Abstract
A few dozen genes are known on the human Y chromosome. The completion of the human genome sequence will allow identification of the remaining loci, which should shed further light on the function and evolution of this peculiar chromosome.
In humans, sex is determined by the presence or absence of the Y chromosome, which encodes the SRY gene necessary for testis development [1]. The Y chromosome is very unusual: it harbors just a few dozen genes, does not recombine over most of its length, is largely heterochromatic, and is riddled with repetitive DNA. Of all human chromosomes, the Y is probably the most intractable to genetic and molecular analysis; because it does not recombine during meiosis, classical linkage mapping is impossible, and the high density of repeated sequences makes physical mapping and sequencing difficult. In a recent article [2], David Page and colleagues describe the first detailed map of the non-recombining region of the human Y chromosome, providing a foundation for the sequencing and further analysis of this chromosome.
Evolution of the human Y chromosome
The human Y chromosome is small and mostly devoid of genes, while the X chromosome, its meiotic pairing partner, contains several thousand genes [3]. Comparative studies strongly suggest that the X and Y chromosomes in mammals are descended from a homologous pair of autosomes [3,4,5]; this hypothesis is further supported by the existence of regions of homology between the two sex chromosomes [6].
But how did the X and Y chromosomes evolve to become so different? A likely scenario is that the pair of autosomes that would eventually become the sex chromosomes acquired a sex-determining role, and suppression of recombination between the nascent Y and X chromosomes allowed them to evolve independently [5]. The X chromosome can still recombine in females, where two X chromosomes can pair, while most of the Y chromosome is completely sheltered from crossing over. The lack of recombination over most of the Y chromosome means that natural selection is less effective in preventing the accumulation of deleterious mutations and in driving the fixation of beneficial ones, resulting in the genetic erosion of the Y chromosome [5]. In response, dosage compensation evolves to restore equality of the dosage of gene products from X-linked loci in males and females [5]. In eutherian mammals, this is achieved by the inactivation of one of the two X chromosomes in females.
In mammals, Lahn and Page [6] have found evidence that recombination ceased along the sex chromosomes in an unexpected, stepwise fashion. First, a block of DNA surrounding the SRY gene stopped recombining, and then discrete non-recombining blocks evolved along most of the chromosome length (Figure 1). By comparing X-Y nucleotide divergence at 19 homologous genes located in the non-recombining region of the X and Y chromosomes, they identified four 'evolutionary strata' along the human X chromosome. Within each stratum, the X and Y copies of genes differ by about the same amount, indicating that recombination ceased at the same time for all the genes in question. But the different groups clearly originated at different time points [6]. Large inversions, which are known to suppress recombination, could account for this stepwise pattern. Interestingly, one such potentially important polymorphic inversion (3.5 megabases) has been identified on the human Y chromosome [2].
Figure 1.
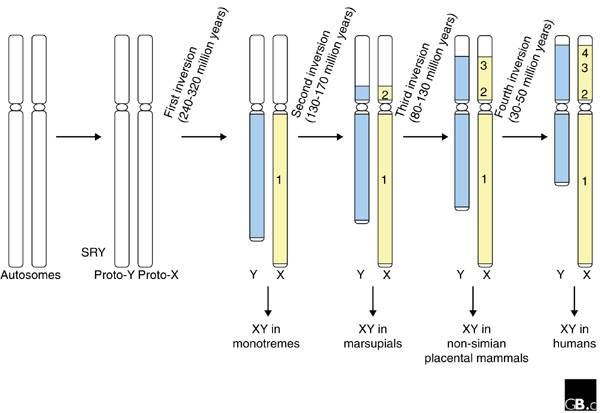
A proposed path for the evolution of the human sex chromosomes. Lahn and Page [6] postulate four inversions on the human Y chromosome, which suppressed recombination between the 'proto' sex chromosomes. Each inversion (designated 1-4) is thought to have reduced the size of the pseudoautosomal region (white) and enlarged the non-recombining portions of the X (yellow) and Y (blue) chromosomes. Time points at which the human X and Y may have diverged from the sex chromosomes of other mammals are indicated.
