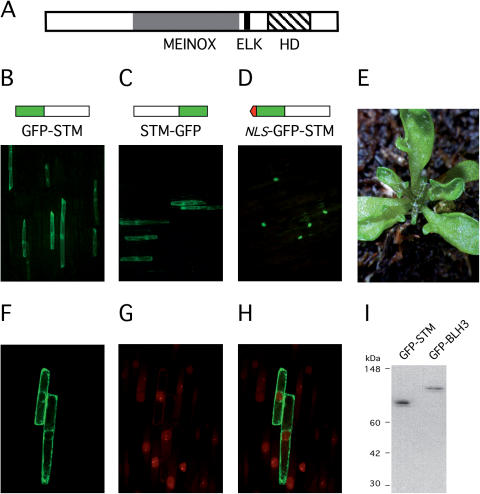
The method of choice to prove a physical interaction between candidate partner proteins in plant cells is BiFC (31,36). As a prerequisite to substantiate interactions between potential partner proteins identified in a yeast two-hybrid screen, C- and N-terminal fusions between the ORFs of STM and the GFP were constructed and expressed in leek or onion epidermal cells after particle bombardment. As a potential transcription factor, STM was expected to direct GFP fluorescence into the plant cell nucleus. However, both fusion proteins, GFP-STM and STM-GFP remained in the cytoplasm (Figure 1B and C). The same result was obtained in epidermal cells of the Arabidopsis hypocotyl. In contrast, the addition of the virD2 NLS (33) in front of the chimeric GFP-STM ORF efficiently directed the chimeric NLS-GFP-STM protein into the plant cell nucleus (Figure 1D). An inappropriate folding of the N- or C-terminal GFP fusion proteins is unlikely, because expression of both chimeric proteins (GFP-STM + STM-GFP) in transgenic Arabidopsis plants behind the CaMV35S promoter yielded the characteristic STM overexpression phenotype (data not shown). However, in contrast to ectopic STM activity affecting the SAM or causing meristems on leaves (37), the expression of a nuclear targeted NLS-STM version from the CaMV 35S promoter in transgenic Arabidopsis plant predominantly resulted in a lobed leaf phenotype (Figure 1E), which is reminiscent of KNAT1 overexpression (38) rather than STM over expression (37). Lobed leaves were detected in 187 (76%) from 247 primary transformants (T1), whereas only 44 (18%) T1 plants showed meristem defects including stm phenocopies, presumably due to co-suppression. A nuclear-targeted STM protein version therefore affects leaf development more than SAM function.
Figure 1.
The STM protein in non-nuclear. (A) Domain structure of the STM protein. (B and C) Cellular staining of GFP-STM or STM-GFP. (D) Nuclear incorporation of NLS-GFP-STM. (E) Lobed leaf phenotype obtained in CaMV 35S NLS-STM overexpression plants. (F–H) Z-stack projection of a median series through the nucleus performed with confocal laser scanning microscope in leak epidermal cells. (F) Empty plant cell nuclei in GFP-STM expressing leek epidermal cells. (G) Same projection as in (F), nuclei stained with propidium iodide. (H) Merge of F and G stacks confirming absence of GFP-STM in the nuclear compartment. (I) Stability of GFP-STM and GFP-BLH3 fusion proteins in leak epidermal cells visualized by a GFP antibody.
The cytoplasmic retention of the chimeric GFP-STM protein was further substantiated by confocal laser microscopy scanning and staining of nuclei with propidium iodide. The comparison of a median Z-stack projection through the nucleus either analysed for GFP-STM fluorescence (Figure 1F), the corresponding propidium iodine stains (Figure 1G) and their overlay in Figure 1H confirm exclusion of GFP fluorescence from the nuclear compartment. In contrast to the median sections depicted in Figure 1F, the same scanning series shows GFP-STM fluorescence surrounding the nucleus, which would explain fluorescence at the position of the nucleus in whole mounts depicted in Figure 1B or C. Protein extracts prepard from leek epidermal cells after particle bombardement showed no degradation of the GFP-STM fusion protein with an antibody directed against the GFP (Figure 1I). Taken together, the data lead to the conclusion that STM is devoid of an efficient NLS. Additional support for nuclear exclusion was obtained by several STM deletion constructs, none of which targeted GFP fluorescence to the plant cell nucleus (data not shown). This nuclear import deficiency is noteworthy since the ELK domain conserved between the MEINOX domain and the homeodomain in plant KNOX proteins (Figure 1A) has been assumed to provide a functional NLS (39).
Isolation of MEINOX-mediated STM partners and co-immunoprecipitation of full-length BLH proteins
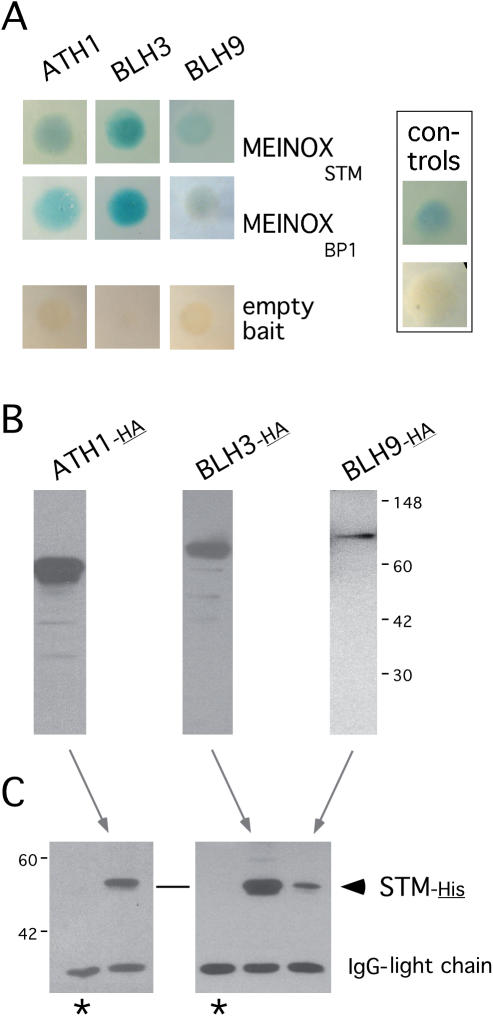
Potential STM protein interaction partners were identified in a yeast two-hybrid screen performed with a meristem-enriched cDNA library (Materials and Methods) established in the vector pACT2 (prey) and the MEINOXSTM domain as a bait expressed from the pGDKT7 vector. In total 223 positive clones were identified and all were annotated as ORFs in the Arabidopsis genome. Besides known false positives, proteins of unknown function or metabolic enzymes, a major class was membrane-associated proteins including transporters or ion-channels, however, any significance of this finding remains to be established. Amongst 31 cDNAs encoding genes related to transcriptional control, 3 BLH proteins, ATH1, BLH3 and BLH9, are subject to a detailed analysis here. Although BLH9 has been implicated to interact with STM (18,19) it was included in the in-depth analysis described here for comparison concerning a function in the SAM.
