Short abstract
The sole function of cellulose synthases, which are found in plants bacteria, fungi, and animals, is to produce the biopolymer cellulose. Although no crystal structure has yet been solved, a considerable amount is known about their structure, function and evolution.
Abstract
Cellulose, an aggregate of unbranched polymers of β-1,4-linked glucose residues, is the major component of wood and thus paper, and is synthesized by plants, most algae, some bacteria and fungi, and even some animals. The genes that synthesize cellulose in higher plants differ greatly from the well-characterized genes found in Acetobacter and Agrobacterium sp. More correctly designated as 'cellulose synthase catalytic subunits', plant cellulose synthase (CesA) proteins are integral membrane proteins, approximately 1,000 amino acids in length. The sequences for more than 20 full-length CesA genes are available, and they show high similarity to one another across the entire length of the encoded protein, except for two small regions of variability. There are a number of highly conserved residues, including several motifs shown to be necessary for processive glycosyltransferase activity. No crystal structure is known for cellulose synthase proteins, and the exact enzymatic mechanism is unknown. There are a number of mutations in cellulose synthase genes in the model organism Arabidopsis thaliana. Some of these mutants show altered morphology due to the lack of a properly developed primary or secondary cell wall. Others show resistance to well-characterized cellulose biosynthesis inhibitors.
Gene organization and evolutionary history
Gross gene structure
A number of cellulose synthase (CesA) genes have been cloned from a variety of plant species, the first in 1996 [1]. The most information is known about the Arabidopsis thaliana CesA gene family, as the Arabidopsis genome sequence is nearly finished. CesA genes range in size from 3.5 to 5.5 kb, with 9-13 small introns (Figure 1). They produce transcripts ranging in size from 3.0 to 3.5 kb, encoding proteins 985 to 1,088 amino acids in length. The intron-exon boundaries are highly conserved, with differences in gene structure primarily due to the loss of introns.
Figure 1.
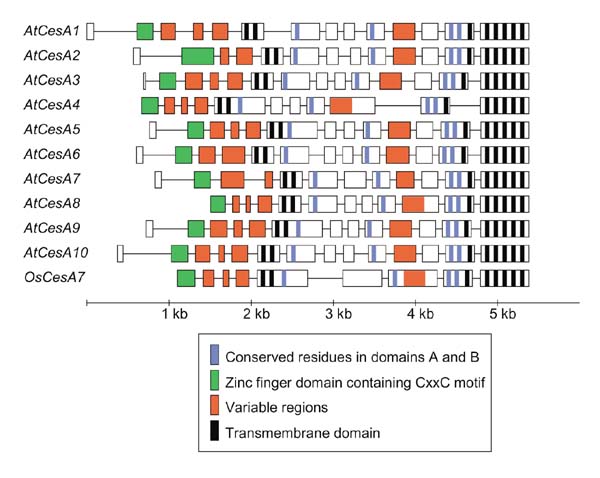
Gene structure of the Arabidopsis CesA gene family and the rice CesA7 gene, the only CesA genes for which full genomic sequence is available. At, Arabidopsis thaliana; Os, Oryza sativa. Exons are represented by boxes and introns by connecting lines. Exons or portions of exons encoding the domains shown in Figure 3 are colored as indicated.
Chromosomal location and organization
In Arabidopsis thaliana, there are at least ten cellulose synthase genes. These are scattered throughout the genome, with no apparent recent duplication events. Unlike bacterial cellulose synthase genes, there are no functionally linked genes in close proximity to one another. Sequence data indicate that the CesA gene family is as large, or larger, in other plant species.
Evolutionary history
Plant cellulose synthases belong to family 2 of processive glycosyltransferases [2,3], a large family of enzymes with members from viruses, bacteria, fungi, and all other eukary-otes. The proteins in this family are inverting processive glycosyltransferases that make β linkages. Cellulose synthases synthesize β-1,4-glucans, homogeneous strands of glucose residues. In addition to higher plants, cellulose is synthesized by a number of bacterial species (i.e. Acetobacter, Agrobacterium, and Rhizobium), algae and lower eukaryotes (i.e. tunicates). While the end product is the same, there is little similarity at the amino-acid level between these genes and CesA genes from higher plants.
