Abstract
We have used the in vitro selection method to search for catalytically active variants of the antigenomic delta ribozyme with mutations in the regions that constitute the ribozyme active site: L3, J1/4 and J4/2. In the initial combinatorial library 16 nt positions were randomized and the library contained a full representation of all possible sequences. Following ten cycles of selection-amplification several catalytically active ribozyme variants were identified. It turned out that one-third of the variants contained only single mutation G80U and their activity was similar to that of the wild-type ribozyme. Unexpectedly, in the next one-third of the variants the C76 residue, which was proposed to play a crucial role in the ribozyme cleavage mechanism, was mutated. In these variants, however, a cytosine residue was present in a neighboring position to the polynucleotide chain. It shows that the ribozyme catalytic core possesses substantial ‘structural plasticity’ and the capacity of functional adaptation. Four selected ribozyme variants were subjected to more detailed analysis. It turned out that the variants differed in their relative preferences towards Mg2+, Ca2+ and Mn2+ ions. Thus, the functional properties of the variants were dependent on both the structure of their catalytic sites and divalent metal ions performing catalysis.
INTRODUCTION
In the genomic RNA strand of the hepatitis delta virus (HDV) as well as in its antigenomic counterpart generated during virus replication via the double rolling circle mechanism there are two sequences with ribozyme activities, called the delta ribozymes (1–5). These ribozymes are required for self-cleavage of polymeric RNA transcripts into monomeric units. The crystal structure of genomic delta ribozyme 3′ cleavage product (6,7) has depicted the ribozyme as a highly compact molecule with the active site buried deeply within its tertiary structure. Importantly, no divalent metal ions were found close to the cleavage site although such ions are absolutely required for catalysis in physiological conditions. It was also suggested that C75, one of the ribozyme cytosine residues located in the single-stranded J4/2 region, might actively participate in the cleavage mechanism (6,7). The results of the subsequent studies performed in different laboratories (8,9) have opened a debate whether C75 plays the role of a base or an acid in general acid–base catalysis, which has been proposed to take place in the delta ribozymes.
Recently, the crystal structure of the genomic delta ribozyme in its pre-cleavage state has been solved and compared to that of the ribozyme 3′ cleavage product (10). The results reveal that in the pre-cleavage state the catalytically critical divalent metal ion is present but after cleavage this ion is ejected from the active site. Moreover, a significant RNA conformational change seems to accompany the catalysis (10). Changes in the overall conformation of the delta ribozyme in different stages of the reaction have also been observed by FRET measurements (11–13) or chemical probing (14,15). Very recently, in order to investigate the role of the critical C75 residue, an approach has been used which merges phosphorothiolate substitution with enzyme kinetics (16). The authors have concluded that the cytidine provides general acid catalysis, mediating proton transfer to the 5′ OH leaving group through a protonated N3-imino nitrogen, while the ionized hydrate of magnesium ion serves as a base in the cleavage mechanism. Thus the delta ribozymes belong to the metalloenzyme class, in which nucleobases function similarly to catalytic histidine residues in protein enzymes (16–18).
Despite large progress in elucidation of the structure and mechanism of catalysis of the delta ribozymes, the existing data do not, however, reveal how their catalytic centers might have evolved. They may have been naturally selected as the most effective in performing catalysis. Alternatively, the cores of the wild-type ribozymes of genomic and antigenomic strand origin are among several similarly behaving arrangements of nucleotides. This is particularly intriguing since it has been shown that the surrounding viral sequences can greatly influence folding of delta ribozymes and modulate catalysis (4,19). Hence, it is interesting to know whether a similar or better ribozyme performance could be achieved by catalytic centers that are composed of nucleotides other than the wild-type residues. High sequence conservation of ribozyme regions of viral RNAs precludes answering this question. The existing data on catalytic activity of genomic and antigenomic ribozyme variants with single mutations introduced in their catalytic cores (1,20,21) can be interpreted only in terms of importance of the particular nucleotides for the performance of these ribozymes. Simultaneous testing of a very large number of ribozyme variants with multiple mutations is, however, possible with the use of the in vitro selection methodology. This approach, with a simplified protocol designed to identify catalytically inactive mutants, has revealed nucleotide preferences in the L3 region of the genomic ribozyme (22). In another selection experiment, a doped library of the genomic ribozyme with randomized 44 positions, including those forming the catalytic core, has been used (23). Interestingly, no variant was found with catalytic activity higher than the wild-type ribozyme and the selection was dominated by a variant with changes that resembled the antigenomic variant. However, in this experiment the initial library was highly under represented thus searching of the entire ‘sequence space’ was not possible.
Here we present the results of an in vitro selection experiment that aimed to obtain catalytically active variants of antigenomic delta ribozyme from the fully represented combinatorial library, in which three ribozyme regions: L3, J1/4 and J4/2 were randomized. These regions are directly involved in the formation of the ribozyme catalytic core. We expected that mutations might disturb the organization of the ribozyme spatial structure and that the catalytic activity as well as ribozyme preference towards divalent metal ions promoting catalysis might change.
MATERIALS AND METHODS
Materials
Polynucleotide kinase T4, T7 RNA polymerase, RNase inhibitor and T4 DNA ligase were purchased from MBI Fermentas. AmpliTaq polymerase was from Perkin Elmer. M-MuLV reverse transcriptase was from MBI Fermentas or Promega. The chemicals were purchased from Fluka or Serva. [γ-32P]ATP (4000 Ci/mmol) was from ICN. Chemically synthesized oligodeoxyribonucleotides were from Sigma-Ark and oligoribonucleotides were from Dharmacon (Lafayette, USA).
Synthesis of ribozyme combinatorial library
The dsDNA transcription template, encoding combinatorial library of ribozyme variants acting in cis, was prepared as follows (24,25): equimolar amounts of two oligomers (400 pmol of each): A3 (5′-GAAAAGTGGCTCTCCNNNNNNCATCCGAGTGCTCGGATG NNNAGGTCGGACCNNNNNNNGGTGGAGATGCCC-3′) and B3 (5′-TAATACGACTCACTATAGGGCGGGTCGGGGGCATCTCCACC-3′) [N denotes random nucleotides (4 nt were mixed in equal proportions by a DNA synthesizer), the T7 RNA polymerase promoter is marked in italics and the complementary sequences are underlined] were annealed in the total volume of 400 µl and the template was generated by PCR. The reaction mixture contained 1 µM of both DNA oligomers, 10 mM Tris–HCl (pH 8.3), 2 mM MgCl2, 50 mM KCl, 200 µM of each dNTP and 25 U/ml AmpliTaq polymerase. The reaction was performed in five cycles: 30 s at 94°C, 30 s at 46°C and 2 min at 72°C. The dsDNA template was recovered by phenol: chloroform extraction (1:1), ethanol precipitated and dissolved in TE buffer.