Besides obvious controls in yeast such as retransformation of bait or prey plasmids and the combination with the empty bait vector, we included the MEINOXBp1 domain of the BP1 gene as bait as it encodes the closest relative to the MEINOXSTM domain in the Arabidopsis genome. All three BLH proteins interact with both the MEINOXSTM and the MEINOXBp1 domain and thus recognize sequence or structural features common to the MEINOX domains of BP1 or STM (Figure 2A). In addition to the interactions in yeast, we performed co-immunoprecipitation experiments with epitope-tagged full-length proteins. The HA-tagged BLH proteins shown in Figure 2B were used to co-precipitate epitope-tagged STM-His (Figure 2C). The detection of the STM-His protein was strictly dependent on the presence of BLH partner proteins. The co-immunoprecipitation experiments therefore confirm that the full-length STM protein interacts with full-length ATH1, BLH3 and BLH9 proteins in vitro and substantiate the affinity of BLH/STM interactions.
Figure 2.
STM/BLH interactions in yeast and co-immunoprecipitations. (A) Galactosidase activity mediated by the interaction of the MEINOX domain cloned into pGBKT7 (bait) with BLH partner proteins expressed in pACT2. No signal is observed with the empty bait vector. The positive controls upper panel to the left shows the interaction between p53 and the SV40 T-antigen (pGBKT7-53 and pADT7-T, respectively, provided in the MATCHMAKER Biosensor kit). (B) In vitro translated full-length BLH proteins tagged with the hemaglutine (HA) epitope which were used in co-precipitation experiments with His-tagged STM protein and visualized by an anti-HA HRP-conjugated antibody. (C) Co-precipitated STM-His protein detected by a penta-His antibody. Asterisks mark control lanes showing that STM-His is undetectable in immunoprecipitations in the absence of BLH partner proteins.
Expression domains of BLH proteins in the SAM
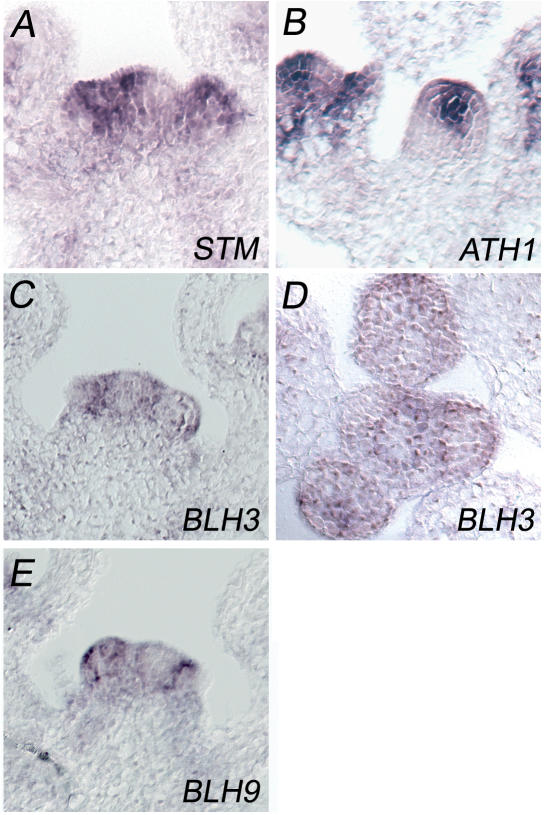
A further step towards proof of biological relevance was the confirmation of a spatial overlap between the expression domain of STM and those of candidate partner BLH genes by performing RNA in situ hybridizations. All three BLH genes are transcribed in the early Arabidopsis seedling, whereas the ATH1 and BLH9 expression patterns have been analysed previously (21,22). RT–PCR experiments readily detected BLH3 transcripts at the 2–4 leaf seedling stage (data not shown). Combined in Figure 3 are the cellular transcription patterns in immature inflorescences, where the indeterminate inflorescence meristem (IM) and determinate FMs can be compared. All three BLH genes are transcribed in IMs and FMs. High levels of ATH1 transcripts are generally detected in the center of the IM or FM and an additional stripe of ATH1 expression is found in the FM (to the left in Figure 3B), which is adjacent to a lateral organ primordium. In contrast, BLH3 is transcribed in a cup-shaped expression domain enclosing the ATH1 domain, whereas BLH3 transcripts are absent in the centre of the IM (Figure 3C and D). The BLH9 transcription pattern has been analysed on the cellular level previously (22); a section is included in Figure 3 to show its preference at the SAM periphery (Figure 3E). The in situ hybridizations with ATH1, BLH3 and BLH9 probes therefore suggest a zonation of the IM, from the center to the periphery. In contrast, STM (Figure 3A) is ubiquitously transcribed throughout the SAM and only down-regulated when cells acquire primordial fate at the SAM periphery.