In Arabidopsis thaliana, there are a total of six families of genes, designated 'cellulose synthase-like' (Csl), that appear to be related to the CesA family, on the basis of sequence similarity, conserved protein domains, and overall gene structure [4,5] (Figure 2). The function of these families is not yet known; it is possible that one or more of these families is also part of the cellulose synthesis pathway.
Figure 2.
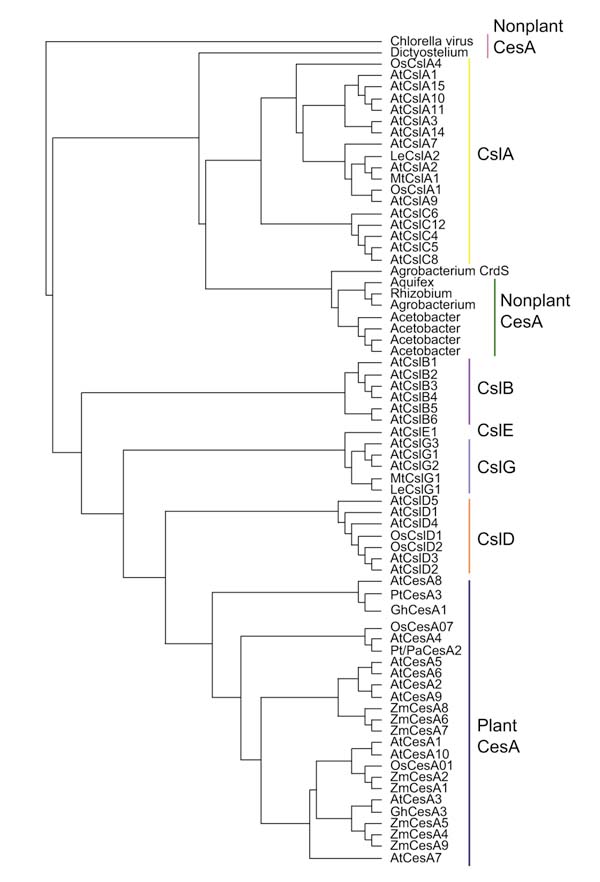
A cladogram of the plant CesA superfamily and related non-plant proteins. ClustalX (version 1.8) was used to create an alignment of the protein sequences that was then bootstrapped (n = 1000 trials) to create the final tree. Subfamilies are indicated with colored bars on the right. At, Arabidopsis thaliana (thale cress); Gh, Gossypium hirsutum (cotton); Le, Lycopersicon esculentum (tomato); Mt, Medicago truncatula (barrel medic); Os, Oryza sativa (rice); Pt, Populus tremuloides (quaking aspen); Pt/Pa, Populus alba x Populus tremula (gray poplar); Zm, Zea mays (maize).
Characteristic structural features
Overall structural organization
All cellulose synthases described to date have a number of conserved structural features. It is thought that CesA acts as a member of a protein complex that can be visualized by electron microscopy on the surface of the plasma membrane in structures called 'rosettes'. These appear to consist of six large subunits arranged in a hexagonal pattern, each approximately 9 nm in size. At the amino terminus of the CesA protein is an amino acid domain that bears some resemblance to a zinc finger or LIM transcription factor. It is thought that this domain might play a role in protein-protein interactions in the CesA complex. Within this domain is a strictly conserved sequence motif, the 'CxxC' motif, beginning 10-40 amino acids from the amino terminus: Cx2Cx12FxACx2Cx2PxCx2Cx-Ex5GX3Cx2C in the single-letter amino-acid code, where x is any amino acid.
