Abstract
Comparative genome analyses, including chromosome painting in over 40 diverse mammalian species, ordered gene maps from several representatives of different mammalian and vertebrate orders, and large-scale sequencing of the human and mouse genomes are beginning to provide insight into the rates and patterns of chromosomal evolution on a whole-genome scale, as well as into the forces that have sculpted the genomes of extant mammalian species.
As the Human Genome Project enters a phase focusing on comparative and functional genomics, further emphasis will be placed on understanding our genes, their regulation and expression and their complex interactions, primarily as they pertain to human health and medicine. It will be important to resolve the evolutionary history of the human genome, as this has affected both genome organization and chromosomal architecture. Mapping and sequencing projects in several vertebrate species have recently begun to reveal the conservative, though dynamic, nature of genome organization [1,2].
The achievement of the first drafts of the full human genome sequence this year [3,4] leaves many researchers anxiously anticipating full genome sequences of other mammalian and vertebrate species, which are expected to add extra dimensions to our understanding of genome organization and evolution. Using features of genomic organization to better understand patterns and rates of chromosomal evolution, and applying these to reveal the processes involved in reshaping genomes, are some of the forthcoming challenges for comparative genome analysis. To illustrate these potential applications, we summarize here our current knowledge of comparative mammalian genome organization and describe how this field has benefited from cytogenetic, genetic and physical gene mapping technologies. We discuss important questions that remain to be answered with these technologies, and suggest some anticipated benefits that would derive from future comparisons of whole-genome sequences.
Approaches to understanding genome evolution
Genome organization has traditionally been inferred using two approaches: cytogenetic mapping and genetic-linkage or physical mapping. Comparisons of G-banded chromosome patterns were first used to infer homologies of whole chromosomes or subregions between species and even across mammalian orders [5,6,7]. Gene mapping using somatic cell hybrids subsequently confirmed that large tracts of mammalian genomes are remarkably conserved [8,9,10,11], suggesting that transferring information from species such as human and mouse, which have gene-rich maps, to the gene-poor developing maps of domestic animals might be feasible [11,12,13].
With the advent of chromosome painting (or 'zoo' fluorescent in situ hybridization, Zoo-FISH) in the early 1990s, entire chromosomal homologies could be readily visualized across mammalian orders [14,15,16], and in certain cases (the X chromosome) between placental and marsupial mammals [17]. In this procedure, individual chromosomes from a given species are physically isolated using fluorescence-activated cell sorting. DNA is extracted from each of these limited pools of chromosomes and each is labeled with a particular fluorescent dye using degenerate oligonucleotide primed (DOP)-PCR. The amplified chromosome pools (paints) are then hybridized to metaphase chromosomes of a different species (for an example, see Figure 1). Zoo-FISH has been most intensively applied to primate genomes [16,18], though Zoo-FISH analyses have now been performed for over 40 mammalian species from nine placental orders, usually relative to the human genome (Table 1) [1,2,19].
Figure 1.
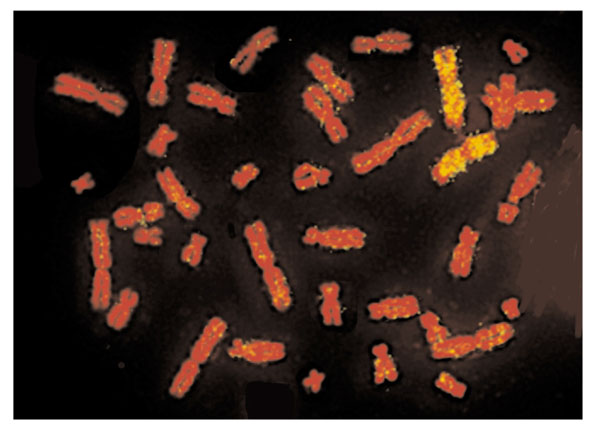
In situ hybridization of a human chromosome 3 paint to a metaphase spread of a flying fox (Pteropus poliocephalus). The human chromosome paint was made by flow-sorting and subsequent degenerate-oligonucleotide-primed (DOP) PCR. The probe was labeled with biotin and detected with avidin-fluorescein isothiocyanate (FITC). Note that only two signals are detected, revealing that human chromosome 3 is entirely syntenic with a single flying fox chromosome. The unhybridized section of this chromosome in the flying fox is homologous to human 21. The association of human chromosomes 3+21 is ancestral for placental mammals and is the largest commonly conserved region of synteny in the mammalian genome.
Table 1.
