Abstract
Proteins of the Rab and SNARE families target vesicles to their intracellular destinations. A comparison of these families from the budding yeast, fission yeast, nematode and fruitfly genomes has implications for the organization of membrane traffic in different organisms.
Much has been said and written about the impact of the availability of complete genome sequences on biology. Rather less emphasis has been placed on the potential use of comparing genomes, particularly eukaryotic ones - not only to illuminate the evolutionary relations between organisms, but also to understand the organization of basic biological processes. Genomic information alone can allow the formulation, and even testing, of quite specific hypotheses. When the genomes are from organisms that are readily amenable to experiments, the potential to test such hypotheses is obviously much greater. At the time of writing, the protein-coding parts of four genomes of experimentally amenable eukaryotes are substantially available: the budding yeast Saccharomyces cerevisiae, the nematode Caenorhabditis elegans, the fruitfly Drosophila melanogaster and, with rather less fanfare thus far, the fission yeast Schizosaccharomyces pombe [1]. Here, I offer a preliminary glimpse at how these four genomes can influence our view of the cellular processes of membrane trafficking.
Eukaryotic cells comprise a collection of discrete membrane-enclosed organelles with different functions, and hence distinct complements of proteins. Given that nearly all of these proteins are made by the same translation apparatus, the cell requires mechanisms to send different proteins to, and between, different organelles. Movement between organelles occurs by means of vesicles: patches of membrane that pinch off one organelle, taking a selected group of its proteins, and fuse with another. To preserve the identity of the organelles, each vesicle must know its destination. The members of two different protein families, the Rabs and the SNAREs, have been implicated in targeting different vesicles to distinct organelles. How large are these families in different eukaryotes? And can the differences between the complements of Rabs and SNAREs be correlated to differences in intracellular organization, and sophistication, between the four organisms whose genomes are available?
The Rab proteins
The Rabs are a group of GTP-binding proteins that attach reversibly to the cytoplasmic side of different vesicular and organellar membranes [2,3]. Because this is the side where targeting and fusion occur, the Rabs are ideally placed to control these processes. In general, the different Rabs function in different trafficking steps, and individual Rabs are widely supposed to regulate the fusion of distinct vesicle types. So how many Rab genes are there?
The budding yeast S. cerevisiae has eleven Rabs [4], but three of these are apparently redundant copies. Two have no homologs in other organisms, and their disruption has no obvious effect on the yeast cell [4]. Comparing the four available genomes (Table 1) reinforces the view that a core of only six Rabs is conserved among eukaryotes. On the face of it, this is quite a surprise. In the simplest cell, membrane traffic out of the cell occurs from the endoplasmic reticulum, through several stages within the Golgi complex, to the cell surface. Endocytosis proceeds from the surface, through intermediates, to the lysosomes; new lysosomal proteins are diverted from the Golgi to the lysosomes; and various recycling steps allow essential machinery to return to its original place (Figure 1). It is a struggle to add these together and make only six distinct steps for regulation by Rabs. So genomic comparison alone indicates what much recent work has tended to show: that membrane traffic cannot be neatly boxed up, with each targeting protein acting at only one stage.
Table 1.
Homologous Rab proteins in four eukaryotic genomes
| S. cerevisiae [4] | S. pombe | C. elegans | D. melanogaster |
| Ypt1p | Ypt1 (S10025) | Rab1 (AAC692) | AAF55873 |
| Sec4p | Ypt2 (S12790) | Rab8 (AAC78494) | AAF49101, AAF56345 |
| Ypt31p, Ypt32p | Ypt3 (S10026) | Rab11 (AAB54158, CAB07678) | AAF55850 |
| Vps21p, Ypt52p, Ypt53p | Ypt5 (CAB11737) | Rab5 (CAB04205) | AAF51265, AAF47018 |
| Ypt6p | Ryh1 (CAA36715) | Rab6 (P34213, AAC69020) | AAF53168 |
| Ypt7p | Ypt7 (CAB38603) | Rab7 (CAA91357) | AAF56218 |
| Ypt10p | |||
| Ypt11p | |||
| Ypt4 (CAB11239) | |||
| Rab2 (AAB52431, CAB07357) | AAF57381 | ||
| Rab3 (AAB16980) | AAF58762 | ||
| Rab9 (AAF53798) | |||
| Rab10 (AAC48203) | AAF50924 | ||
| Rab4 (AAF57831) | |||
| Rab14 (CAB01884) | AAF53390 | ||
| Rab18 (AAF60884) | AAF46057 | ||
| Rab19 (CAB60605) | AAF50452 | ||
| Rab21 (CAA91296) | AAF45341 | ||
| Rab23 (AAF51970) | |||
| Rab26 (AAB52888) | AAF51708 | ||
| Rab27 (CAB54484) | AAF45634, AAF45635 | ||
| Rab30 (CAA21489) | AAF52477 | ||
| Rab32 (AAF58970) | |||
| Rab 33 (Q20365) | |||
| Rab35 (AAF45371) | |||
| RabX (CAA87774) | AAF46271 |
C. elegans and D. melanogaster proteins are named according to their close mammalian homologs. (RabX has no clear mammalian homolog at present.) Numbers shown in parentheses after gene names are GenBank accession numbers.