Also within the amino terminus of the protein is a region of about 150 amino acids originally designated as a 'hypervariable' region. It is clear, however, as additional full-length protein sequences have become available, that this region is more conserved than was previously thought. This region is rich in acidic amino acids. The contribution of this region to the overall function of the enzyme is unknown.
Following the ammo-terminal domains are two predicted transmembrane domains, near positions 270 and 300 in the Arabidopsis CesA proteins (Figure 3). The carboxy-terminal portion of the protein, extending from approximately amino acid position 850, contains six additional predicted transmembrane domains. The region between the sets of transmembrane domains is often designated as the globular domain or the soluble domain. Consisting of around 550 amino acids, it is thought to form a loop that extends into the cytoplasm. Within this domain are several characteristic conserved regions. There is a second variable region, approximately 50 residues in length, beginning near position 650. Also within the globular domain are the motifs indicative of processive glycosyltransferases. The first motif (Domain A) consists of several widely spaced aspartic acid residues; a single D followed by a DxD (see Figures 2,3). These residues are thought to bind the UDP-glucose substrate, and are found in both processive and non-processive enzymes. Processive enzymes catalyze the addition of many sugar residues to a growing chain. Non-processive enzymes catalyze the addition of only a single sugar residue to an acceptor molecule. The second motif (Domain B) is found only in processive enzymes. It consists of a third conserved aspartic acid residue and three conserved amino acids, QxxRW, which are thought to be part of the catalytic site. There are many conserved residues found around these motifs in the plant cellulose synthase proteins.
Figure 3.
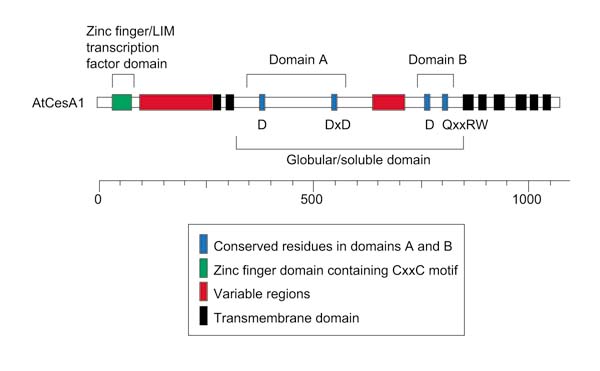
Protein features characteristic of plant cellulose synthase proteins, shown using the Arabidopsis CesA1 protein as a paradigm. Regions indicated above and below are described within the text and domains are colored as indicated.
The various members of the plant CesA family range in size from 985 to 1,088 amino acids and can vary in sequence identity from 53% to 98%. Care must be taken to avoid confusing the cellulose synthase genes with members of the cellulose synthase-like families, especially the CslD family. The most distinguishing feature is the first 250 amino acids before the first predicted transmembrane domains; only the CesA proteins contain the CxxC motif.
Localization and function
Cellulose synthase has been localized to the plasma membrane by immunocytochemistry. As cellulose is a major component of all higher plant cell walls, CesA proteins are expressed in all tissues and cell types of the plant. Studies indicate, however, that the various members of the family in each species are differentially expressed - in tissue types and in primary versus secondary cell wall formation. For example, the AtCesA1 (RSW1) protein is responsible for primary cell wall biosynthesis throughout the plant, while the AtCesA7 (IRX3) protein functions only in secondary cell wall biosynthesis in the stem.
The sole function of cellulose synthase is the production of the biopolymer cellulose, a β-1,4-glucan chain, ranging in size from 2,000 to 25,000 glucose residues. Cellulose is found as fibrils in plants, most often consisting of 36 glucan chains, although some cellulosic algae have very large microfibrils consisting of more than 1,200 glucan chains.