Figure 3.
RNA in situ hybridization patterns of STM and partner protein genes in the IM. (A) STM, (B) ATH1, (C) BLH3, (D) BLH3 transverse, (E) BLH9. [Median longitudinal sections except (D)].
Heterodimerization with BLH partners directs STM into the plant cell nucleus
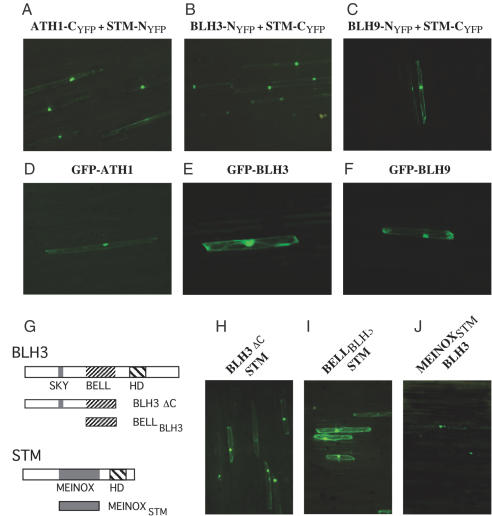
The principle of BiFC is the formation of a fluorescent complex by fragments of the enhanced YFP brought together by the association of two interaction partners (31,36). To verify STM interaction partners we constructed translational fusions between the N- or the C-terminal YFP sub-domains and the full-length STM protein or individual full-length BLH partner proteins. Optimal combinations of chimeric gene constructs are indicated in Figure 4 above each corresponding experiment. Chimeric genes were co-delivered into leek epidermal cells by bombardment of tungsten particles double-loaded with two appropriate chimeric gene constructs. Any detectable fluorescence was dependent on the combination of the C-terminal with the N-terminal YFP sub-domain fused to STM or BLH partner proteins. In contrast, the fusion of the C- or the N-terminal YFP sub-domain to the STM protein and their co-bombardment in plant cells gave no fluorescence (data not shown). BiFC therefore provided no evidence for the formation of STM homodimers but substantiated the formation of STM/BLH heterodimers.
Figure 4.
Nuclear import of STM by interaction with BLH partner proteins. (A–C) BiFC staining of the nucleus obtained after coexpression of STM/BLH constructs as indicated above each photograph in leek epidermal cells. (D) GFP-ATH1. (E) GFP-BLH3. (F) GFP-BLH9. (G) Schematic representation of BLH3 and STM deletion constructs fused to the YFP NYFP- or CYFP- sub-domains. Deletion polypeptides were co-expressed with the full-length STM of BLH3 proteins fused to the complementaryYFP sub-domain. (H) Interaction between BLH3ΔC-NYFP and STM-CYFP. (I) Combination of BLH3BELL-NYFP and STM-CYFP. (J) MEINOXSTM-CYFP combined with the full-length BLH3-NYFP. Note the fluorescence preference for the nuclear compartment in (H) and (J) versus the cytoplasmic signal in (I).
As shown in Figure 4A–C, co-expression of ATH1-CYFP/STM-NYFP, BLH3-NYFP/STM-CYFP and BLH9-NYFP/STM-CYFP resulted in reconstitution of YFP fluorescence. In contrast to results obtained with STM-GFP fusions, however, the BiFC signal exhibited a high preference for the nucleus whereas fluorescence was low in the cytoplasm. The BiFC results therefore, confirm that full-length STM and BLH proteins interact in plant cells, but more notably, that BLH/STM heterodimers are efficiently incorporated into the nuclear compartment. Expectedly, the GFP-ATH1, GFP-BLH3 and GFP-BLH9 fusion proteins exerted a preference for the nuclear compartment (Figure 4D–F). The nuclear import of GFP-BLH9 in leek epidermal cells appears to be in contrast to previous results obtained in tobacco leaves after agroinfection with a C-terminal BLH9-GFP fusion construct. However, without analysing the cellular localization of the STM protein, Bhatt et al. (19) arrive at the same conclusion that STM/BLH9 heterodimers are efficiently imported into the plant cell nucleus. In contrast to bombardments with the ATH1 and BLH3 constructs, the number of BiFC-positive cells obtained with the BLH9/STM combination (e.g. BLH9-NYFP/STM-CYFP in Figure 4C) was generally low. This may indicate a weak interaction between STM and BLH9 as suggested by the yeast two-hybrid results, that chimeric BLH9-NYFP protein levels remain low in leak epidermal cells, or that steric constraints interfere with a reconstitution of YFP fluorescence. Reminiscent, however, of the MEIS/PBC situation in animals, the GFP and BiFC data show that the TALE-HD transcription factor STM is targeted into the nuclear compartment through interaction with three BLH partner proteins, which are members of the non-MEINOX class of TALE-HD proteins in plants.
The BELL domain is essential for the interaction of STM with BLH partner proteins. BiFC experiments with BLH3 deletion polypeptides (BLH3 ΔC or BELLBLH3 see schematic drawing in Figure 4G) demonstrated that amino acid sequences upstream and downstream of the BELL domain including the homeodomain are dispensable for heterodimerization with STM (Figure 4H and I). BiFC fluorescence still exerted a preference for the nucleus if sequences C-terminal to the BELL domain including the DNA-binding homeodomain were deleted (BLH3 ΔC, Figure 4H). In contrast, YFP fluorescence reconstituted with the isolated BELL domain (BELLBLH3 in Figure 4I) remained mainly cytoplasmic. An effective NLS therefore must be located in the BLH3 protein N-terminal to its BELL domain. In the STM protein, the MEINOXSTM domain is sufficient to interact with full-length BLH3 (Figure 4J). However, we have been unable to reconstitute YFP fluorescence in BiFC experiments with the isolated MEINOXSTM and BELLBLH3 domains although several compatible N- or C-terminal combinations with each YFP sub-domain were tested. One explanation for this may be that the relative arrangement of YFP sub-domains sterically interferes with their functional association. This assumption is supported by the fact that the MEINOXSTM domain is precipitated via the BELLBLH3 domain in co-immunoprecipitation experiments (data not shown).