The in vitro transcription reaction was performed in the total volume of 400 µl and contained: 200 pmol dsDNA template, 40 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 0.001% Triton X-100, 2 mM spermidine, 5 mM DTT, 1 mM each NTP, 5 mM guanosine 5′-monophosphorothioate (5′-GMPS),750 U/ml RNase inhibitor and 2000 U/ml T7 RNA polymerase. The transcription mixture was incubated at 4°C for 48 h. Subsequently, EDTA solution was added to final concentration of 15 mM and the transcription products were purified by electrophoresis on 8% polyacrylamide gels containing 1 mM EDTA and 7 M urea. The band corresponding to the non-cleaved ribozyme was localized by UV shadowing, cut out and RNA was eluted from the gel with 0.3 M sodium acetate (pH 5.2), 1 mM EDTA, ethanol precipitated and dissolved in sterile water containing 0.1 mM EDTA.
In vitro selection
A total of 100 µl SulfoLink® Coupling Gel (Pierce) was washed three times with 100 µl buffer BP: 20 mM Na2HPO4 (pH 8.9), 1 mM EDTA and 40% methanol. Subsequently, the gel suspension was shaken with the same buffer containing 5 µg crude carrier tRNA (Sigma) for 30 min, and finally, it was washed again three times with buffer BP.
The combinatorial RNA library (50 pmol sample was taken from the RNA pool which was transcribed using 200 pmol dsDNA template; this amount of RNA contained 4.3 × 109 unique sequences each of them present in ∼7000 copies, on average) was dissolved in 10 µl buffer BH: 3M NaCl, 50 mM Tris–HCl (pH 7.5) and 1 mM EDTA, supplemented with sterile water containing 0.1 mM EDTA to the final volume of 100 µl, then ethanol precipitated and dissolved in 100 µl buffer BP. The library was incubated at 65°C for 3 min, cooled to room temperature for 10 min and mixed with SulfoLink® Coupling Gel, which was prepared as described above. The suspension was subjected to mild shaking at 25°C for 2 h. Subsequently, the gel was washed three times with 200 µl buffer BH and four times with 200 µl buffer BR: 200 mM NaCl, 50 mM Tris–HCl (pH 7.5), 0.1 mM EDTA (26). In buffer BR the self-cleavage reaction was initiated by adding Mg2+ ions to the final concentration of 5 mM and the reaction proceeded at 25°C. After 1 h the suspension was centrifuged for 1 min at 14 000 r.p.m. in an Eppendorf’s centifuge. Subsequently, RNA molecules that underwent self-cleavage were collected with liquid phase, the RNA was ethanol precipitated in the presence of glycogen and 0.3 M AcONa (pH 5.2) and finally it was dissolved in 34 µl sterile water containing 0.1 mM EDTA.
Prior to reverse transcription, the RNA from the step described above was annealed with 25 pmol of primer C3: 5′-GAAAAGTGGCTCTCC-3′ at 65°C for 3 min and cooled on ice for 10 min. The reaction was carried out at 42°C for 1.5 h, in 50 mM Tris–HCl (pH 8.0), 65 mM KCl, 10 mM MgCl2, 0.5 mM of each dNTPs, 4 mM DTT, 600 U/ml M-MuLV reverse transcriptase in the final volume of 70 µl (24). The cDNA was then amplified by PCR with 0.5 µM of C3 and B3 primers, in the presence of 75 mM Tris–HCl (pH 8.8), 2 mM MgCl2, 20 mM (NH4)2SO4, 0.01% Tween-20, 0.2 mM of each dNTPs, 20 U/ml Taq DNA polymerase in the final volume of 400 µl. The number of cycles of PCR was optimized for each pool. Conditions were as follows: of 30 s at 94°C, 30 s at 46°C and 2 min at 72°C. Subsequently, the DNA was recovered by phenol:chloroform extraction (1:1), ethanol precipitated and dissolved in 60 µl TE buffer.
The DNA library from the final selection step was digested with HindIII and SacI and ligated into pUC19 vector. Escherichia coli JM 109 cells were transformed with the ligation mixture and plasmids from individual clones were sequenced using standard dideoxy sequencing procedure (24).
Synthetic ribozyme variants
The dsDNA templates for the in vitro transcription of the R20, R25, R37 and R51 ribozymes were prepared as follows (24,25). Two DNA oligomers were synthesized: A2: 5′-GAAAAGTGGCTCTCC(CTTAGC)CATCCGAGTGCTCGGATG(CCC)AGGTCGGACC (GCGAGGA)GGTGGAGATGCCC-3′ and B2: 5′-TAATACGACTCACTATA GGGCATCTCCACC-3′ (in oligomer A2 letters in parentheses denote nucleotides of the wild-type sequence, which were changed to appropriate residues present in the ribozyme variants as shown in Figure 4; in both oligomers the complementary sequences are underlined, and in oligomer B2 letters in italics mark the T7 RNA polymerase promoter). Equimolar amounts of both oligomers (A2, B2) were annealed and dsDNA templates were generated by PCR according to the same procedure as described for the synthesis of the initial ribozyme combinatorial library. The in vitro transcription reactions were performed in standard conditions using 0.4 µM dsDNA templates at 37°C for 4 h. The RNA transcripts were purified on 8% denaturing polyacrylamide gels.
Cleavage reaction
Cleavage activity of RNA pools was examined after each step of selection. In order to avoid technical problems with laborious transcription reaction of cis-acting variants (at 4°C for 48 h), the dsDNA templates were re-amplified by PCR with primers B2 and C3. As a result, dsDNA was obtained, which encoded ribozymes shortened by 11 nt from the 5′ ends, acting in trans. The transcription reaction was performed in standard conditions at 37°C for 4 h. The cleavage assay was carried out with oligonucleotide substrate Sub: 5′-CUUCGGGUCGG-3′ as described below in the presence of 10 mM Mg2+ ions at 37°C.
The trans-acting ribozymes were prepared by mixing the 5′ end-32P-labeled oligonucleotide substrate Sub (∼50 000 c.p.m., 0.1 pmol of RNA in 50 µl reaction volume) with the analyzed ribozyme or ribozyme library in the standard reaction buffer: 50 mM Tris–HCl (pH 7.5), 0.1 mM EDTA to obtain the final RNA concentration of <2 nM and 700 nM, for the substrate and ribozyme component, respectively. The mixtures were subjected to a denaturation–renaturation procedure by incubating at 100°C for 2 min, 0°C for 10 min and finally at 37°C for 10 min. The cleavage reactions were initiated by adding an appropriate divalent metal chloride solution and the reactions proceeded at 37°C. Aliquots of the reaction mixtures (5 µl) were removed at specified time points and quenched with equal volumes of 20 mM EDTA/7M urea mixture containing electrophoresis dyes (25,27).