Enzyme mechanism
The mechanism by which cellulose synthase creates a β-1,4-glucan chain is not yet known. Although putative substrate binding sites and catalytic residues have been identified, it is not clear whether cellulose chains are synthesized by the addition of single sugars or disaccharides. The β-1,4-linkage in cellulose requires that each glucose residue be flipped nearly 180° with respect to its neighbors. To make this chain one sugar residue at a time would require either the glucan chain or the synthase to rotate 180°, or the sugar residues to be added, then rotated into the proper orientation by another factor associated with the catalytic subunit. Reorientation problems are eliminated when models invoking two sugar-binding sites are used, however. But at present there is no experimental evidence for either model. Having the structure for cellulose synthase would be likely to help answer some of the mechanistic questions, but as yet there is no crystal structure available for any cellulose synthase or closely related enzyme. One of the hypothetical three-dimensional structures for the CesA proteins has the eight transmembrane helices forming a pore in the plasma membrane, through which the growing glucan chain passes to reach the newly forming cell wall [6] (Figure 4). The amino terminus, with the putative protein-protein interaction domain, would reside in the cytoplasm, free to make contact with other proteins or factors necessary for activity.
Figure 4.
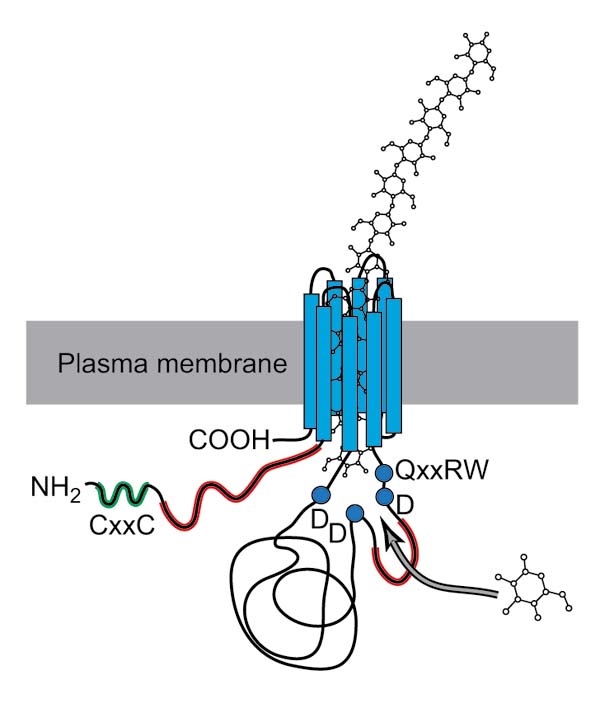
Hypothetical three-dimensional structure of CesA proteins. The transmembrane helices of the CesA protein are thought to form a pore in the plasma membrane through which the growing glucan chain passes. Regions are colored to follow those shown in Figure 2. This figure is adapted from [6].
Important mutants
There are a number of mutants currently known in plant cellulose synthase genes. The rsw1 temperature-sensitive mutation in AtCesA1, when grown at the non-permissive temperature, causes a specific reduction in cellulose synthesis, the accumulation of noncrystalline β-1,4-glucan, disassembly of cellulose synthase, and widespread morphological abnormalities [7]. The irx3 (irregular xylem 3) point mutation in AtCesA7 shows a defect in secondary cell wall formation in xylem. As a result, the tracheary elements in the irx3 mutant have weakened walls and collapse upon themselves [8,9]. The irx1 point mutation in AtCesA8 is a member of the same family of mutants as irx3 (N. Taylor and S. Turner, personal communication). Not to be confused with irx is ixr1 (isoxaben resistance). There are two mutant alleles, ixr1-1 and ixr1-2, that confer resistance to the cellulose biosynthesis inhibitor isoxaben. Both alleles are point mutations in the AtCesA3 gene (W. Scheible and C. Somerville, personal communication). Another mutation that confers resistance to isoxaben is ixr2; ixr2-1 is a point mutation in the AtCesA6 gene (H. Höfte, personal communication). Finally, procuste1 is one of a class of mutants that show decreased elongation and increased radial expansion in hypocotyls in Arabidopsis; procuste1 is mutation in the AtCesA6 gene, the same gene as ixr2 (H. Höfte, personal communication).