Functional synergism between STM and BLH proteins in planta
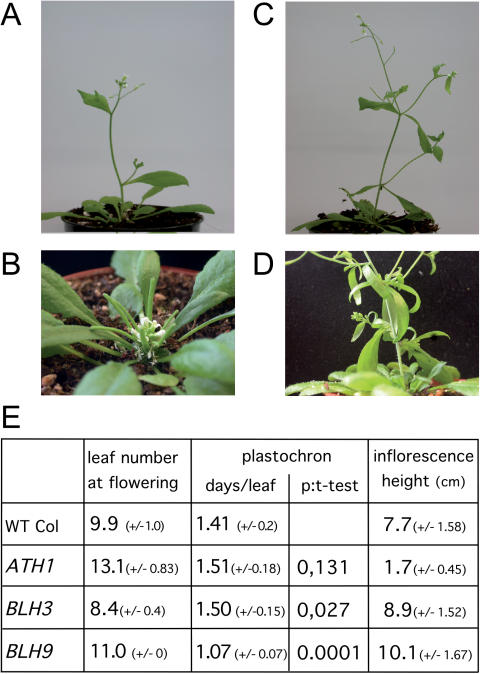
We attempted to substantiate the functional relevance of STM heterodimers in Arabidopsis by combining an inducible STM-GR effector line (35) with lines constitutively overexpressing BLH3, BLH9 or ATH1. For this purpose, we first raised and analysed transgenic lines expressing individual BLH genes behind the CaMV 35S promoter. The most evident phenotype in primary transformants (T1) was observed with ectopic ATH1 gene activity, which affected the elongation of internodes in inflorescence shoots. The severest phenotype, essentially an absence of internode elongation was observed in 40 from total of 240 BASTA-resistent progeny (compare Figure 5A with B). Normal and fertile flowers emerged in an appropriate phyllotaxy from a stunted, compact inflorescence, a phenotype inherited up to the T4 generation. Some reduction in inflorescence height (∼20–30%) was also observed in T1 BLH9 overexpressing plants (30 from total of 65 transgenic seedlings); however, this phenotype was not heritable in subsequent generations. In addition, BLH9 overexpression caused alterations in phyllotaxy and phenotypes (Figure 5C and D) reminiscent of those described for loss-of-function alleles e.g. multiple cauline leaves subtending second order inflorescence shoots, multiple axillary shoots emerging from a single inflorescence node or irregular elongation of internodes [bellringer, pennywise, replumless, vaamana (18,19,22,23)]. In contrast, T1 BLH3 overexpressing plants (total >200) appeared similar to wild type, although independent lines in the T2 generation were early flowering compared with wild-type plants.
Figure 5.
Phenotypes of STM partner protein overexpression. (A) Wild-type inflorescence shoot for comparison to BLH gene overexpression lines at the same stage. (B) Stunted inflorescence of an ATH1 overexpressing plant, note maturing siliques at the base close to flowers at the tip. (C) BLH9 overexpression line with multiple cauline leaves emerging from single nodes. (D) Close-up of another BLH9 overexpression inflorescence. (E) Number of leaves at flowering, plastochron and inflorescence length in T3 progeny of ATH1, BLH3 or BLH9, overexpressing lines used for crosses into the STM-GR background (WT Col: Columbia control plants). T-test P-values in the plastochron column are relative to wild type and substantiate the significance of the shorter plastochron in BLH9 overexpressing plants. All numbers are based on two independent lines (BLH9: 3 lines) and 6–10 progeny each.
For each BLH candidate partner protein gene two representative T2 overexpressing lines were crossed into the STM-GR background. To discriminate single from double-transgenic plants, T3 progeny of these lines were subjected to a detailed phenotypic analysis (Figure 5E). Overexpression of either BLH3 or BLH9 caused early flowering compared with Columbia wild-type controls, whereas ATH1 transgenic progeny flowered much later. With P-values ranging from 0.002 (BLH9) or 0.00001 (BLH3) to 0.000002 (ATH1) in the student's t-test, these differences in flowering time are highly significant. In BLH3 overexpressing plants early flowering is reflected by a reduced leaf number compared with Columbia control plants (8 or 10 leaves on average, respectively; Figure 5E). In contrast, BLH9 transgenic plants flowered at the same time as BLH3 overexpressing plants, but developed more rosette leaves. BHL9 overexpressing plants flowered three days earlier than wild-type controls and produced on average one more leaf than wild type. The plastochron was estimated to be 1.41 for wild type, whereas BLH9 overexpressing plants developed a new leaf approximately every day. This difference was confirmed in a third BLH9 transgenic line and is highly significant (P = 0.00013). In contrast no significant deviation in the plastochron was observed in 35S::BLH3 or 35S::ATH1 transgenic plants compared with the wild type (Figure 5E). However, transgenic ATH1 plants flowered on average 4 days later than wild-type Columbia control plants and developed on average three more leaves than the wild type. The overexpression of three BLH genes, which are potential interaction partners of STM in the SAM therefore consistently affected flowering time either by altering the number of vegetative leaves (BLH3/ATH1) or the plastochron (BLH9).
F1 seeds obtained from crosses between STM-GR and BLH3, BLH9 or ATH1 overexpressing lines were sown on soil and monitored for transmission of the potential STM partner protein transgene by selection for BASTA resistance. The surviving seedlings of the individual crosses and lines were subdivided into three groups and either sprayed with Dexamethasone (Dex) at the 2–3 or 5–6 leaf stage, or third group serving as an untreated control. Since the 35S::STM-GR effector transgene is propagated heterozygously, only 50% of the progeny can respond to the hormone treatment by releasing the chimeric STM-GR protein from cytoplasmic retention (40). When induced at the early 2–3 leaf stage, STM-GR control plants remained in the vegetative phase with newly initiated leaflets which were small and remained closely associated with the SAM (Figure 6A).
