Abstract
A recent study, comparing the maize SILKY1 gene to its well-characterized homolog APETALA3 from Arabidopsis, has provided some of the first evidence pointing to conservation of homeotic gene function between monocots and dicots.
The genomics era has heralded the accumulation of an unprecedented amount of sequence information from a vast array of species. With this wealth of information, the issue at hand is to determine to what extent homologous genes from different species function in a similar manner, as well as the extent to which their roles have diversified. Gene expression and loss-of-function studies are now paving the way for comparative functional studies in several model species, and may soon provide us with mechanistic explanations for how different morphologies have evolved.
The angiosperms (the flowering plants) arose about 130 million years ago and gave rise to over 250,000 extant species that contain a remarkable diversity of floral forms. Although flowers have dramatically different forms in different species, there are some basic structural similarities. Flowers contain stamens (male reproductive organs) and carpels (female reproductive organs) surrounded by sterile perianth organs. In many species, the perianth is composed of distinct petals and sepals, while in other species the petals and sepals are indistinguishable and are referred to as tepals.
How did the vast array of different floral morphologies arise? Recent evidence is converging to support the idea that the primitive angiosperm flower consisted of reproductive organs, with few or no perianth organs. Several recent phylogenetic analyses have independently provided support for placing Amborella, with its diminutive flowers containing reproductive organs and just a few tepals, at the base of the angiosperm tree [1,2,3,4]. This evidence suggests that the primitive angiosperm flower was small and few-parted, in contrast to the more traditional view that the earliest angiosperms had large, multiparted flowers similar to present day magnolias [5,6]. In addition, the fossil evidence, although fragmentary, also supports the idea that the primitive angiosperm flower lacked perianth organs [7,8,9]. If the earliest angiosperm flower indeed consisted of just stamens and carpels, then perianth organs must have arisen during the course of angiosperm evolution. Within the angiosperms, two monophyletic groups have been identified, the monocots and the eudicots, and these are contained within a basal assemblage of magnoliid dicots (Figure 1). Petals are thought to have arisen multiple times in different angiosperm lineages and, in particular, monocot and core eudicot petals are thought to have arisen independently [10]. This would imply that all petals are not homologous organs and has important implications for comparing the roles of the floral homeotic genes in different species.
Figure 1.
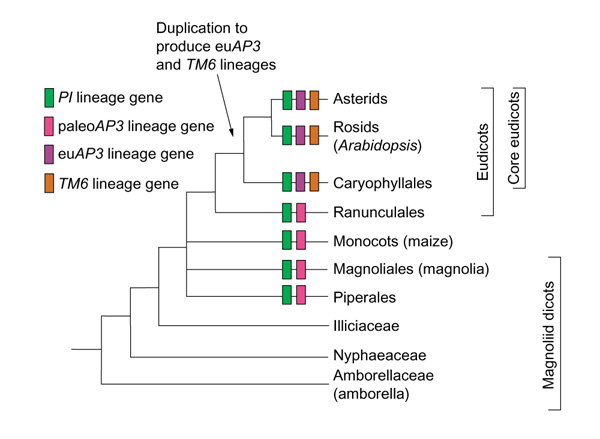
Simplified tree of the angiosperms, based on [2]. Common names of representatives of selected lineages in parentheses. A duplication event in the AP3 lineage gave rise to the euAP3 and TM6 lineages in core eudicots [22]. Clades in which one or more examples of a particular gene lineage have been found are marked with a colored box.
Extensive experimental studies on the roles of the floral homeotic genes in Arabidopsis and other core eudicot species have led to the formulation of the ABC model of floral development [11,12]. This model posits that three classes of floral homeotic genes, termed A, B and C, function in overlapping domains to give rise to the different floral organs: the sepals, petals, stamens, and carpels (Figure 2). In Arabidopsis, the B-group genes, APETALA3 (AP3) and PISTILLATA (PI), act together to specify petal and stamen identities. These two genes encode MADS-box-containing DNA-binding proteins and presumably act by regulating the transcription of downstream genes responsible for petal and stamen morphogenesis and cell-type specific differentiation [13,14].
Figure 2.