Figure 1.
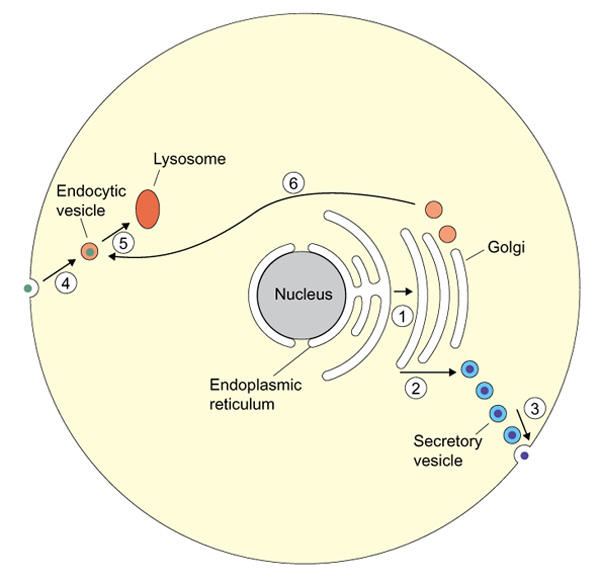
Even in a simplified cell, outward membrane traffic occurs from the endoplasmic reticulum (1), through several stages of the Golgi complex (2), to the cell surface (3). Inward traffic - endocytosis - proceeds from the surface (4) to the lysosomes (5), and lysosomal proteins are delivered from the Golgi (6).
What of the additional metazoan Rabs? Rab4 and Rab9, both of which are found in Drosophila, appear to function in endocytic recycling and endosome-to-Golgi traffic respectively [5,6]. Not only do the yeast genomes fail to accommodate these apparently useful functions: the nematode also lacks them. Thus, a comparison of these stages in the different organisms might prove illuminating. In some tissues, larger organisms require specialized vesicles into which small molecules (such as neurotransmitters) are packaged, for release in response to an external signal; Rab3 and its homologs are implicated in this: so it is perhaps reasonable that no Rab3 has been found in either yeast. Metazoans also have polarized epithelia, in which different parts of the plasma membrane acquire different protein compositions by selective vesicle targeting. Indeed, metazoans might appear to carry out a large collection of additional trafficking processes in particular cell types. If so, they manage with only a relatively modest increase in the number of Rabs when compared to unicellular eukaryotes (Table 1). The same theme emerges from comparing the total sizes of the yeast, fly and worm genomes - additional complexity seems to have been acquired without as large an increase in gene number as might have been predicted [7].
The SNARE proteins
Like Rabs, the SNARE group of proteins are also anchored to the cytoplasmic face of vesicle and organelle membranes. They have the propensity to form α-helical coiled-coil complexes with each other; when this happens between SNAREs on a vesicle and a target organelle, the two membranes are brought in close apposition, and their subsequent fusion is likely or inevitable [8]. Again, different SNAREs tend to function at different places in the cell. As a family, however, their degree of sequence conservation is not always sufficient to make them immediately identifiable, or distinguishable from other coiled-coil proteins. Nevertheless a preliminary estimate has been made of the number of vesicle SNAREs (v-SNAREs, or synaptobrevins) and target SNAREs (t-SNAREs, or syntaxins) in the fly [7]: 20 in total, which is barely more than are found in budding yeast.
Analysis of the SNAREs in budding and fission yeasts, however, starts to illustrate the potential of genomics for comparative biology (Table 2). The t-SNAREs of the secretory pathway are quite well conserved, but those implicated in traffic to the lysosomes (known as vacuoles in yeasts) are less so. In particular, homologs of Vam3p and Nyv1p, a pair of SNAREs involved in fusion between vacuoles in S. cerevisiae [9], are not yet obvious in S. pombe. One morphological difference between the yeasts is that S. cerevisiae has one or a few large vacuoles; it grows by budding, and hence the bud must acquire fragments of any organelle which is inherited. A function originally proposed for vacuole-vacuole fusion is to stick the vacuole fragments back together in the bud [10]. S. pombe, in contrast, has many small vacuoles [11], and grows at either end of its rodshaped cell. Hence it has no obvious need for a mechanism to fragment and reassemble its vacuoles during cell division. So, the cell biology of the two yeasts and their respective gene complements may make sense when considered together.
Table 2.