Figure 6.
Functional interaction between STM-GR and partner proteins in transgenic Arabidopsis plants. (A) STM-GR, (B) ATH1/STM-GR, (C) BLH3/STM-GR, (D) BLH9/STM-GR. The double-transgene combinations ATH1/STM-GR and BLH3/STM-GR respond with floral induction [compare (B and C) with (A)] or leaf lobing [compare (D) with (A)]. In contrast the parental STM-GR line remained in the vegetative phase and developed multiple small vegetative leaflets. All plants were sprayed at the 2–3 leaf stage and photographed 16 days after Dex application.
In contrast, BLH3/STM-GR and ATH1/STM-GR double-transgene combinations ceased producing leaf primordia and instead initiated FMs (Figure 6B and C). Young leaflets already initiated at the time of Dex application expanded but lacked petioles and in consequence remained in close association to the SAM. The release of STM-GR from cytoplasmic retention in two independent lines providing either ectopic ATH1 or ectopic BLH3 activity therefore transforms meristem identity from vegetative to reproductive. A different response to Dex application was observed in the combination BLH9/STM-GR; these plants remained in the vegetative phase but initiated new leaves which were severely lobed (Figure 6D). Plants sprayed later, at the 5 or 6 leaf-stage also responded to Dex application. However, some STM-GR control plants started flowering shortly thereafter, as did each double-transgene combination. Floral induction at this late stage, therefore, may be biased by STM-GR overexpression. Noteworthy is that cauline leaves in BLH9/STM-GR transgenic plants remained severely lobed, thus confirming specificity of this additive leaf phenotype. Characteristic for STM-GR flowers are abnormalities such as a size reduction in outer whorl organs and a prominent carpel surrounded by few stamens (32). These features are shared by flowers emerging from early or late Dex-induced double-transgene combinations (see also Figure 6C). Although this phenotype provides evidence for STM-GR expression, the co-activity of both transgenes was confirmed by RNA-gel blot experiments with pooled explant material for each transgene combination (data not shown). No additional phenotype was observed in the no-DEX control group, single-transgene STM-GR plants or untreated double-transgene combinations. In conclusion, these additive combinatorial transgene results provide strong evidence that the overexpression of three BLH partner proteins in transgenic Arabidopsis plants creates novel functional competencies, which are realized after STM-GR is released from the HSP90 chaperonin. Two discrete phenotypes, floral induction and leaf lobing indicate response specificity, and more importantly imply that individual BLH proteins may contribute different functions through their interaction with STM.
DISCUSSION
Domain specificity of STM heterodimers in the SAM and functional differences
Although expressed throughout the Arabidopsis SAM, we have shown here that the homeodomain transcription factor STM is devoid of an efficient NLS and does not enter the nucleus by default. Our results are compatible with previous data showing that the chimeric STM-GFP protein moves from the L1 layer to subtending L2 or L3 meristem layers (41) as an obligatory nuclear import of the STM transcription factor would interfere with intercellular trafficking. According to our BiFC results, however, STM is targeted into the nucleus as a heterodimer with ATH1, BLH3 and BLH9. A passive co-transport is also not in contradiction to the nuclear localization of STM-GFP in the L2 or L3 layers of the SAM (41) if nuclear-targeting partner proteins such as ATH1, BLH3 and BLH9 are readily available. However, an intriguing aspect of BLH gene expression in the SAM is their confinement to discrete SAM sub-domains. Disregarding possible post-transcriptional control mechanisms, the STM transcription factor encounters ATH1 preferentially in the centre of the SAM, followed by BLH3 and finally BLH9 moving towards the IM periphery.
All three BLH partner protein genes are already transcriptionally active during the vegetetative phase [(21,22), BLH3 data not shown]; however, ectopic expression of ATH1, BLH3 and BLH9 behind the CaMV 35S promoter altered flowering time. BLH9 and BLH3 overexpression caused early flowering, whereas ectopic ATH1 activity caused a delay. The link between BLH gene activity and flowering time control is further strengthened by the additive phenotype observed after induction of STM-GR in the pre-conditioned ectopic ATH1 or BLH3 background resulting in floral induction. ATH1 and BLH3 both are expressed in the center of the IM whereas BLH9 is transcribed at the periphery and in combination with BLH9 the release of STM-GR by Dex caused leaf lobing. In ligand blot experiments, BHL9 reportedly interacts with the MEINOXBP1 domain and only weakly with the MEINOXSTM domain (18). However, double-mutant analyses (stm-2/bellringer) showed that BLH9 can compensate for reduced STM activity (22), although another allele pennywise appears to act synergistically with bp1 (18). The phenotype obtained here after Dex induction of STM-GR in the ectopic BLH9 overexpression background is reminiscent of BP1/KNAT1 overexpression (38), which is mimicked by the ectopic expression of a nuclear targeted NLS-STM protein version (Figure 1E). Although leaf lobing does not necessarily relate to SAM function, this novel phenotype depends on the combination of the STM-GR and BHL9 proteins. Leaf-lobing is not observed in single transgenic lines and one possibility compatible with the NLS-STM results is that BLH9 mediates nuclear import of STM-GR in leaf cells (compare Figure 1E with Figure 6D).
The transition from the vegetative to reproductive phase is controlled by environmental and/or intrinsic developmental cues that converge at the SAM (1,2). In consequence, the vegetative meristem undergoes morphological changes and new growth patterns required for flowering are established. An essential contribution of BLH genes to floral induction has been shown in Arabidopsis plants containing null mutations in two paralogous BLH genes, PENNYWISE (pny a BLH9 allele) and POUND-FOOLISH [pnf a BLH8 allele; (42)]. pny/pnf double mutants are completely unable to flower even though the SAM displays morphological and molecular changes that are consistent with it having received floral inductive signals. Our gain-of-function data with BLH3 and ATH1 not only provide further evidence that BLH gene products influence floral induction pathways, but suggest that MEINOX proteins such as STM may be contributing partners. In contrast to the pny/pnf double-mutant data, no direct support for a role for BLH9 in floral induction was obtained during this study, however, based on genetic data, the native partner of BLH9 may be BP1 not STM (18,22).
