Abstract
Ever since DNA microarrays were first applied to the quantitation of RNA levels, there has been considerable interest in generating a protein homolog that can be used to assay cellular protein expression. A recent paper describes the first microarray that can be used for such protein profiling.
For the past 10 years there has been considerable interest in the creation of high-density arrays of nucleic acids for use in genomics and 'transcriptomics'. Since the first model experiments in academic laboratories [1], the number of spots on each array has steadily grown whilst the overall size of the arrays has shrunk (see [2,3,4]). Today's DNA microarray is the size of a thumbnail and can contain over 10,000 different oligonucleotides. The pages of scientific journals are peppered with advertisements touting robots for probing DNA microarrays, machines for imaging DNA microarrays, software for analyzing DNA microarrays and, of course, the DNA microarrays themselves; the estimated annual market for the technology is in excess of $527 million [5]. The application of DNA microarrays is equally widespread, ranging from DNA sequencing [6,7] to the genotyping of disease genes [8,9,10].
It has long been the goal of molecular biologists to develop a technology that can quantify, in a reliable and reproducible manner, the expression level of every individual protein in a tissue sample. DNA microarrays have been used extensively for the analysis of RNA in cellular extracts, from which the expression of individual proteins can be inferred by assessing the levels of their corresponding mRNAs [11,12,13] (see [14] for a review). Changes at the mRNA level, however, are not necessarily proportional to changes at the protein level because of differences in rates of protein translation and degradation. Furthermore, nucleotide screens are unable to provide information on the post-translational modifications of a protein, which may be critical for a protein's function. After all, it is the protein and not the mRNA that provides cellular function, whether it be for communication, metabolism or building cellular architecture.
Although two-dimensional pulse-field gel electrophoresis (2D PFGE) can be used to analyze the proteins expressed in different cellular extracts directly, it requires both a high degree of technical skill and sophisticated computational analysis to identify protein spots that are present on the gel or blot from one extract but not on the other. Even then, it is still necessary to determine the identity of the corresponding protein. The Holy Grail for such proteomic analyses is a highly sensitive protein array screen, in which the strength of the signal at each point on the array provides a readout of the expression level of each protein in the human body. The paper by Haab, Dunham and Brown in this issue of Genome Biology [15] indicates that such a screen may be close at hand.
The Brown group was among the pioneers of DNA microarrays [11,16], developing many of the robotic systems and protocols that are currently used for screening at the nucleic-acid level [17]. They have now applied the same techniques to the creation and screening of protein microarrays, consisting of either spotted antibodies or spotted protein antigens [15] (see Figure 1). As each of the antibody-antigen pairs is mutually exclusive - that is, each antibody only binds to a single cognate antigen and vice versa (a feature that makes antibodies particularly well suited to the task of protein profiling) - a single microarray comprising 115 antibodies can be used to assay the presence of different target antigens in a complex mixture. Although the concept of using antibody arrays to profile protein expression is not new, the screen described by Haab et al. [15] achieves, for the first time, the kind of sensitivity and reproducibility that would be required to detect the vast majority of proteins present in a human cell lysate. Furthermore, the protocol is centered on well-established technology that could easily be reproduced by any researcher well versed in the art of DNA arrays. Indeed, the same glass slides, robotic gridders, dye labels, slide readers and image analysis software have simply been adapted for use with proteins rather than with DNA.
Figure 1.
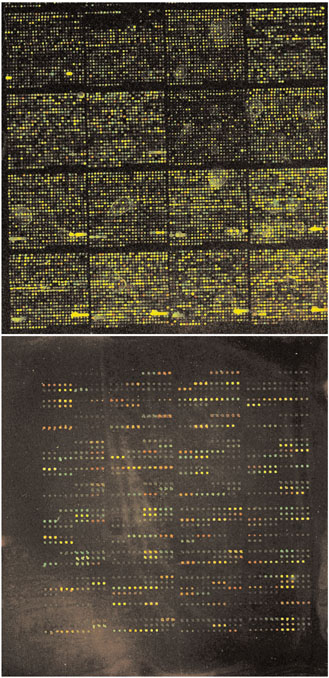
From DNA to protein. Although the appearance of the final product is very similar, DNA microarrays (top) are a world apart from protein microarrays (bottom). Images courtesy of Brian Haab.
Over the past three years there has been considerable excitement about the impending availability of protein arrays, yet remarkably few publications to justify it [18,19]. Of course, protein is quite different from DNA. Whereas the immobilization of DNA to a solid support requires a simple charge interaction, a protein must be attached in such a way as to maintain its folded (and thereby functional) conformation. Although the nucleotide sequence of one piece of DNA differs from the next, they generally have similar chemical and physical properties. Two proteins, on the other hand, may be quite dissimilar, having different sizes, charges, stabilities and solvent solubilities. The binding of one protein to another is an even more complicated affair. The strength of the interaction between a single strand of DNA and its complementary strand is mainly dictated by the length of contiguous base pairing, which on a DNA array can be controlled by laying down different oligonucleotides of the same length. Each interaction that occurs on the surface of a DNA array therefore has a similar (high) affinity. By contrast, different protein-protein interactions may have affinities ranging from femtomolar to micromolar - 12 orders of magnitude difference.