Products of the catalytic cleavage were analyzed by electrophoresis on 12% polyacrylamide, 0.75% bis-acrylamide, 8M urea gels. Autoradiography was performed at −70°C with an intensifying screen. For quantitative analysis, radioactive bands were quantified using phosphorimaging screens, the Typhoon 8600 Imager and ImageQuant software (Molecular Dynamics). First-order rate constants (kobs) were calculated from data fitted to the single-exponential equation: [P]t = [EP](1−e−kobs × t), where kobs is the first-order rate constant, [P]t and [EP] are the fraction cleaved at time t and the reaction end point, respectively. Reactions of the same ribozyme variant in the presence of various divalent metal ions were usually performed on a gel side by side and at least two independent data sets were collected.
Structural probing with Pb2+ ions
For structural probing the trans-acting ribozyme variants were prepared by mixing the 5′ end-32P-labeled ribozyme components: R20, R25, R37 and R51 (∼100 000 c.p.m. in 50 µl reaction volume) with the oligonucleotide substrate Sub in the buffer: 50 mM NaCl, 10 mM Tris–HCl (pH 7.2), 10 mM MgCl2 to obtain the final RNA concentration of 200 nM and <2 nM, for the substrate and ribozyme component, respectively. The mixtures were subjected to a denaturation–renaturation procedure by incubating at 100°C for 2 min, 0°C for 10 min and finally at 37°C for 10 min. Subsequently, lead acetate solution was added and the reactions proceeded at 25°C for 10 min (28,29). The reactions were terminated by mixing their aliquots with 8 M urea/dyes and 20 mM EDTA solution and samples were loaded on 12% polyacrylamide, 0.75% bis-acrylamide and 7 M urea gels. Electrophoresis was performed at 2000 V for 2 to 3 h, followed by autoradiography at −70°C with an intensifying screen or phosphorimaging with Typhoon 8600 analyzer (Molecular Dynamics).
In order to assign the cleavage sites, products of metal ion-induced cleavage were run along with the products of alkaline RNA hydrolysis and limited T1 nuclease digestion of the same RNA. An alkaline hydrolysis ladder was generated by incubation of 5′-end-32P-labeled RNA with 5 vol of formamide at 100°C for 10 min. Partial T1 nuclease digestion was performed in 50 mM sodium citrate (pH 5.3), 7 M urea with 0.2 U of the enzyme at 55°C for 10 min.
RESULTS
In vitro selection
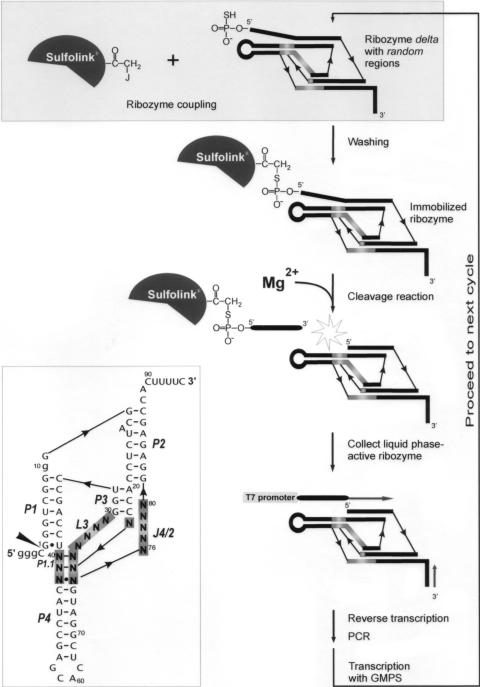
The combinatorial library of the antigenomic delta ribozyme, which was used in an in vitro selection experiment, is shown in Figure 1. The ribozyme variants differed in three regions: L3, J1/4 and J4/2, which are known to participate in the formation of ribozyme catalytic core, and 16 nt positions were randomized in total. In order to minimize the tendency for adapting alternative RNA folds, a characteristic typical of the wild-type sequence, the P4 stem was shortened. This domain does not participate in catalysis but is largely responsible for ribozyme conformational heterogeneity (1,19,25). Additionally, the sequence was modified in the J1/2 region. Nucleosides C8 and A9 were deleted and U10 was mutated to guanosine. The minimal ribozyme sequence was extended at its 3′ end by additional few nucleotides, which resulted in a constant stretch of 15 nt. The stretch was sufficiently long for binding an oligonucleotide primer used in the selection procedure but too short to interfere with the functioning of the ribozyme (4). The library was transcribed in the presence of thio-analog of GMP at 4°C for 48 h in order to minimize self-cleavage of catalytically active variants. Under these conditions the extent of self-cleavage of the wild-type ribozyme accounts for ∼30% (M. Łęgiewicz and J. Ciesiołka, unpublished data).
Figure 1.
Combinatorial library of cis-acting antigenomic delta ribozyme (inset) and a scheme of the selection-amplification procedure used to search for its catalytically active variants. The randomized regions are boxed. Numbering of nucleotides corresponds to the wild-type ribozyme sequence and nucleotide changes are shown in lower case letters. Base paired segments are denoted P1–P4 and the catalytic cleavage site is marked by the filled triangle.
The in vitro selection protocol, which is schematically underlined in Figure 1, followed that used by Pace and coworkers (26) for selection of catalytically active variants of M1 RNA. In the first step, the ribozyme variants with SH groups introduced at their 5′ ends during transcription were covalently attached to SulfoLink® Coupling Gel (Pierce) activated with iodoacetyl groups. The initial RNA pool contained 4.3 × 109 unique sequences each of them present in ∼7000 copies, on average. The coupling reaction was carried out for 120 min at 25°C. In such conditions ∼65% of SH-modified RNA molecules binds to this matrix (26). We also tested Bromoacetyl-Cellulose (Sigma), however, in that case the fraction of covalently bound RNA did not exceed 35%. The immobilized ribozymes were subjected to self-cleavage reaction in the presence of 5 mM Mg2+ for 1 h at 25°C. The liquid phase with catalytically active variants was collected, and subsequently, the RNA pool was reverse transcribed, amplified by PCR, and transcribed with T7 RNA polymerase. The obtained new RNA pool was used in the next selection cycle (24).
It is known that conditions in which an in vitro selection experiment is performed impose intended and unintended selection pressures on the evolving population of variants and as a consequence favor the development of molecules that are optimized for that situation—‘you get what you select for’ (30). In our experiment each ribozyme library was transcribed under conditions in which large fractions of very active variants might be cleaved. Such variants would be underrepresented in the libraries and after several cycles might escape selection. On the other hand, self-cleavage reactions were performed in the presence of 5 mM Mg2+ ions for 1 h at 25°C, that is at a relatively weak selection stringency. Such conditions enabled even less active variants to proceed to the next cycles. Survival of sequences with sub-optimal properties was particularly important since it allowed to explore the entire ‘sequence space’ consisting of ribozyme variants that differ in the catalytic core.