Gene content of the human Y chromosome
X-Y homologous regions are located at either end of the Y chromosome; these pair and recombine with the X chromosome during male meiosis [7]. Because genes that map to these regions do not show strict sex linkage, they are called 'pseudoautosomal regions' (PARs, see Figure 2). The PARs contain several genes that are active on the Y chromosome and not subject to X inactivation in females [8].
Figure 2.
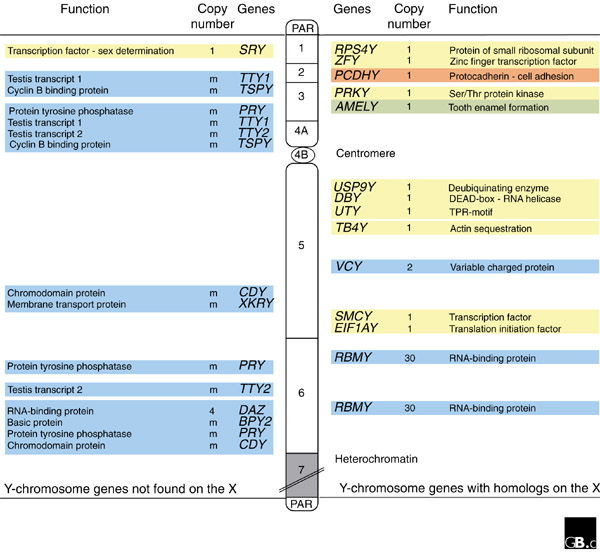
Genetic map of the non-recombining region of the human Y chromosome. The human Y chromosome consists of a large non-recombining region (NRY, which consists roughly of 24 megabases (Mb) euchromatin and 30 Mb heterochromatin) flanked by short pseudoautosomal regions (PAR, about 2.6 Mb and 0.4 Mb, respectively). Genes specific to the Y chromosome are indicated on the left-hand side of the diagram, and genes with homologs on the X chromosome are on the right-hand side. Ubiquitously expressed genes are shown in yellow, testis-specific genes are in blue, tooth-bud-specific genes in green, and brain-specific genes in red. Some testis-specific gene families have additional locations on the NRY not shown (m: multiple copies). The extensive region of Y-chromosomal heterochromatin is indicated by the grey area. The NRY is divided into intervals (as indicated by numbers on the chromosome), defined by naturally occurring deletions.
In humans, about 95% of the Y chromosome forms the non-recombining region (NRY). The NRY was long thought to be a functional wasteland, but reports in 1959 of XO females and XXY males established the existence of a sex-determining gene on the human Y chromosome [9], and in 1976 a factor required for spermatogenesis was mapped to the human Y chromosome [10]. The search for the testis determinant resulted in the cloning of the SRY gene in 1990 [1]. Now, more than 20 active genes or gene families have been identified. These genes fall into two main classes (Figure 2), unlike the situation on other chromosomes, whose gene content is relatively haphazard [11]. Genes in the first class are expressed in many tissues; these housekeeping genes have homologs on the X chromosome that escape X inactivation in females. Genes in the second group have a testis-specific function and have no X-linked homologs. Nearly all the testis-specific genes are present in multiple copies.
What are the evolutionary forces responsible for this bipartite division of genes on the Y chromosome? Selection will oppose the degeneration of genes that have critical functions in males and lack X homologs. Accordingly, most of the Y-specific genes are transcribed solely in the testis, indicating a function in spermatogenesis, and several deletions on the human Y chromosome are associated with male infertility [12]. The presence of these genes in multiple copies may reflect selection for high levels of their products. The loci shared between X and Y chromosomes are probably genes whose Y-linked copies have simply not degenerated. Most of these genes are located in the 'younger' strata of the Y chromosome [6], suggesting that they have not had sufficient time to degenerate. Many pseudogenes on the Y chromosome also have active homologs on the X, most of which are located in the younger strata as well (for example, STSP, KALP and GYG2P). Some of the housekeeping genes that are active on the human Y chromosome are pseudogenes in other eutherian lineages. For example, RPS4Y can be detected only in primates, where the homologous X-linked locus RPS4X escapes X inactivation [13]. In non-primate lineages, RPS4X has not retained a Y-linked homolog and is subject to X inactivation in females [13]. Similarly, ZFX, which has a strongly conserved homolog, ZFY, on the human Y chromosome, has lost its Y-linked copy in myomorph rodents [13]. The ubiquitin-activating enzyme locus UBE1X, in contrast, has a homolog on the squirrel-monkey Y chromosome, but no Y-linked copy can be detected in humans [14].