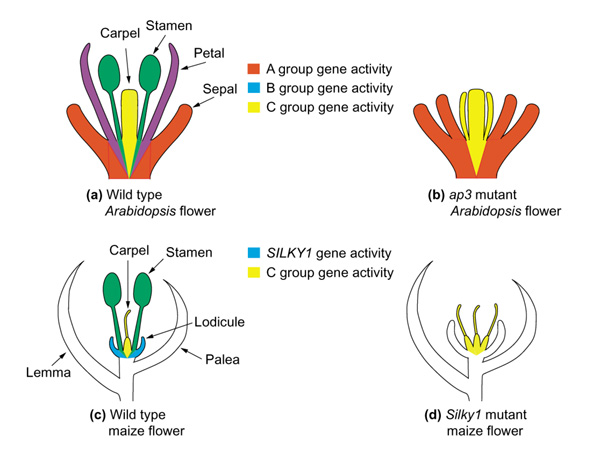
(a)Arabidopsis, like other core eudicots, has flowers that contain four whorls of floral organs: sepals, petals, stamens and carpels. A combination of A, B and C group floral homeotic gene activities results in specification of different organ identities [11]. Petals result from a combination of A+B activities, and stamens result from a combination of B+C activities. (b) Mutation of the B group gene AP3 results in a loss of B group gene activity, resulting in a transformation of petals to sepals and of stamens to carpels. (c) The maize flower is composed of a lemma, a palea, lodicules and the reproductive organs. During maize flower differentiation, abortion of the carpels results in functionally male flowers, while abortion of the stamens results in functionally female flowers. (d) Mutations in the SILKY1 gene result in a transformation of stamens to carpels and the replacement of lodicules with structures that resemble paleas or lemmas [23].
Cloning and characterization of B group genes from a wide array of other species has indicated that the AP3 and PI gene lineages arose prior to the diversification of the angiosperms, suggesting that ancestral AP3- and PI-like genes were present before flowers evolved [15,16,17,18]. A few gymnosperm AP3- and PI-like genes have been identified and shown to be expressed in male reproductive organs [15,17,19,20]. Since gymnosperms do not have perianths, this supports the contention that the ancestral role of the B-group genes was in specifying male reproductive organ development.
When the angiosperms arose and diversified, how did the role(s) of the B-group genes change? How, and in which lineages, did the B-group genes acquire an additional role in specifying petal development? One way in which B-group genes may have acquired new functions is through gene duplication. An ancient duplication event occurred in the AP3 lineage at the base of the core eudicots (Figure 1) and may be associated with the independent origin of petals in this group [21]. The evolution of the 'euAP3' lineage with new sequence characteristics may reflect the acquisition of new functions that include the specification of core eudicot petal identity. The monocots, on the other hand, contain AP3-like genes that are more similar in sequence to the ancestral 'paleoAP3' lineage genes [22].
Characterization of the maize Silky1 gene, a member of the paleoAP3 lineage, has begun to shed light on the similarities and differences in the roles of the eudicot euAP3 lineage genes and the monocot paleoAP3 lineage genes. Maize, like other grasses, has flowers which are highly derived and contain stamens and carpels surrounded by sterile organs known as paleas, lemmas and lodicules. Silky1 is expressed in lodicules and stamens [23]. Mutations in Silky1 result in a transformation of the stamens to carpel-like structures complete with the characteristic long silk, but also result in lodicules being replaced by organs with characteristics of lemmas and paleas [23]. In comparison, mutations in the Arabidopsis AP3 gene result in homeotic transformations of stamens into carpel-like structures and petals into sepal-like structures (Figure 2) [24].
How do the roles of the Arabidopsis AP3 gene and the maize Silky1 gene compare? Since all stamens are homologous (that is to say, they have a common evolutionary origin), it is perhaps not surprising that these Arabidopsis 'euAP3' and the maize 'paleoAP3' lineage genes are both required for stamen identity. It is unclear though if AP3 and Silky1 have similar roles in perianth development. Petals are thought to have evolved independently in the core eudicots and in the monocots; and furthermore, in the grasses, it is unclear as to whether lodicules are perianth organs or represent modified sterile stamens [25,26,27]. The fact that Silky1 mutations cause a transformation of the lodicule has been used to support the idea that lodicules are homologous to petals, but this does not take into account the fact that homology implies common descent [23,28]. An alternative possibility is that the lodicule represents another organ type that has no counterpart in the eudicot flower [25].
The roles of the Arabidopsis AP3 gene in petal development and the maize Silky1 gene in lodicule development could result from parallel evolution [29]. In other words, similar developmental modifications may have occurred independently in the eudicots and in the monocots. In the case of the eudicots, the euAP3 gene lineage appears to have been recruited to specifying petal identity in addition to stamen identity. In the monocots, a similar scenario may have taken place independently, such that the paleoAP3 lineage genes may have been recruited to a new role in lodicule development.
The question of how AP3-like genes are involved in specifying non-reproductive structures may have as many solutions as there have been independent origins of perianth parts. Critically comparing the roles of homologous homeotic genes in different species will require an understanding of their evolutionary relationships, as well as genetic tests of function. The tools needed are already in place: sequence information is providing the basis for developing robust angiosperm phylogenies, and the potential exists to genetically manipulate a wide array of plant species using Agrobacterium-mediated transformation. By analyzing the roles of the homeotic genes in a wide range of angiosperm species, we should soon be able to understand how the evolution of developmental mechanisms is causally linked to changes in floral morphology.