Selection progress and identification of ribozyme variants
After each selection cycle, the catalytic activity of every new generation of ribozyme variants was examined (Figure 2). In order to simplify the analysis we changed the system in which ribozymes were operating from cis- to trans-acting. Double-stranded DNA libraries were re-amplified with the use of appropriately designed primers (details are described in Materials and Methods) and the newly synthesized templates were transcribed generating trans-acting ribozymes which began with G10 at their 5′ ends. The 11 nt-long Sub oligomer was used as a substrate in the cleavage reactions.
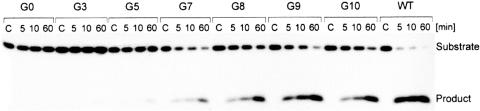
Figure 2.
Cleavage activity of the successive generations of delta ribozyme variants obtained during in vitro selection. The cleavage assays were performed with trans-acting forms of the ribozyme pools and 5′ end-32P-labeled oligonucleotide substrate (see the text for further details). The reactions were carried out in the presence of 10 mM Mg2+ for 5, 10 and 60 min. WT; -wild-type ribozyme, C; control lines.
It turned out that already after five cycles of selection-amplification the library showed measurable catalytic activity, while after ten cycles it reached an activity comparable to that of the wild-type ribozyme. This library was cloned using pUC19 vector and E.coli cells and 37 individual sequences of ribozyme variants were determined. The sequences were classified into a few families taking into account two major criteria: the possibility of formation of P1.1 segment, which is present in the wild-type ribozyme and the nucleotide composition of region J4/2 with the catalytic cytosine in position 76 (Figure 3).
Figure 3.
Nucleotide sequences and cleavage activities of delta ribozyme variants obtained by in vitro selection. Regions that were randomized in the initial ribozyme library are marked in bold. All nucleotides that differ from those found at corresponding positions of the wild-type ribozyme sequence (WT) are denoted by red fonts. Cleavage extents (in percentages) were determined for trans-acting forms of ribozymes after 10 min in the presence of 10 mM Mg2+.
We determined the relative catalytic activities of several selected ribozyme variants, in their trans-acting forms, in the presence of 10 mM Mg2+ ions (Figure 3). The cleavage extents were measured after 5, 10 and 60 min. Based on these data it was evident that all tested variants showed activity similar or lower than the wild-type ribozymes. Roughly estimated kobs values were in the range of 0.05–0.3 min−1 while the wild-type ribozyme was cleaved with kobs 0.44–0.47 min−1 in the same conditions. Unexpectedly, three representative variants of the fifth family (nine sequences not included in Figure 3), showed no measurable activity. No cleavage was also observed when the variants were synthesized and assayed in their cis-acting forms (data not shown). It is unclear whether these sequences were selected as having marginally low activity, which was not detected in the conditions of our assay or they were inactive but survived in the selection process due to some unintended properties. Interestingly, in most of these variants the L3 and J4/2 regions were exceptionally rich in adenosine residues but the reason for that remains unclear.
Analysis of catalytic activity and structure of four selected ribozymes
Based on the results of the initial screening of catalytic activity of several selected ribozymes (Figure 3) four variants were chosen for more detailed analysis. Variant #37 represented the first family of sequences and its activity was similar to that of the wild-type ribozyme. The other three variants, #20 and #25 from the second and #51 from the third family, were moderately active. We synthesized four ribozymes: R37, R20, R25 and R51 which corresponded to the cloned sequences (Figure 4). The ribozymes were obtained in their trans-acting forms with the use of chemically synthesized oligodeoxyribonucleotides for constructing DNA templates for in vitro transcription with T7 RNA polymerase. Moreover, all mutations, which had been introduced during selection in the constant stretches of the initial ribozyme library, were replaced by nucleotides present in the wild-type sequence. A mutant of the R51 ribozyme in which C77 had been replaced by a guanosine residue was also synthesized. The mutant showed no measurable activity in the presence of 10 mM Mg2+ (data not shown). This observation supports the suggestion that the C77 residue may play a catalytic role corresponding to that of C76 in the wild-type ribozyme (we would like to thank an anonymous Referee for suggesting this experiment).
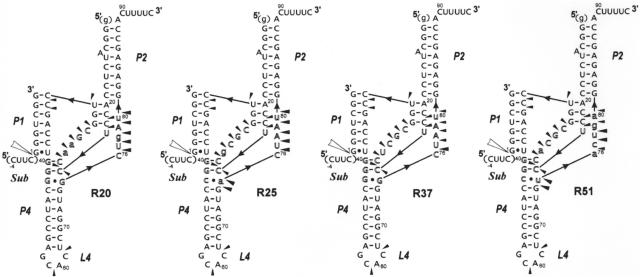
Figure 4.
Secondary structure models of trans-acting ribozymes: R20, R25, R37 and R51 and the results of their structural probing by Pb2+-induced cleavage method. The models derive from the consensus delta ribozyme pseudoknot structure. Nucleotides different than those found at corresponding positions of the wild-type sequence are marked by lower case letters. Outlined letters denote nucleotides present in the regions, which were randomized in the initial combinatorial library that was used in the selection of the ribozymes shown in the figure. Cleavages induced by Pb2+ ions are denoted by black triangles and the data derived from the autoradiograms which are shown in Supplementary Figure 1. The ribozyme components that were used for structural probing were deprived of the 5′-terminal guanosine residue in order to increase the homogeneity of RNA transcripts at their 5′ ends.