Finally, a truly high-throughput array requires a wide range of binding molecules, each of which has a unique specificity for a given target present in the cell lysate. For DNA arrays, oligonucleotides can be synthesized that correspond to specific nucleotide sequences known to be present at the RNA level. For each protein in the lysate, however, a cognate binder must be first isolated and then tested to confirm its contextual specificity. Thus, if a human cell lysate is to be probed, each binder in the array must specifically recognize a single protein in that lysate and not cross-react with any others. This is where antibodies come into their own. Highly specific monoclonal antibodies can be generated either in vivo by mouse immunization [20] or in vitro using techniques such as phage display [21,22]. Ideally, the antibodies are coupled or adsorbed to a solid support and then probed with a labeled lysate. Several model systems have demonstrated the feasibility of this approach using a handful of antibodies [23,24,25,26]. The publication by Haab et al. [15] takes these experiments one step further, using over a hundred different rodent monoclonal antibodies to simultaneously assay the levels of their respective cognate proteins in a complex antigen mixture.
So what comes next? Obviously, for an antibody array to profile an entire human proteome there needs to be at least one specific antibody directed against each human protein product. As there are estimated to be 50,000 human genes, with each giving rise to an average of perhaps five functional variants (by differential splicing, phosphorylation, glycosylation and so on), 250,000 different antibodies would be required. Because, in practice, it would be useful to have several antibodies against each protein variant, this equates to a total of more than a million different (highly specific) antibodies. Obtaining them by mouse immunization is likely to be very costly, not to mention the ethical considerations that may be encountered. In vitro methods of selection are therefore increasingly regarded as the key to isolating monoclonal antibodies for such proteomic screening [27,28]. The use of such recombinant antibodies together with the protein-microarray technology described by Haab et al. [15] may lead to the creation of vast antibody microarrays that can rapidly determine the protein profile of any organism, no matter how complex.
Contributor Information
Ian M Tomlinson, Email: imt@mrc-lmb.cam.ac.uk.
Lucy J Holt, Email: lhi@mrc-lmb.cam.ac.uk.
References
- Southern EM, Maskos U, Elder JK. Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics. 1992;13:1008–1017. doi: 10.1016/0888-7543(92)90014-j. [DOI] [PubMed] [Google Scholar]
- Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Southern E, Mir K, Shchepinov M. Molecular interactions on microarrays. Nat Genet. 1999;21:5–9. doi: 10.1038/4429. [DOI] [PubMed] [Google Scholar]
- Robertson D. Affymetrix license valid, rules court. Nat Biotechnol. 2001;19:13–14. doi: 10.1038/83443. [DOI] [PubMed] [Google Scholar]
- Cantor CR, Mirzabekov A, Southern E. Report on the sequencing by hybridization workshop. Genomics. 1992;13:1378–1383. doi: 10.1016/0888-7543(92)90079-8. [DOI] [PubMed] [Google Scholar]
- Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SP. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- Cronin MT, Fucini RV, Kim SM, Masino RS, Wespi RM, Miyada CG. Cystic fibrosis mutation detection by hybridization to light-generated DNA probe arrays. Hum Mutat. 1996;7:244–255. doi: 10.1002/(SICI)1098-1004(1996)7:3<244::AID-HUMU9>3.3.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Favis R, Day JP, Gerry NP, Phelan C, Narod S, Barany F. Universal DNA array detection of small insertions and deletions in BRCA1 and BRCA2. Nat Biotechnol. 2000;18:561–564. doi: 10.1038/75452. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet. 1999;21:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biology. 2001;2:research0004.1–0004.12. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalon D, Smith SJ, Brown PO. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- The Brown Lab http://cmgm.stanford.edu/pbrown/
- Luekling A, Horn M, Eickhoff H, Bussow K, Lehrach H, Walter G. Protein microarrays for gene expression and antibody screening. Anal Biochem. 1999;270:103–111. doi: 10.1006/abio.1999.4063. [DOI] [PubMed] [Google Scholar]
- MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- Silzel JW, Cercek B, Dodson C, Tsay T, Obremski RJ. Mass-sensing, multianalyte microarray immunoassay with imaging detection. Clin Chem. 1998;44:2036–2043. [PubMed] [Google Scholar]
- Mendoza LG, McQuary P, Mongan A, Gangadharan R, Brignac S, Eggers M. High-throughput microarray-based enzyme-linked immunosorbent assay (ELISA). Biotechniques. 1999;27:778–788. doi: 10.2144/99274rr01. [DOI] [PubMed] [Google Scholar]
- Rowe CA, Tender LM, Feldstein MJ, Golden JP, Scruggs SB, MacCraith BD, Cras JJ, Ligler FS. Array biosensor for simultaneous identification of bacterial, viral, and protein analytes. Anal Chem. 1999;71:3846–3852. doi: 10.1021/ac981425v. [DOI] [PubMed] [Google Scholar]
- Arenkov P, Kukhtin A, Gemmell A, Voloschu KS, Chupeeva V, Mizabekov A. Protein microchips: use for immunoassay and enzymatic reactions. Anal Biochem. 2000;278:123–131. doi: 10.1006/abio.1999.4363. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Enever C, de Wildt RM, Tomlinson IM. The use of recombinant antibodies in proteomics. Curr Opin Biotechnol. 2000;11:445–449. doi: 10.1016/s0958-1669(00)00133-6. [DOI] [PubMed] [Google Scholar]
- De Wildt RM, Mundy CR, Gorick BD, Tomlinson IM. Antibody arrays for high-throughput screening of antibody-antigen interactions. Nat Biotechnol. 2000;18:989–994. doi: 10.1038/79494. [DOI] [PubMed] [Google Scholar]