Genome characteristics of selected placental mammalian species studied to date
| Order | Species | Common name | Haploid chromosome | CSUs | CSOs | Citation |
| number | ||||||
| Primates | Pan troglodytes | Chimpanzee | 24 | 24 | [61] | |
| Gorilla gorilla | Gorilla | 24 | 26 | [61] | ||
| Pongo pygmaeus | Orangutan | 24 | 24 | [61] | ||
| Cercopithecus aethiops | African green monkey | 30 | 34 | [61] | ||
| Cebus capucinus | Capuchin monkey | 27 | 33 | [61] | ||
| Papio hamadryas | Baboon | 21 | 24 | ~ 49 autosomal | [62] | |
| Macaca fuscata | Macaque | 21 | 24 | [61] | ||
| Callithrix jacchus | marmoset | 23 | 32 | [61] | ||
| Ateles geoffroyi | Black-handed spider monkey | 17 | 49 | [61] | ||
| Hylobates lar | Gibbon | 22 | 52 | [61] | ||
| Hylobates concolor | Concolor gibbon | 25 | 70 | [61] | ||
| Hylobates syndactylus | Siamang | 25 | 59 | [61] | ||
| Presbytis cristata | Silvered leaf monkey | 23 | 34 | [61] | ||
| Eulemur fulvus | Lemur | 30 | 40 | [61] | ||
| Scandentia | Tupaia belangeri | Tree shrew | 31 | 42 | [20] | |
| Dermoptera | Flying lemurs | No data | No data | No data | ||
| Lagomorpha | Oryctolagus cuniculus | Rabbit | 22 | 39 | [63] | |
| Rodentia | Mus musculus | Mouse | 20 | 180 | >200* | [3,23] |
| Rattus norvegicus | Rat | 21 | 152* | 190 | [26,64] | |
| Carnivora | Canis familiaris | Dog | 38 | 68 | [34] | |
| Felis catus | Cat | 19 | 34 | 100-102* | [22] | |
| Mustela putorius | Ferret | 20 | 34 | [33] | ||
| Mustela vison | Mink | 15 | 33 | [2] | ||
| Phoca vitulina | Harbor seal | 16 | 31 | [47] | ||
| Ailuropoda melanoleuca | Giant panda | 42 | 44 | [43] | ||
| Pholidota | Pangolins | No data | No data | No data | ||
| Cetartiodactyla | Bos taurus | Cow | 30 | 50 | 149 | [28] |
| Ovis aries | Sheep | 27 | 40 | [1] | ||
| Capra hircus | Goat | 30 | ~ 50 | 107 | [39] | |
| Muntiacus muntjak v. | Indian muntjak | 4 | 50 | [2] | ||
| Sus scrofa | Pig | 19 | 47 | [1] | ||
| Tursiops truncatus | Dolphin | 22 | 29 | [65] | ||
| Perissodactyla | Equus caballus | Horse | 32 | 46 | [1] | |
| Chiroptera | Glossophaga soricina | Bat | 16 | 42 | [66] | |
| Eulipotyphla | Sorex araneus | Common shrew | 10 | 33 | [1] | |
| Xenarthra | Sloths, anteaters, armadillos | No data | No data | |||
| Macroscelidea | Elephant shrews | No data | No data | |||
| Afrosoricida | Tenrecs, golden moles | No data | No data | |||
| Tubulidentata | Aardvark | No data | No data | |||
| Proboscidea | Elephants | No data | No data | |||
| Sirenia | Manatees | No data | No data | |||
| Hyracoidea | Hyraxes | No data | No data | |||
| Galliformes | Gallus gallus | Chicken | 37 | 96* | [37] | |
| Atheriniformes | Danio rerio | Zebrafish | 25 | 418* | [32] |
Chicken and zebrafish are included for outgroup comparison. For a complete list of the 25 primate species analyzed by Zoo-FISH, see [61]. CSUs, conserved segments with unknown gene order; CSOs, conserved segments with known gene order; *predicted using methods of Nadeau and Taylor [67].CSUs and CSOs are based on comparison to human chromosomes.
Although they are extremely powerful for identifying conserved syntenies, somatic cell hybrid mapping and chromosome painting approaches are limited by their inability to discern gene order within conserved segments. Gene orders can be determined using subregional probes derived from whole-chromosome paints from species with highly fragmented genomes, which can then be used to define sub-chromosomal homologies in reciprocal painting experiments. Alternatively, probes derived from chromosome microdis-section or large-insert clones such as cosmids, bacterial artificial chromosomes, and yeast artificial chromosomes allow even greater precision [20,21]. These methods cannot, however, resolve smaller rearrangements or inversions within chromosome segments; this limitation might lead to systematic undercounting of the number of chromosomal rearrangements between two species.
Even greater precision in identifying conserved segments in which the gene order is known (CSOs) [22] can be achieved through gene mapping approaches using genetic linkage or radiation hybrid mapping technologies, which lend themselves to automation and high throughput. Genetic linkage maps are best suited to ordering polymorphic microsatellite markers (also called Type II markers), but are much less efficient for developing comparative maps; this is not surprising given the limited degree of coding locus (Type I) polymorphism observed within most intraspecies crosses [12]. The major exception to this is the mouse genome maps, which have been largely developed using interspecies crosses [23]. Interspecies backcrosses have the advantage of accumulated genetic differences between two evolutionary divergent species, particularly in less polymorphic coding gene loci [12].