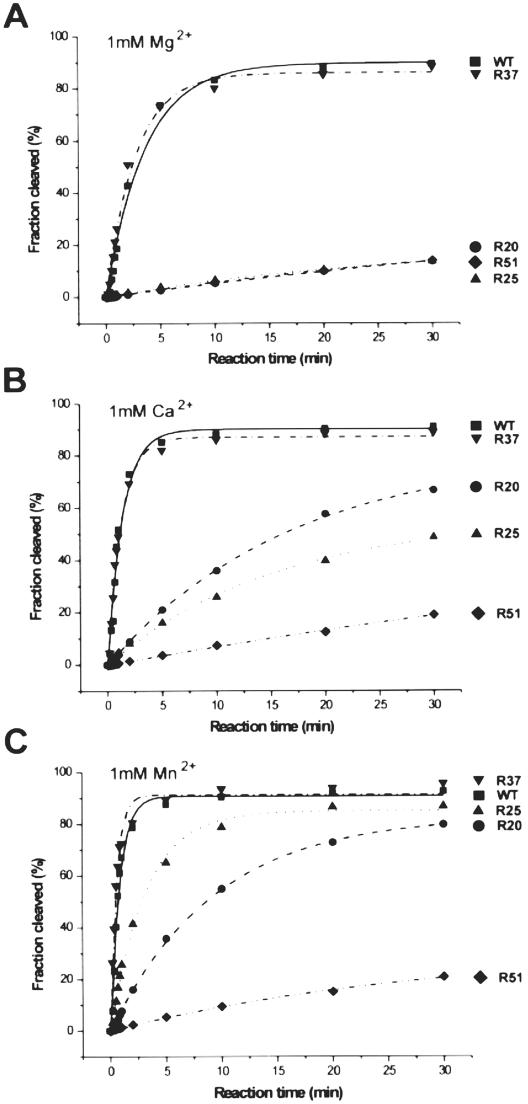
Catalytic activities of the ribozymes: R37, R20, R25 and R51 were determined at 1 mM concentration of Mg2+, Ca2+ and Mn2+ ions (Figure 5 and Table 1). The cleavage extents of R37 reached ∼90% after 30 min in the presence of all three metal ions, similarly as in the case of the wild-type variant (25,27). Unexpectedly, the R20, R25 and R51 ribozymes were cleaved in Mg2+ to only 13–14%. In the presence of Ca2+ and Mn2+ the cleavage extents were much more differentiated. In both these metal ions R51 was cleaved to only ∼20%. In Ca2+ R20 and R25 were cleaved to 67 and 49%, respectively, but in Mn2+ the cleavage extents were higher, reaching 80 and 87%, respectively. The ribozymes also differed in their cleavage rates. The cleavage rate constants kobs were calculated from the linear parts of the reaction curves and are shown in Table 1. When compared with the kobs values determined for the wild-type ribozyme, the said values for the R37 ribozyme observed in the presence of Mg2+ and Mn2+ were slightly higher while in the presence of Ca2+ they were similar. The cleavage rate constants for the R20, R25 and R51 ribozymes in Mg2+ were almost identical and ∼40-fold lower than that determined for the wild-type variant. These three ribozymes were, however, cleaved differently in the presence of Ca2+ and Mn2+. For R51 both kobs values were similar to that determined in Mg2+. In comparison with the wild-type ribozyme this variant was less active by factors of 100 and 150, in Ca2+ and Mn2+, but only by the factor of 40 in the presence of Mg2+. Thus, variant R51 seem to prefer Mg2+ ions in catalysis, unlike the R25 ribozyme, which prefers Mn2+ ions and in the presence of these ions this ribozyme was only 4-fold less active than the wild-type variant. Finally, comparing the kobs values for R20 with those determined for the wild-type ribozyme, this variant turned out to be 40-fold less active in Mg2+ but only 20- and 13-fold in Ca2+ and Mn2+, respectively.
Figure 5.
Cleavage kinetics of the trans-acting ribozymes: WT, R20, R25, R37 and R51 in the presence of Mg2+ (A), Ca2+ (B) and Mn2+ (C) ions at 1 mM ion concentration.
Table 1.
Summary of kinetic data
| Ribozyme | Mg2+ | Ca2+ | Mn2+ | |||
|---|---|---|---|---|---|---|
| kobs [min−1] | Fr30 [%] | kobs [min−1] | Fr30 [%] | kobs [min−1] | Fr30 [%] | |
| WT | 0.2199 ± 0.0231 | (89) | 0.7566 ± 0.0840 | (91) | 1.1885 ± 0.0554 | (93) |
| R37 | 0.3199 ± 0.0257 | (88) | 0.7243 ± 0.3948 | (89) | 1.3437 ± 0.0917 | (96) |
| R20 | 0.0050 ± 0.0001 | (14) | 0.0386 ± 0.0012 | (67) | 0.0907 ± 0.0012 | (80) |
| R25 | 0.0049 ± 0.0002 | (13) | 0.0228 ± 0.0009 | (49) | 0.3038 ± 0.0128 | (87) |
| R51 | 0.0051 ± 0.0001 | (14) | 0.0069 ± 0.0001 | (19) | 0.0078 ± 0.0002 | (21) |
In order to confirm the predicted secondary structures of trans-acting ribozymes: R37, R20, R25 and R51 the Pb2+-induced cleavage method was used (28,29). The ribozyme components were labeled with 32P at the 5′ ends, and subsequently, the molecules were hybridized in the presence of 10 mM Mg2+ to the oligonucleotide substrate Sub, which was used in excess of 100-fold. The complexes were treated with Pb2+ ions at 25°C for 10 min. In parallel, the same ribozyme components but with no oligomer Sub hybridized, were also subjected to Pb2+ probing. Representative cleavage patterns from this experiment are shown in Supplementary Figure 1 and the cleavage sites are displayed on the anticipated secondary structures of trans-acting ribozymes in Figure 4. In the applied conditions ∼95% of ribozyme R37 and ∼50% of ribozymes: R20, R25 and R51 had been self-cleaved before Pb2+ probing. Thus we expected that the Pb2+-induced cleavage patterns would be a superposition of (i) the patterns reflecting the structure of the ribozyme components hybridizing with the 7 nt-long cleavage product and (ii) that characteristic of the ribozyme—11 nt substrate complexes. It turned out, however, that despite different cleavage extents of the ribozyme variants, the Pb2+-induced patterns were very similar. However, they were clearly different from those obtained for the ribozymes without the substrate hybridized, in which a series of cleavages were observed also in the P1(3′) regions confirming single-stranded character of these stretches (in Supplementary Figure 1, as an example, the Pb2+-induced patterns for R37 and R37 + Sub are shown for comparison).
DISCUSSION
All regions of the antigenomic delta ribozyme, which were randomized in the combinatorial library used in the in vitro selection (Figure 1, inset) are highly conserved in natural isolates of viral HDV RNAs (1,4). In the L3 region only U26 has been found to be replaced by a cytosine residue. Entirely conserved are three guanosines in positions 40–42 of J4/1. A few changes are possible in the J4/2 region, where U77 can be replaced by cytosine or adenosine, and A78 by guanosine (1,4). Importantly, these changes concern only single-nucleotide substitutions i.e. in each variant no more than one residue is changed simultaneously. Strong conservation of these regions in natural viral isolates makes impossible any discussion on correlation of sequence variations with organization of the ribozyme catalytic core.
The aim of our in vitro selection experiment was to determine to what extent the ribozyme core could be changed without losing its catalytic properties. Therefore, it was very important to begin the selection process with a library consisting of a full representation of the possible variants. The separation step relied on the ability of the variants covalently linked to agarose matrix to cleave off self-catalytically after the addition of Mg2+ ions. After 10 cycles of selection-amplification procedure the enriched library showed an activity which was lower but comparable to that of the wild-type ribozyme (Figure 2). Several individual members of the final library were identified and classified into four families based on sequence similarities (Figure 3).