Admittedly, all the combinatorial phenotypes result from ectopic protein activity throughout the plant and by no means reflect functional interactions restricted to the SAM. However, the release of STM-GR from cytoplasmic retention in the presence of ectopic BLH gene activity gains an additional phenotype with all three possible partner combinations. This combinatorial effect suggests a functional interaction of STM with BLH proteins in planta, but both the leaf lobing and the floral induction phenotype must be interpreted cautiously with respect to overexpression. On the spatial level, the STM expression domain in the Arabidopsis SAM overlaps with the transcription patterns of all three BLH genes and the distinct phenotypes observed with ATH1 and BLH3 or BLH9 overexpression therefore suggest that STM may provide different functional contributions at the centre of the apex than at its periphery.
Heterodimerization to control nuclear import: an ancient paradigm in the TALE HD super gene family
BLH proteins comprise one of two TALE-HD protein subfamilies in plants (10), STM is the most prominent member of the MEINOX protein sub-branch in Arabidopsis. Our findings that STM resides in the cytoplasm but is nuclear in heterodimers with BLH partners is reminiscent of the situation in animals, where nuclear import of MEIS proteins is directed by heterodimerization with PBC gene products. For example, the Drosophila exd (extradenticle) gene is transcribed and translated in most embryonic cells but its function is regulated through sub-cellular localization. EXD sub-cellular distribution is regulated by heterodimerization with the product of the homothorax (hth) gene product, an interaction which depends on the presence of the MEINOXHTH domain. In Drosophila leg imaginal discs, the EXD protein is nuclear localized in proximal regions, which correspond to the expression domain of the HTH protein. Conversely, in distal parts, where HTH is not present, EXD is cytoplasmic [(12,16,14), reviewed in (43)]. The accepted scheme of nuclear import control of MEINOX TALE-HD proteins through interaction with the second class of TALE-HD proteins, BLH or PBC in plants and animals, respectively, suggests an ancient and conserved mechanism. In animals, the interaction between PBC and MEINOX proteins requires integrity of the bipartite PBC-A and B domains in the PBC and the MEINOX domain in MEIS proteins (13). Co-immunoprecipitation and BiFC experiments performed in the course of this study show that the MEINOXSTM and the BELLBLH3 domains are sufficient for interaction in vitro and in planta.
Based on protein–protein interaction data, the animal PBC-A/B domain finds its counterpart in the BELL domain of plant BLH proteins although conservation at the amino acid level is weak. As this is in strong contrast to the high conservation of the MEINOX domain it has been suggested that the separation of the plant and animal lineages started from a single ancestral gene and subsequently, independent gene duplications occured in the plant or animal lineages resulting in the relatively recently evolved paralogous pairs of MEIS and PBC genes in animals and KNOX and BLH genes in plants (10). However, the finding that nuclear import of STM is mediated by heterodimerization with BLH partner proteins in plants strengthens the alternative hypothesis, that KNOX/MEIS and BLH/PBC precursor genes may have already existed in a common ancestor of animals and plants (11,17).
This assumption implies a significant tolerance for divergence of the BELL and PBC-A/B domain other than for the MEINOX partner domain. Part of the explanation could reside in the conservation of secondary structure, since the BELL and PBC-A/B domains still share a pronounced bipartite helical organization. Another possibility is that the BELL or PBC-A/B domains may be more prone to acquisition of novel functions than the MEINOX domain. In the zebrafish Prep1 protein, the MEINOX domain is sufficient to provide MEIS activity to a PBX/HOX complex (13). Its DNA-binding homeodomain is dispensable as in Arabidopsis, where overexpression of the MEINOXSTM or MEINOXBP1 domains trans-dominantly phenocopy stm or bp1 loss-of function mutants, respectively (27). MEIS/PBX heterodimers in animals interact with various other transcription factors, including HOX or bHLH gene products (13,44,45) and improve specificity through formation of multimeric complexes with PBX/MEIS proteins, which is important for patterning of the anterior/posterior axis (46). If the MEINOX domain provides a central backbone for an assembly of multimeric complexes in animals as in plants, such functional constraints could easily explain its conservation during evolution. Alternatively, an intriguing aspect of animal PBC protein function is its regulation at the post-translational level, a mechanism that also plays a crucial role in animal patterning processes [(47), reviewed in (48)]. The PBC-B domain contains several conserved phosphorylation sites for Ser/Thr kinases and PBX1 sub-cellular localization correlates with the phosphorylation state of these residues whose dephosphorylation induces nuclear export (49). Any freedom in the PBC domain to tolerate mutations better than the MEINOX domain would allow the acquisition of new functions and therefore explain differences between the plant BLH and animal PBC lineages.
The evolutionary conservation of a functional interaction between members of two families of TALE-HD gene products, MEIS/PBX and KNOX/BLH proteins in animals or plants, respectively, suggests an ancient and possibly extant function. In the multimeric complexes involved in animal patterning processes, PBX proteins reportedly interact with transcriptional corepressors such as histone-deacetylases or N-CoR/SMRT (50). Recent data demonstrate that PBC/MEIS heterodimers penetrate repressive chromatin to mark specific genes for activation e.g. by myoD (44). The finding that dominant mutations in the miRNA target site of PHABULOSA and PHAVOLUTA, which effect adaxial/abaxial patterning of the Arabidopsis leaf correlate with changes of DNA methylation outside the SAM (51) or that the floral repressor FLC is prone to epigenetic silencing by histone methylation (52) indicates that chromatin changes accompany developmental decisions such as primordial cell fate or flowering time in plants. An extant aspect of STM/BLH function thus could be the activation or the repression of transcriptionally competent chromatin. According to their transcription patterns, STM could account for qualitative differences between meristematic and primordial cells whereas BLH partner proteins could implement positional cues from the center to the SAM periphery.