Comparative studies across mammalian taxa have demonstrated that the Y chromosome in humans has undergone multiple rearrangements [15]. Some genes such as SRY, RPS4Y or SMCY, were presumably originally Y-linked and escaped degeneration, as indicated by their presence on the sex chromosomes of marsupials, which diverged from a common ancestor with eutherians around 170 million years ago [16]. Others are derived from direct additions of autosomal genes, like the Y-specific DAZ gene cluster [17] or the CDY gene family [18]. CDY is intronless, suggesting that it originated by retrotransposition, while DAZ contains introns. Another source of Y-linked genes is provided by originally autosomal genes that were incorporated via the pseudoautosomal region. STS and ANT3, which are autosomal in marsupials, have been added to the PAR in eutherians, as revealed by their locations in sheep and dogs [15]. In humans, ANT3 is part of the PAR, while STS is incorporated into the non-recombining region of the sex chromosomes, where its Y-linked homolog (STSP) has become a pseudogene.
Non-coding and repetitive DNA
One of the greatest challenges for mapping the human Y chromosome was presented by massive, NRY-specific amplified regions, which comprise about one third of the euchromatic NRY [2]. These euchromatic amplified regions are diverse in composition, size, copy number and orientation, with some occurring as tandem repeats, others as inverted repeats, and still others dispersed throughout both arms of the chromosome [2]. Most Y-specific genes are found in these amplified regions [2].
The Y chromosome also contains highly abundant Alu and LINE repetitive elements, which are found throughout the entire human genome [19]. Much of the human Y chromosome is composed of repetitive satellite DNA, the bulk of which is located in the block of heterochromatin in the long arm (roughly 30 Mb in size; see Figure 2). This heterochromatic region mostly consists of the 3.4 kilobase (kb) DYZ1 and the 2.5 kb DYZ2 repeat families, which together account for around 50-70% of the DNA content of the human Y chromosome [20]. Heterochromatic regions, which are assumed to have no genes, are generally ignored in mapping and sequencing projects, because of the difficulties in cloning and sequencing them. Several genes have been identified in the heterochromatic Y chromosome of Drosophila [21], however, suggesting that there may be some genes to discover in the heterochromatic block of the human Y chromosome.
It is interesting to note some differences between Y-linked genes in Drosophila and humans. All known functional genes on the D. melanogaster Y chromosome have essential male-specific functions. In contrast to the human Y-specific genes, almost all are believed to be single-copy [21]. In Drosophila, the dynein genes present on the Y chromosome have huge introns up to several megabases in size [22]. These introns contain satellite- and transposable-element-derived DNA. Non-recombining genomic regions, such as the Y chromosome, are predicted to accumulate transposable elements and satellite repeat sequences [23]. In humans, no systematic trend towards larger introns can be seen in the genes characterized on the Y chromosome; this may be associated with the reduced activity of transposable elements in the human lineage [19].
What future insights may be provided by the human Y chromosome sequence? Probably the major difference between the X and the Y chromosomes from an evolutionary perspective is the lack of recombination over most of the Y chromosome. Evolutionary biologists are uncertain about the nature of the advantage gained from recombination [24]; the grisly fate of a large piece of non-recombining genome is demonstrated by the degenerate Y chromosome, however. Sequence analysis will provide a better understanding of the evolution of this bizarre chromosome.
But a more practical benefit may also be derived. As many as 7.5% of men are infertile, and one estimate suggests that in up to a quarter of these men infertility is caused by a Y-chromosomal defect [12]. Discoveries of Y-chromosome genes that influence reproductive capacity could lead to new treatments for men who lack those genes or have defective copies.