In the first family, which accounted for one-third of the identified variants, only 1 nt change was found in the initially randomized regions, i.e. guanosine in position 80 was replaced by a uridine residue. These variants showed high catalytic activity similar to that of the wild-type ribozyme. It has been observed earlier that deletion of G80 results in only 2-fold loss of ribozyme activity (21). It is unclear why G80U mutation was so beneficial in the selection-amplification process and why the wild-type sequence was not found among the cloned individuals. Most likely, some other factor than the intended selection criteria caused the occurrence of this particular nucleotide substitution. While excluding this single-nucleotide mutation it turned out that one-third of all the identified variants shared the catalytic core with that of the wild-type sequence. Therefore, in the applied selection conditions the native ribozyme core seemed to perform catalysis most efficiently.
The ribozyme variants belonging to the second family shared the ‘correct’ P1.1 helix and a cytosine residue in position 76 while having mutations in at least two out of three initially randomized regions. In particular, in the L3 region, uridine in position 26 was mutated to adenosine or guanosine. It has been shown that mutation U26A leads to less than 2-fold reduction of ribozyme activity (21). Interestingly, only cytosine was not found in position 26 of the selected variants whereas this residue has been observed to occur in a natural virus isolate (4). In three variants the non-standard G42–G75 bp at the top of P4 stem was replaced by AA or CA interactions. Standard base pairing in this region has been shown to be detrimental for cleavage. For example, mutations G42C or G75C result in ∼1000-fold reduction of the activity while mutation G42U, which creates a GU base pair, decreases the activity by factor of 100 (21). Finally, in two variants A78 was replaced by guanosine and such residue has also been found in this position in viral RNA isolates (1,4).
The ribozyme variants classified to the third family comprised one-third of the selected sequences thus the family was as large as the first family with variants bearing single-nucleotide substitutions. In the L3 region the only change was that in almost all cases, uridine was replaced by adenosine in position 26. Only in one case, a guanosine residue was present in this position. But this particular substitution seemed to decrease the catalytic activity, since variants #56 and #57 were more active than variant #45. In all the variants base pairing between positions 24 and 41 as well as 25 and 40 was preserved thus the ‘correct’ P1.1 segment could be formed. The most significant changes, however, included the non-standard G42–G75 interaction at the top of helix P4 as well as the nucleotide composition of the J4/2 region. In all but one selected variants of the third family a uridine residue was present in position 75. In variants #40 and #55 this would result in A42–U75 bp. However, it has been shown that standard base pairs in this region are highly detrimental for cleavage (21). Unexpectedly, the variants did not possess a cytosine residue in position 76. In single-substituted mutants the presence of adenosine in this position has resulted in 400-fold less active ribozyme while guanosine or uridine caused complete loss of activity (21).
Despite the absence of C76 in all the variants of the third family a cytosine residue was present in the neighboring 77 position of the polynucleotide chain. We propose that a substantial rearrangement of the catalytic core takes place in these variants. In the wild-type ribozyme the guanosine residue that precedes C76 is involved in a non-standard interaction with G42 at the top of helix P4. In all variants of this family, except variant #19, uridine is present in position 75, but likely, this nucleotide is looped out and base pairing occurs between the nucleotides in positions 42 and 76. These are either CA or AG interactions. In this way a cytosine residue present in all of the variants in position 77 may play a catalytic role corresponding to that of C76 in the wild-type ribozyme. The downstream stretches are 78-UGAG-81 or 78-UGGA-81. In comparison with the wild-type sequence of this region, i.e. 77-UAAG-80, it turns out that in the former stretch the only difference concerns the A to G substitution in the second position and in the latter stretch also the order of the last 2 nt is reversed. Summarizing, it seems that the structural elements of the J4/2 region crucial for ribozyme performance might be found in structurally equivalent places of the selected variants despite their different primary structures. Thus the delta ribozyme catalytic core shows a substantial degree of ‘structural plasticity'. Importantly, this feature could not be revealed by studying ribozyme variants with single-point mutations.
In the fourth family, four ribozyme variants were classified. Although the catalytic C76 was preserved in three variants and in one variant a cytosine residue was present in the neighboring 77 position, the variants showed a relatively low catalytic activity. Strikingly, the L3 and J4/1 regions were exceptionally rich in adenosine residues. Possibly, these residues are necessary for the formation of sufficiently stable helix P1.1. The helix is composed of 2 bp involving 24–41 and 25–40 nt. In these positions at least one AA pair occurred in three variants of the family. Additionally, in two variants GC, in two GA, and in one variant AC interactions might contribute to helix stability. Potentially, in any of the non-standard AA, GA or CA base pairs two H-bonds could be formed (31). Thus even in variants #31 and #32, which are deprived of GC base pairing in this region, the P1.1 helix could attain sufficient stability.
Extensive analysis of variants of delta ribozyme in terms of the possible P1.1 interactions has suggested that at least one GC base pair (three H-bonds) is necessary for the minimal ribozyme activity (32). Stable interactions between 25 and 40 nt seem to be most important and the data show that in position 40 a purine residue is preferred. The last preference is seen in all the variants selected in our studies. In other studies (21) it has been shown that two cytosine residues in positions 24 and 25 are not structurally equal. Only 10-fold loss of activity was observed in C25A single-nucleotide mutant while C24A mutation resulted in 2000-fold decrease in activity despite the fact that in both cases AG interactions were formed.
The Pb2+-induced cleavage patterns of four selected ribozymes, which were analyzed in their trans-acting forms (Figure 4), were consistent with the consensus delta ribozyme secondary structure and known specificity of Pb2+ ions (28,29,33–36). Moreover, the patterns were essentially independent of the different levels of ribozyme self-cleavage that had occurred prior to Pb2+ probing. This suggests that low final cleavage extents of some selected variants in the presence of Mg2+ were not a consequence of their overall folds being substantially different than the wild-type ribozyme structure. Hence, much subtle structural differences must be responsible for the incomplete cleavage. Most likely, these are the non-optimal arrangement of nucleotides constituting the catalytic core and/or different abilities of ribozymes to undergo conformational changes necessary to occur on the cleavage reaction trajectory.