In conclusion, the data presented here show that STM is targeted to the nuclear compartment through interaction with BLH partner proteins. Discrete BLH gene expression domains in the SAM imply that STM interfaces with different BLH partner proteins from the center of the SAM to the periphery and inducible co-expression experiments in transgenic Arabidopsis plants suggest that individual STM/BLH heterodimers may contribute distinct functions. In ectopic overexpression experiments, a common switch to reproductive phase observed in double BLH3/STM-GR or ATH1/STM-GR transgenics after Dex treatment implicates a role for STM in floral induction.
Acknowledgments
We are grateful to Dr Rüdiger Simon (Universität Düsseldorf) for gift of the inducible STM-GR line and Dr Klaus Harter (Universität Tübingen) for providing BiFC vectors before publication. We also thank Dr John Chandler for experimental suggestions concerning flowering time and for critical reading and improving the manuscript. This project was funded by the Deutsche Forschungsgemeinschaft (DFG) through SFB572. Funding to pay the Open Access publication charges for this article was provided by the SFB 572.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sussex I.M. Developmental programming of the shoot meristem. Cell. 1989;56:225–229. doi: 10.1016/0092-8674(89)90895-7. [DOI] [PubMed] [Google Scholar]
- 2.Lyndon R.F., Francis D. Plant and organ development. Plant Mol. Biol. 1992;19:51–68. doi: 10.1007/BF00015606. [DOI] [PubMed] [Google Scholar]
- 3.Traas J., Doonan J.H. Cellular basis of shoot apical meristem development. Int. Rev. Cytol. 2001;208:161–206. doi: 10.1016/s0074-7696(01)08004-4. [DOI] [PubMed] [Google Scholar]
- 4.Barton M.K., Poethig R.S. Formation of the shoot apicalmeristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development. 1993;119:823–831. [Google Scholar]
- 5.Long J.A., Moan E.I., Medford J.I., Barton M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 6.Clark S.E., Jacobsen S.E., Levin J.Z., Meyerowitz E.M. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development. 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- 7.Endrizzi K., Moussian B., Haecker A., Levin J.Z., Laux T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 1996;10:967–979. doi: 10.1046/j.1365-313x.1996.10060967.x. [DOI] [PubMed] [Google Scholar]
- 8.Tsiantis M., Hay A. Comparative plant development: the time of the leaf? Nature Rev. Genet. 2003;4:169–180. doi: 10.1038/nrg1002. [DOI] [PubMed] [Google Scholar]
- 9.Hay A., Craft J., Tsiantis M. Plant hormones and homeoboxes: bridging the gap? Bioessays. 2004;26:395–404. doi: 10.1002/bies.20016. [DOI] [PubMed] [Google Scholar]
- 10.Burglin T.R. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burglin T.R. The PBC domain contains a MEINOX domain: coevolution of Hox and TALE homeobox genes? Dev. Genes Evol. 1998;208:113–116. doi: 10.1007/s004270050161. [DOI] [PubMed] [Google Scholar]
- 12.Rieckhof G.E., Casares F., Ryoo H.D., Abu-Shaar M., Mann R.S. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 13.Berthelsen J., Zappavigna V., Ferretti E., Mavilio F., Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaw T.J., You L.R., Knoepfler P.S., Yao L.C., Pai C.Y., Tang C.Y., Chang L.P., Berthelsen J., Blasi F., Kamps M.P., et al. Direct interaction of two homeoproteins, homothorax and extradenticle, is essential for EXD nuclear localization and function. Mech. Dev. 2000;91:279–291. doi: 10.1016/s0925-4773(99)00316-0. [DOI] [PubMed] [Google Scholar]
- 15.Penkov D., Tanaka S., Di Rocco G., Berthelsen J., Blasi F., Ramirez F. Cooperative interactions between PBX, PREP, and HOX proteins modulate the activity of the alpha 2(V) collagen (COL5A2) promoter. J. Biol. Chem. 2000;275:16681–16689. doi: 10.1074/jbc.M909345199. [DOI] [PubMed] [Google Scholar]
- 16.Abu-Shaar M., Ryoo H.D., Mann R.S. Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 1999;13:935–945. doi: 10.1101/gad.13.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellaoui M., Pidkowich M.S., Samach A., Kushalappa K., Kohalmi S.E., Modrusan Z., Crosby W.L., Haughn G.W. The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell. 2001;13:2455–2470. doi: 10.1105/tpc.010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith H.M., Hake S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell. 2003;15:1717–1727. doi: 10.1105/tpc.012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhatt A.M., Etchells J.P., Canales C., Lagodienko A., Dickinson H. VAAMANA–a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene. 2004;328:103–111. doi: 10.1016/j.gene.2003.12.033. [DOI] [PubMed] [Google Scholar]
- 20.Modrusan Z., Reiser L., Feldmann K.A., Fischer R.L., Haughn G.W. Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell. 1994;6:333–349. doi: 10.1105/tpc.6.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quaedvlieg N., Dockx J., Rook F., Weisbeek P., Smeekens S. The homeobox gene ATH1 of Arabidopsis is derepressed in the photomorphogenic mutants cop1 and det1. Plant Cell. 1995;7:117–129. doi: 10.1105/tpc.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byrne M.E., Groover A.T., Fontana J.R., Martienssen R.A. Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development. 2003;130:3941–3950. doi: 10.1242/dev.00620. [DOI] [PubMed] [Google Scholar]