TSNAREs in S. cerevisiae and S. pombe
Rabs to SNAREs
Rab proteins act 'upstream' of SNAREs, apparently controlling their ability to form complexes between membranes. But they do not do this directly: recent evidence has led to the surprising conclusion that the protein complexes mediating these interactions are quite different for each trafficking step [3,8]. One example involves the Sec4 protein of S. cerevisiae, which functions in the delivery of vesicles to the cell surface. Its downstream 'effector' is a protein complex called the 'exocyst' [12], which in turn attaches to the Sec3 protein on the membrane. All of the exocyst components have homologs in other genomes; but Sec3 apparently does not. But Sec3 does more than allow the vesicle to find the membrane: it is pivotal in defining the sites on the membrane where vesicles are delivered [13]. For most of the cell cycle, vesicles are delivered to the growing bud. This process is quite different from exocytosis in fission yeast and metazoans; hence, it seems reasonable that these organisms should have a different (and as yet unidentified) target for the exocyst. Fundamentally, the delivery of vesicles to the plasma membrane is what allows cells to grow, so the analog of Sec3 will be an interesting protein to find.
Do any general lessons emerge from this primitive exercise in intergenomic cell biology? One key idea is the notion of 'core' proteins, meaning anything conserved between S. cerevisiae and metazoans and thus likely to be needed for a basic ancestral function [7]. S. pombe is widely diverged from S. cerevisiae. Its admirers argue that it has evolved less rapidly than S. cerevisiae; hence, it is believed to be closer to the common ancestor of yeasts and metazoans, and potentially a better 'model' organism [14]. By evolving rapidly, S. cerevisiae may have developed its own ways of accomplishing 'core' functions, such as the role of Sec3 in membrane growth. So, four eukaryotic genomes may offer much more insight than three, in membrane traffic and in every other process undertaken by all of these organisms. Clearly, genomic approaches will have a large part to play in understanding each of these processes, and how they operate in another well-known eukaryote whose complete genome is due soon to emerge, particularly as it is so much harder to do genetic manipulation on ourselves.
References
- The Schizosaccharomyces pombe Genome Sequencing Project. http://www.sanger.ac.uk/Projects/S_pombe/
- Schimmoeller F, Simon I, Pfeffer SR. Rab GTPases, directors of vesicle docking. . J Biol Chem. 1998;73:22161–221643. doi: 10.1074/jbc.273.35.22161. [DOI] [PubMed] [Google Scholar]
- Armstrong J. How do Rab proteins function in membrane traffic? Int J Biochem Cell Biol. 2000;32:303–307. doi: 10.1016/s1357-2725(99)00112-0. [DOI] [PubMed] [Google Scholar]
- Lazar T, Gotte M, Gallwitz D. Vesicular transport: how many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem Sci. 1997;22:468–472. doi: 10.1016/s0968-0004(97)01150-x. [DOI] [PubMed] [Google Scholar]
- Van der Sluijs P, Hull M, Webster P, Male P, Goud B, Mellman I. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. . Cell. 1992;70:729–740. doi: 10.1016/0092-8674(92)90307-x. [DOI] [PubMed] [Google Scholar]
- Lombardi D, Soldati T, Riederer MA, Goda Y, Zerial M, Pfeffer SR. Rab9 functions in transport between late endosomes and the trans Golgi network. . EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Miklos GLG, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W. Comparative genomics of the eukaryotes. . Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR. Transport-vesicle targeting: tethers before SNAREs. . Nat Cell Biol. 1999;1:E17–E22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- Nichols BJ, Ungermann C, Pelham HR, Wickner WT, Haas A. Homotypic vacuolar fusion mediated by t- and v-SNAREs. . Nature. 1997;387:199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- Weisman LS, Bacallao R, Wickner W. Multiple methods of visualizing the yeast vacuole permit evaluation of its morphology and inheritance during the cell cycle. . J Cell Biol. 1987;105:1539–1547. doi: 10.1083/jcb.105.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone N, Millar JB, Toda T, Armstrong J. Regulated vacuole fusion and fission in Schizosaccharomyces pombe: an osmotic response dependent on MAP kinases. . Curr Biol. 1998;8:135–144. doi: 10.1016/s0960-9822(98)00060-8. [DOI] [PubMed] [Google Scholar]
- Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. . EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger FP, Hughes TE, Novick P. Sec3p is a spatial landmark for polarized secretion in budding yeast. . Cell. 1998;92:559–571. doi: 10.1016/s0092-8674(00)80948-4. [DOI] [PubMed] [Google Scholar]
- Sipiczki M. Taxonomy and phylogenesis. . In Molecular Biology of the Fission Yeast Edited by Nasim A, Young P, Johnson BF San Diego: Academic Press. 1989:431–452. [Google Scholar]