Detailed kinetic analysis of the four selected ribozyme variants in the presence of 1 mM Mg2+, Ca2+ and Mn2+ ions showed that the variants differentiate in the preferential use of catalytic divalent metal ions (Figure 5 and Table 1). Recently, the crystal structures of genomic delta ribozyme in its pre- and post-cleaved states have revealed that the divalent metal ion bound in the precursor is released from the catalytic site after cleavage (10). Moreover, the precursor that had been crystallized in the absence of divalent metal ions had the same structure as the ribozyme with the ion found in the catalytic center. It might suggest that these ions were not required for folding of the genomic delta ribozyme (10). In the antigenomic variant, however, Mg2+ ions seem to be required for the formation of helix P1.1 (37). These two, seemingly inconsistent, propositions may be explained by the different roles of metal ions in the formation of ribozymes’ active centers. In the case of the genomic variant, its overall folding is directed by the formation of helix P2 (38). The first 82 nt from the cleavage site, which correspond to 4 bp in the P2 stem, are required for ribozyme catalytic activity (39). A shorter polynucleotide chain consisting of 78 nt, thus ending on the last nucleotide of the J4/2 region, is unable to form the catalytic core since the region corresponding to the P3/L3 stem–loop motif folds into another hairpin structure (38). Similar rearrangement of ribozyme secondary structure does not occur in the antigenomic variant (J. Wrzesinski and J. Ciesiołka, unpublished data). In that case already the first 79 nt are sufficient for ribozyme activity (40) thus clearly the presence of helix P2 is not absolutely required for cleavage. One may speculate that although the catalytic ion might not be required for genomic ribozyme folding, the same ion (or ions located in close proximity) would be much more important for the formation of the catalytic center in the antigenomic variant. In that case much higher constraints of such metal ion/ions coordination could be expected. This assumption seem to be supported by very small variations of the antigenomic viral sequences in the L3 region (1,4) possibly involved in metal ion coordination. This region was also exceptionally conservative in our selection experiment while in the J4/2 region much larger variations were observed. Summarizing, the differences in the catalytic activity of the selected ribozyme variants seem to be a consequence of the different abilities of various metal ions both to perform a chemical reaction as well as to aid the formation of ribozyme structural core.
While this paper was in the reviewing process Nehdi and Perreault (41) published the results of the in vitro selection experiments in which they used two libraries of variants of the antigenomic delta ribozyme. In the first library, 6 nt of the J4/2 region were randomized. In the other, they randomized 25 nt positions which were involved in the formation of ribozyme catalytic center although the catalytic C76 and the nucleotides forming helix P1.1 were left unchanged. These two libraries as well as experimental protocols used in their selections differed from those applied in our studies. Nevertheless, some observations were similar; e.g. while using the first library they selected ribozyme variants which were deprived of C76 but a cytosine residue was present in the neighboring 77 position. On the other hand, nucleotide variations within helix P1.1 could be revealed only in our studies since in Perreault's libraries these positions were not randomized. Thus, the independently obtained results of two laboratories supplement each other giving a more comprehensive picture of the architecture of the catalytic center of the antigenomic delta ribozyme.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Barbara Smólska for her excellent technical assistance and members of our laboratory for valuable comments on the manuscript. This work was supported by the Polish Ministry of Scientific Research and Information Technology (grants no. 6P04B01720 and 2P04A05128). Funding to pay the Open Access publication charges for this article was provided by the Polish Ministry of Scientific Research and Information Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Been M.D., Wickham G.S. Self-cleaving ribozymes of hepatitis delta virus RNA. Eur. J. Biochem. 1997;247:741–753. doi: 10.1111/j.1432-1033.1997.00741.x. [DOI] [PubMed] [Google Scholar]
- 2.Ciesiołka J., Wrzesinski J., Łęgiewicz M., Smólska B., Dutkiewicz M. Ribozymes of the hepatitis delta virus: recent findings on their structure, mechanism of catalysis and possible applications. Acta. Biochim. Pol. 2001;48:409–418. [PubMed] [Google Scholar]
- 3.Shih I.H., Been M.D. Catalytic strategies of the hepatitis delta virus ribozymes. Annu. Rev. Biochem. 2002;71:887–917. doi: 10.1146/annurev.biochem.71.110601.135349. [DOI] [PubMed] [Google Scholar]
- 4.Wadkins T.S., Been M.D. Ribozyme activity in the genomic and antigenomic RNA strands of hepatitis delta virus. Cell. Mol. Life Sci. 2002;59:112–125. doi: 10.1007/s00018-002-8409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lilley D.M.J. Structure, folding and mechanisms of ribozymes. Curr. Opin. Struct. Biol. 2005;15:313–323. doi: 10.1016/j.sbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Ferré-D'Amaré A.R., Zhou K., Doudna J.A. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 7.Ferré-D'Amaré A.R., Doudna J.A. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J. Mol. Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- 8.Perrotta A.T., Shih I., Been M.D. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- 9.Nakano S., Chadalavada D.M., Bevilacqua P.C. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 10.Ke A., Zhou K., Ding F., Cate J.H.D., Doudna J.A. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 11.Harris D.A., Rueda D., Walter N.G. Local conformational changes in the trans-acting hepatitis delta virus ribozyme accompany catalysis. Biochemistry. 2002;41:12051–12061. doi: 10.1021/bi026101m. [DOI] [PubMed] [Google Scholar]
- 12.Pereira M.J.B., Harris D.A., Rueda D., Walter N.G. Reaction pathway of the trans-acting hepatitis delta virus ribozyme: a conformational change accompanies catalysis. Biochemistry. 2002;41:730–740. doi: 10.1021/bi011963t. [DOI] [PubMed] [Google Scholar]
- 13.Tinsley R.A., Harris D.A., Walter N.G. Magnesium dependence of the amplified conformational switch in the trans-acting hepatitis delta virus ribozyme. Biochemistry. 2004;43:8935–8945. doi: 10.1021/bi049471e. [DOI] [PubMed] [Google Scholar]