References
- Qiu YL, Lee J, Bernasconi-Quadroni F, Soltis DE, Soltis PS, Zanis M, Zimmer EA, Chen Z, Savolainen V, Chase MW. The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes. Nature. 1999;402:404–407. doi: 10.1038/46536. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE, Chase MW. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature. 1999;402:402–404. doi: 10.1016/S0168-9002(97)00880-2. [DOI] [PubMed] [Google Scholar]
- Mathews S, Donoghue MJ. The root of angiosperm phylogeny inferred from duplicate phytochrome genes. Science. 1999;286:947–950. doi: 10.1126/science.286.5441.947. [DOI] [PubMed] [Google Scholar]
- Parkinson CL, Adams KL, Palmer JD. Multigene analyses identify three earliest lineages of extant flowering plants. Curr Biol. 1999;9:1485–1488. doi: 10.1016/s0960-9822(00)80119-0. [DOI] [PubMed] [Google Scholar]
- Endress PK. Floral structure and evolution of primitive angiosperms: recent advances. PI Syst Evol. 1994;192:79–97. [Google Scholar]
- Crane PR, Friis EM, Pedersen KR. The origin and early diversification of angiosperms. Nature. 1995;374:27–33. [Google Scholar]
- Taylor DW, Hickey LJ. An Aptian plant with attached leaves and flowers: implications for angiosperm origin. Science. 1990;247:702–704. doi: 10.1126/science.247.4943.702. [DOI] [PubMed] [Google Scholar]
- Hickey LJ, Taylor DW. Origin of the angiosperm flower. In Flowering Plant Origin, Evolution and Phylogeny Edited by Taylor DW, Hickey LJ New York: Chapman and Hall, 1995. pp. 176–231.
- Sun G, Dilcher DL, Zheng S, Zhou Z. In search of the first flower: a Jurassic angiosperm, Archefructus, from Northeast China. Science. 1998;282:1692–1695. doi: 10.1126/science.282.5394.1692. [DOI] [PubMed] [Google Scholar]
- Takhtajan A. Evolutionary trends in flowering plants. New York: Columbia University Press. 1991.
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. . Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Sundstrom J, Carlsbecker A, Svenson M, Svensson ME, Engstrom P. MADS box genes active in developing pollen cones of Norway Spruce are homologous to the B class floral homeotic genes in angiosperms. Dev Genet. 1999;25:253–266. doi: 10.1002/(SICI)1520-6408(1999)25:3<253::AID-DVG8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Purugganan MD. The MADS-box floral homeotic gene lineages predate the origin of seed plants: phylogenetic and molecular clock estimates. J Mol Evol. 1997;45:392–396. doi: 10.1007/pl00006244. [DOI] [PubMed] [Google Scholar]
- Winter K-U, Becker A, Munster T, Kirn JT, Saedler H, Theissen G. MADS box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proc Nat Acad Sci USA. 1999;96:7342–7347. doi: 10.1073/pnas.96.13.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Irish VF. Evolution of petal and stamen developmental programs: evidence from comparative studies of the lower eudicots and basal angiosperms. Intl J Plant Sci. 2000.
- Irish VF, Kramer EM. Genetic and molecular analysis of angiosperm flower development. Adv Bot Res. 1998;28:197–230. [Google Scholar]
- Mouradov A, Hamdorf B, Teasdale RD, Kim J, Winter K-U, Theissen G. A DEF/GLO like MADS box gene from a gymnosperm: Pinus radiata contains an ortholog of angiosperm B class floral homeotic genes. Dev Genet. 1999;25:245–252. doi: 10.1002/(SICI)1520-6408(1999)25:3<245::AID-DVG7>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Irish VF. Evolution of genetic mechanisms controlling petal development. Nature. 1999;399:144–148. doi: 10.1038/20172. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. Molecular evolution of petal and stamen development: gene duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics. 1998;149:765–783. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ. Molecular and genetic analyses of the SilkyI gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell. 2000;5:569–579. doi: 10.1016/s1097-2765(00)80450-5. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren RMT, Clifford HT, Yeo PF. The families of the Monocotyledons. New York: Springer Verlag, 1985.
- Cocucci AE, Anton AM. The grass flower: suggestions on its origin and evolution. Flora. 1988;181:353–362. [Google Scholar]
- Clifford HT. Spikelet and floral morphology. In Grass Systematics and Evolution Edited by Soderstrom TR Washington DC: Smithsonian Institution Press, 1988;433:21–30. [Google Scholar]
- Ma H, dePamphilis C. The ABCs of floral evolution. Cell. 2000;101:5–8. doi: 10.1016/S0092-8674(00)80618-2. [DOI] [PubMed] [Google Scholar]
- Meyer A. Homology and homoplasy: the retention of genetic programmes. In Homology Edited by Bock GR, Cardew G West Sussex, England: John Wiley and Sons Ltd, 1999. pp. 141–157. [DOI] [PubMed]