- 23.Roeder A.H., Ferrandiz C., Yanofsky M.F. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 2003;13:1630–1635. doi: 10.1016/j.cub.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 24.Venglat S.P., Dumonceaux T., Rozwadowski K., Parnell L., Babic V., Keller W., Martienssen R., Selvaraj G., Datla R. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl Acad. Sci. USA. 2002;99:4730–4735. doi: 10.1073/pnas.072626099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker A., Bey M., Burglin T.R., Saedler H., Theissen G. Ancestry and diversity of BEL1-like homeobox genes revealed by gymnosperm (Gnetum gnemon) homologs. Dev. Genes Evol. 2002;212:452–457. doi: 10.1007/s00427-002-0259-7. [DOI] [PubMed] [Google Scholar]
- 26.Burglin T.R., Ruvkun G. New motif in PBX genes. Nature Genet. 1992;1:319–320. doi: 10.1038/ng0892-319. [DOI] [PubMed] [Google Scholar]
- 27.Markel H., Chandler J., Werr W. Translational fusions with the engrailed repressor domain efficiently convert plant transcription factors into dominant-negative functions. Nucleic Acids Res. 2002;30:4709–4719. doi: 10.1093/nar/gkf591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakamoto T., Nishimura A., Tamaoki M., Kuba M., Tanaka H., Iwahori S., Matsuoka M. The conserved KNOX domain mediates specificity of tobacco KNOTTED1-type homeodomain proteins. Plant Cell. 1999;11:1419–1432. doi: 10.1105/tpc.11.8.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 30.Bradley D., Carpenter R., Sommer H., Hartley N., Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- 31.Walter M., Chaban C., Schutze K., Batistic O., Weckermann K., Nake C., Blazevic D., Grefen C., Schumacher K., Oecking C., et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 32.Überlacker B., Werr W. Vectors with rare-cutter restriction enzyme sites for expression of open reading frames in transgenic plants. Mol. Breed. 1996;2:293–295. [Google Scholar]
- 33.Howard E.A., Zupan J.R., Ciovsky V., Zambryski P. The VirD2 protein of A.tumefaciens contains a C-terminal bipartit nuclear localisation signal: Implications for nuclear uptake of DNA in plant cells. Cell. 1992;68:109–118. doi: 10.1016/0092-8674(92)90210-4. [DOI] [PubMed] [Google Scholar]
- 34.Bechtold N., Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 35.Brand U., Grunewald M., Hobe M., Simon R. Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 2002;129:565–575. doi: 10.1104/pp.001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. Detection of protein–protein interactions in plants using bimolecular fluorescence complementation. Plant J. 2004;40:419–427. doi: 10.1111/j.1365-313X.2004.02206.x. [DOI] [PubMed] [Google Scholar]
- 37.Williams R.W. Plant homeobox genes: many functions stem from a common motif. Bioessays. 1998;20:280–282. doi: 10.1002/(SICI)1521-1878(199804)20:4<280::AID-BIES2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 38.Lincoln C., Long J., Yamaguchi J., Serikawa K., Hake S. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meisel L., Lam E. The conserved ELK-homeodomain of KNOTTED-1 contains two regions that signal nuclear localization. Plant Mol. Biol. 1996;30:1–14. doi: 10.1007/BF00017799. [DOI] [PubMed] [Google Scholar]
- 40.Schena M., Lloyd A.M., Davis R.W. A steroid-inducible gene expression system for plant cells. Proc. Natl Acad. Sci. USA. 1991;88:10421–10425. doi: 10.1073/pnas.88.23.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J.Y., Yuan Z., Jackson D. Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development. 2003;130:4351–4362. doi: 10.1242/dev.00618. [DOI] [PubMed] [Google Scholar]
- 42.Smith H.M., Campbell B.C., Hake S. Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr. Biol. 2004;14:812–817. doi: 10.1016/j.cub.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 43.Morata G. How Drosophila appendages develop. Nature Rev. Mol. Cell Biol. 2001;2:89–97. doi: 10.1038/35052047. [DOI] [PubMed] [Google Scholar]
- 44.Berkes C.A., Bergstrom D.A., Penn B.H., Seaver K.J., Knoepfler P.S., Tapscott S.J. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- 45.Choe S.K., Vlachakis N., Sagerstrom C.G. Meis family proteins are required for hindbrain development in the zebrafish. Development. 2002;129:585–595. doi: 10.1242/dev.129.3.585. [DOI] [PubMed] [Google Scholar]
- 46.Vlachakis N., Choe S.K., Sagerstrom C.G. Meis3 synergizes with Pbx4 and Hoxb1b in promoting hindbrain fates in the zebrafish. Development. 2001;128:1299–1312. doi: 10.1242/dev.128.8.1299. [DOI] [PubMed] [Google Scholar]
- 47.Vogt T.F., Duboule D. Antagonists go out on a limb. Cell. 1999;99:563–566. doi: 10.1016/s0092-8674(00)81545-7. [DOI] [PubMed] [Google Scholar]
- 48.Affolter M., Marty T., Vigano M.A. Balancing import and export in development. Genes Dev. 1999;13:913–915. doi: 10.1101/gad.13.8.913. [DOI] [PubMed] [Google Scholar]
- 49.Kilstrup-Nielsen C., Alessio M., Zappavigna V. PBX1 nuclear export is regulated independently of PBX-MEINOX interaction by PKA phosphorylation of the PBC-B domain. EMBO J. 2003;22:89–99. doi: 10.1093/emboj/cdg010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saleh M., Rambaldi I., Yang X.J., Featherstone M.S. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 2000;20:8623–8633. doi: 10.1128/mcb.20.22.8623-8633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao N., Lye K.W., Barton M.K. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell. 2004;7:653–662. doi: 10.1016/j.devcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 52.Henderson I.R., Dean C. Control of Arabidopsis flowering: the chill before the bloom. Development. 2004;131:3829–3838. doi: 10.1242/dev.01294. [DOI] [PubMed] [Google Scholar]