- 14.Jeong S., Sefcikova J., Tinsley R.A., Rueda D., Walter N.G. Trans-acting hepatitis delta virus ribozyme: catalytic core and global structure are dependent on the 5′ substrate sequence. Biochemistry. 2003;42:7727–7740. doi: 10.1021/bi034627g. [DOI] [PubMed] [Google Scholar]
- 15.Ouellet J., Perreault J.P. Cross-linking experiments reveal the presence of novel structural features between a hepatitis delta virus ribozyme and its substrate. RNA. 2004;10:1059–1072. doi: 10.1261/rna.7230604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das S.R., Piccirilli J.A. General acid catalysis by the hepatitis delta virus ribozyme. Nature Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 17.Fedor M.J., Williamson J.R. The catalytic diversity of RNAs. Nature Rev. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 18.Doudna J.A., Lorch J.R. Ribozyme catalysis: not different, just worse. Nature Struct. Mol. Biol. 2005;12:395–402. doi: 10.1038/nsmb932. [DOI] [PubMed] [Google Scholar]
- 19.Lazinski D.W., Taylor J.M. Regulation of the hepatitis delta virus ribozymes: to cleave or not to cleave? RNA. 1995;1:225–233. [PMC free article] [PubMed] [Google Scholar]
- 20.Tanner N.K., Schaff S., Thill G., Petit-Koskas E., Crain-Denoyelle A.M., Westhof E. A three-dimensional model of hepatitis delta virus ribozyme based on biochemical and mutational analyses. Curr. Biol. 1994;4:488–498. doi: 10.1016/s0960-9822(00)00109-3. [DOI] [PubMed] [Google Scholar]
- 21.Perrotta A.T., Been M.D. Core sequences and cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. 1996;24:1314–1321. doi: 10.1093/nar/24.7.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawakami J., Kumar P.K.R., Suh Y.A., Nishikawa F., Kawakami K., Taira K., Ohtsuka E., Nishikawa S. Identification of important bases in a single-stranded region (SSrC) of the hepatitis delta (δ) virus ribozyme. Eur. J. Biochem. 1993;217:29–36. doi: 10.1111/j.1432-1033.1993.tb18214.x. [DOI] [PubMed] [Google Scholar]
- 23.Nishikawa F., Kawakami J., Chiba A., Shirai M., Kumar P.K.R., Nishikawa S. Selection in vitro of a trans-acting genomic human hepatitis delta virus (HDV) ribozymes. Eur. J. Biochem. 1996;237:712–718. doi: 10.1111/j.1432-1033.1996.0712p.x. [DOI] [PubMed] [Google Scholar]
- 24.Ciesiolka J., Illangasekare M., Majerfeld I., Nickles T., Welch M., Yarus M., Zinnen S. Affinity selection-amplification from randomized ribooligonucleotide pools. Meth. Enzymol. 1996;267:315–335. doi: 10.1016/s0076-6879(96)67021-9. [DOI] [PubMed] [Google Scholar]
- 25.Wrzesinski J., Łęgiewicz M., Smólska B., Ciesiołka J. Catalytic cleavage of cis-and trans-acting antigenomic delta ribozymes in the presence of various divalent metal ions. Nucleic Acids Res. 2001;29:4482–4492. doi: 10.1093/nar/29.21.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank D.N., Pace N.R. In vitro selection for altered divalent metal specificity in the RNase P RNA. Proc. Natl Acad. Sci. USA. 1997;94:14355–14360. doi: 10.1073/pnas.94.26.14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wichłacz A., Łęgiewicz M., Ciesiołka J. Generating in vitro transcripts with homogenous 3′ ends using trans-acting antigenomic delta ribozyme. Nucleic Acids Res. 2004;32:e39. doi: 10.1093/nar/gnh037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciesiołka J., Michałowski D., Wrzesinski J., Krajewski J., Krzyżosiak W.J. Patterns of cleavages induced by lead ions in defined RNA secondary structure motifs. J. Mol. Biol. 1998;275:211–220. doi: 10.1006/jmbi.1997.1462. [DOI] [PubMed] [Google Scholar]
- 29.Kirsebom L.A., Ciesiołka J. Pb+2-induced cleavage of RNA. In: Hartmann R.K.J., Bindereif A., Schön A., Westhof E., editors. Handbook of RNA Biochemistry. I. Weinheim: WILEY-VCH Verlag GmbH&Co. KGaA; 2005. pp. 214–228. [Google Scholar]
- 30.Joyce G.F. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 31.Burkard M.E., Turner D.H., Tinoco I. Appendix 1: structures of base pairs involving at least two hydrogen bonds. In: Gesteland R.F., Cech T.R., Atkins J.F., editors. The RNA World, Second Edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 675–680. [Google Scholar]
- 32.Deschenes P., Quellet J., Perreault J., Perreault J.P. Formation of the P1.1 pseudoknot is critical for both the cleavage activity and substrate specificity of the antigenomic trans-acting hepatitis delta ribozyme. Nucleic Acids Res. 2003;31:2087–2096. doi: 10.1093/nar/gkg307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Górnicki P., Baudin F., Romby P., Wiewiórowski M., Krzyżosiak W.J., Ebel J.P., Ehresmann C., Ehresmann B. Use of lead(II) to probe the structure of large RNAs. Conformation of the 3′ terminal domain of E.coli 16S rRNA and its involvement in building the tRNA binding sites. J. Biomol. Struct. Dyn. 1989;6:971–984. doi: 10.1080/07391102.1989.10506525. [DOI] [PubMed] [Google Scholar]
- 34.Ciesiołka J., Lorenz S., Erdmann V.A. Structural analysis of three prokaryotic 5S rRNA species and selected 5S rRNA–ribosomal protein complexes by means of Pb(II)-induced hydrolysis. Eur. J. Biochem. 1992;204:575–581. doi: 10.1111/j.1432-1033.1992.tb16670.x. [DOI] [PubMed] [Google Scholar]
- 35.Ciesiołka J., Lorenz S., Erdmann V.A. Different conformational forms of Escherichia coli and rat liver 5S rRNA revealed by Pb(II)-induced hydrolysis. Eur. J. Biochem. 1992;204:583–589. doi: 10.1111/j.1432-1033.1992.tb16671.x. [DOI] [PubMed] [Google Scholar]
- 36.Ciesiołka J. Metal ion-induced cleavages in probing of RNA structure. In: Barciszewski J., Clark B.F.C., editors. RNA Biochemistry and Biotechnology. Vol. 70. Dordrecht: Kluwer Academic Publishers; 1999. pp. 111–121. NATO Science Series, Sub-Series 3: High Technology. [Google Scholar]
- 37.Ananvoranich S., Perreault J.P. The kinetics and magnesium requirements for the folding of antigenomic delta ribozymes. Biochem. Biophys. Res. Commun. 2000;270:600–607. doi: 10.1006/bbrc.2000.2465. [DOI] [PubMed] [Google Scholar]
- 38.Matysiak M., Wrzesinski J., Ciesiołka J. Sequential folding of the genomic ribozyme of the hepatitis delta virus: structural analysis of RNA transcription intermediates. J. Mol. Biol. 1999;291:283–294. doi: 10.1006/jmbi.1999.2955. [DOI] [PubMed] [Google Scholar]
- 39.Perrotta A.T., Been M.D. The self-cleaving domain from the genomic RNA of hepatitis delta virus: sequence requirements and the effects of denaturant. Nucleic Acids Res. 1990;18:6821–6827. doi: 10.1093/nar/18.23.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perrotta A.T., Been M.D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- 41.Nehdi A., Perreault J.P. Unbiased in vitro selection reveals the unique character of the self-cleaving antigenomic HDV RNA sequence. Nucleic Acids Res. 2006;34:584–592. doi: 10.1093/nar/gkj463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.