Frontiers
Issues most studied
There are a number of questions that are currently being addressed in the area of cellulose biosynthesis. Now that the genes for the catalytic subunit of cellulose synthase are known, researchers are interested in the mechanism of synthesis and the regulation of cell wall deposition. Projects have been initiated in several different plant species to look at expression in different tissues and developmental stages using DNA microarrays, immunocytochemistry and RT-PCR. The function of the many CesA genes is being studied using genetic tools, including point mutations, T-DNA insertion lines and transposon lines, and through the use of chemical inhibitors of cellulose biosynthesis. Various groups are attempting to determine the crystal structure of cellulose synthases, a difficult task because these are integral membrane proteins. For a general review of cellulose synthase research, see [10,11].
Major unresolved questions
Key issues that remain concern the enzyme mechanisms, including whether monosaccharides or dissacharides are the substrates for cellulose synthase. And how are substrates delivered to the enzyme? It is not clear whether cellulose biosynthesis requires a primer; if so, what is the primer? How many proteins make up the cellulose synthase complex and what are their individual roles? There is growing evidence that more than one CesA protein may be required in each cell for normal function - do the various CesA proteins interact directly, and, if so, how are they arranged in the subunits of the rosette? Do the transmembrane helices of the CesA protein then form a pore through which the growing glucan chain passes through the membrane? And why are there so many CesA genes in plants? Given that there are so many, how does each plant cell use them to control the synthesis and deposition of cellulose? And is regulation controlled at the transcriptional or the post-translational level? Clearly cellulose are amenable to much more work.
References
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA . 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. This paper describes the cloning of the first plant cellulose synthase gene. Although this paper shows a great deal of experimental evidence that these genes are the cellulose synthase catalytic subunits, the genetic proof was presented by Arioli et al. [7]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–939. doi: 10.1042/bj3260929u. A descriptive paper demonstrating the classification of glycosyltransferases into families based on amino acid sequence. The cellulose synthases belong to family 2, a family containing proteins from many different species of plants, animals, bacteria and fungi (see [3]). The tables from this paper are continually updated (see [12]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glycosyltransferase 2 family http://afmb.cnrs-mrs.fr/~pedro/CAZY/gtf_2.html
- Richmond TA, Somerville CR. The cellulose synthase superfamily. . Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. A description of the family of cellulose synthase and cellulose synthase-like genes in Arabidopsis and some thoughts on the function of these genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellwall.stanford.edu http://cellwall.stanford.edu. A sequence resource for plant cell wall biologists interested in polysaccharide biosynthesis. This site collects, organizes and summarizes sequence information for all cellulose synthase and cellulose synthase-like genes in higher plants.
- Delmer DP. Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. A useful broad review of cellulose biosynthesis. [DOI] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. . Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. This paper provided the critical proof of in vivo function for the cellulosesynthase genes. [DOI] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. Demonstration that mutants with decreased cellulose content can be isolated on the basis of a morphological phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 1999;11:769–780. doi: 10.1105/tpc.11.5.769. Cloning of a cellulose synthase gene involved exclusively in secondary cell wall formation. Wood is composed of secondary cell walls and thus the isolation of a gene with this specificity is very important. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RM, Saxena IM, Kudlicka K. Cellulose biosynthesis in higher plants. Trends Plant Sci. 1996;1:149–156. Although several years old, this review summarizes some of the history of cellulose synthase research. Many of the problems presented are still unsolved and under investigation. [Google Scholar]
- The Delmer lab: compendium of known CesA genes in plants http://www-plb.ucdavis.edu/labs/delmer/genes.html This site has information about the nomenclature for plant cellulose synthase genes and about cellulose synthase genes from other organisms.
- CAZy: Carbohydrate-Active enZYmes http://afmb.cnrs-mrs.fr/~pedro/CAZY/ A very comprehensive site that describes the families of structurally related catalytic domains of enzymes that degrade, modify or create glycosidic bonds.