References
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Tilford CA, Kuroda-Kawaguchi T, Skaletsky H, Rozen S, Brown LG, Rosenberg M, McPherson JD, Wylie K, Sekhon M, Kucaba TA, Waterston RH, Page DC. A physical map of the human Y chromosome. Nature. 2001;409:943–945. doi: 10.1038/35057170. [DOI] [PubMed] [Google Scholar]
- Graves JA. The origin and function of the mammalian Y chromosome and Y-borne genes - an evolving understanding. Bioessays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Ohno S. Sex Chromosomes and Sex Linked Genes. Berlin: Springer, 1967.
- Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1006/bbrc.2001.5480. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS. Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum Genet. 1982;61:85–90. doi: 10.1007/BF00274192. [DOI] [PubMed] [Google Scholar]
- Polani PE. Pairing of X and Y chromosomes, non-inactivation of X-linked genes, and the maleness factor. Hum Genet. 1982;60:207–211. doi: 10.1007/BF00303003. [DOI] [PubMed] [Google Scholar]
- Jacobs PA, Strong JA. Case of human intersexuality having a possible XXY sex-determining mechanism. Nature. 1959;183:302–303. doi: 10.1038/183302a0. [DOI] [PubMed] [Google Scholar]
- Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet. 1976;34:119–134. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- Stuppia L, Gatta V, Calabrese G, Guanciali Franchi P, Morizio E, Bombieri C, Mingarelli R, Sforza V, Frajese G, Tenaglia R, Palka G. A quarter of men with idiopathic oligo-azoospermia display chromosomal abnormalities and microdeletions of different types in interval 6 of Yq11. Hum Genet. 1998;102:566–570. doi: 10.1007/s004390050741. [DOI] [PubMed] [Google Scholar]
- Jegalian K, Page DC. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature. 1998;394:776–780. doi: 10.1038/29522. [DOI] [PubMed] [Google Scholar]
- Mitchell MJ, Wilcox SA, Watson JM, Lerner JL, Woods DR, Scheffler J, Hearn JP, Bishop CE, Graves JA. The origin and loss of the ubiquitin activating enzyme gene on the mammalian Y chromosome. Hum Mol Genet. 1998;7:429–434. doi: 10.1093/hmg/7.3.429. [DOI] [PubMed] [Google Scholar]
- Graves JA, Wakefield MJ, Toder R. The origin and evolution of the pseudoautosomal regions of human sex chromosomes. Hum Mol Genet. 1998;7:1991–1996. doi: 10.1093/hmg/7.13.1991. [DOI] [PubMed] [Google Scholar]
- Toder R, Wakefield MJ, Graves JA. The minimal mammalian Y chromosome - the marsupial Y as a model system. Cytogenet Cell Genet. 2000;91:285–292. doi: 10.1159/000056858. [DOI] [PubMed] [Google Scholar]
- Saxena R, Brown LG, Hawkins T, Alagappan RK, Skaletsky H, Reeve MP, Reijo R, Rozen S, Dinulos MB, Disteche CM, Page DC. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Retroposition of autosomal mRNA yielded testis-specific gene family on human Y chromosome. Nat Genet. 1999;21:429–433. doi: 10.1038/7771. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1086/172716. [DOI] [PubMed] [Google Scholar]
- Cooke HJ, Fantes J, Green D. Structure and evolution of human Y chromosome DNA. Differentiation. 1983;23:S48–55. doi: 10.1007/978-3-642-69150-8_9. [DOI] [PubMed] [Google Scholar]
- Carvalho AB, Lazzaro BP, Clark AG. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc Natl Acad Sci USA. 2000;97:13239–13244. doi: 10.1073/pnas.230438397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reugels AM, Kurek R, Lammermann U, Bunemann H. Mega-introns in the dynein gene DhDhc7(Y) on the heterochromatic Y chromosome give rise to the giant threads loops in primary spermatocytes of Drosophila hydei. Genetics. 2000;154:759–769. doi: 10.1093/genetics/154.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Barton NH, Charlesworth B. Why sex and recombination? Science. 1998;281:1986–1990. doi: 10.1126/science.281.5385.1986. [DOI] [PubMed] [Google Scholar]