Radiation hybrid mapping has proven to be effective for the rapid ordering of evolutionarily conserved Type I coding gene markers over the whole genome of various species [24,25]. In this method, the chromosomes of the species of interest are fragmented using a lethal dose of radiation on cultured cells; the cells are then fused to a thymidine-kinase-deficient recipient hamster cell line and hybrids are selected on HAT medium, on which only cells with active thymidine kinase (which must be hybrids) can grow. DNA from 90-100 radiation hybrid cell clones (each containing a different random assortment of chromosome fragments from the donor (or index) species) are screened for genetic markers from that species, and the markers are then ordered using maximum-likelihood ordering strategies. When several hundred Type I coding genes have been mapped in an index species, their positions can be compared with those of homologous loci from the gene-dense human and mouse Type I maps. Using this approach, whole-genome radiation hybrid gene maps have been constructed for rat [26], dog [27], cat [22], cow [28], pig [29] and zebrafish [30,31,32]. Integrated comparative gene maps for these species are beginning to reveal much about the evolutionary forces reshaping 'modern' genomes.
Rates of genome evolution
Comparative gene-mapping data collected to date indicate that the pattern of genome conservation within mammals appears to be bimodal. The ancestral, or 'default', pattern of genome rearrangement is very slow (roughly one exchange every 10 million years). This slow rate is evident in genomes of species from several orders of mammals, such as cat, mink, ferret, dolphin, and human [1,33]. Nested within some of the same mammalian orders, however, are species whose genomes are two to four times more reshuffled than those of species retaining the more slowly evolving genomes. Rearrangements relative to the ancestral genome are evident in the dog, bear, cow, mouse and rat genomes, and in those of the lesser apes and many New World primates [1,34,35]. Rodent genomes analyzed to date (mouse and rat) show very high rates of genome rearrangement (roughly one rearrangement per million years) relative to other mammal species [36] or when analyzed with respect to an avian outgroup species such as the chicken [37].
Thus far, most of the inferences concerning overall rates of change in mammalian genomes have been made on the basis of unordered segments derived from chromosome-painting experiments [37,38]. The available ordered whole-genome comparisons, however, reveal up to three times as many chromosomal rearrangements as were inferred from unordered comparisons, because intrachromosomal inversions are not detected in the latter (Table 1) [21,22,28,39]. Furthermore, the ordered gene maps of the cat and cow genomes show approximately twice as many intrachromosomal changes (inversions) as interchromosomal changes between species, where the gene order for both species is available [22,28]. These observations are in contrast with predictions from human-mouse comparisons that translocations would exceed inversions [40]. Higher rates of intrachromosomal rearrangements are further indicated by even more distant vertebrate comparisons, such as between the zebrafish and human maps [41]. Analysis of mouse-human comparisons at the sequence level reveal that smaller intrachromosomal rearrangements may be relatively frequent for these species, as well as for other mammalian genomes [42]. The overall contribution of small intrachromosomal rearrangements to genome reorganization will become more apparent with the anticipated sequencing of additional large genomic regions from mammalian species with putatively slow and rapid rates of chromosome change.
Phylogenomics
One promising use of whole-genome analysis is to aid understanding of phylogenetic relationships within a given taxonomic group [1,38,43]. The application of genomic data has yet to be utilized specifically for this purpose in mammals. One approach is to use as a phylogenetic (or cladistic) character associations of genes or segments syntenic to regions on two or more chromosomes of humans - the standard mammalian reference species - within a single chromosome of two or more other species. When the same association is found in two related species, the association was probably inherited from a common ancestor of the two species. A corollary to that assumption would be that disruption of the associations found in some species but not in others are also phylogenomic markers, because chromosomal breaks are probably sufficiently infrequent and near-random in chromosomal position to define unique events in evolutionary history. For example, regions homologous to human chromosomes 14 and 15 are associated together on a single chromosome in most mammalian species, although they are separated into two distinct chromosomes in all hominoids. Given the presence of the 14+15 association in a broad array of placental species [2,19], it is most parsimonious to assume that the 14+15 association is ancestral for placental mammals and that its fission into two chromosomes represents a derived state gained by a more recent common ancestor of humans and extant ape species.
Figure 2 lists nineteen human chromosome segments whose homologous regions are found associated in various species, as analyzed by chromosome painting. Most orders of mammals are characterized by at least one chromosome association; these also provide support for superordinal clades identified by previous phylogenetic studies [44,45]. For example, the order Carnivora retains two chromosome-segment associations not found in any other mammalian species: 2p+20 and 3+19 (Figure 2). A third association, 12q+22+10p, is also found in Carnivora and may be homologous or related phylogenomically (after some inversions that have involved segments of other chromosomes) to the segment associations 22+10+2+15+12+15+14 and 8+12+22+1+10 found in the horse and pig genomes, respectively (Figure 2) [2]. If the 12q+22+10p associations of Carnivora, Perissodactyls (including horses) and Cetartiodactyls (including pigs) are indeed homologs, then the sharing of this putatively ancestral association would be consistent with the phylogenetic association of these orders within the mammalian superordinal clade Fereuungulata [44,45]. The use of specific chromosome-exchange events as phylogenomic cladistic characters may be helpful in providing independent phylogenetic support for relationships previously inferred using morphological or molecular techniques.
Figure 2.
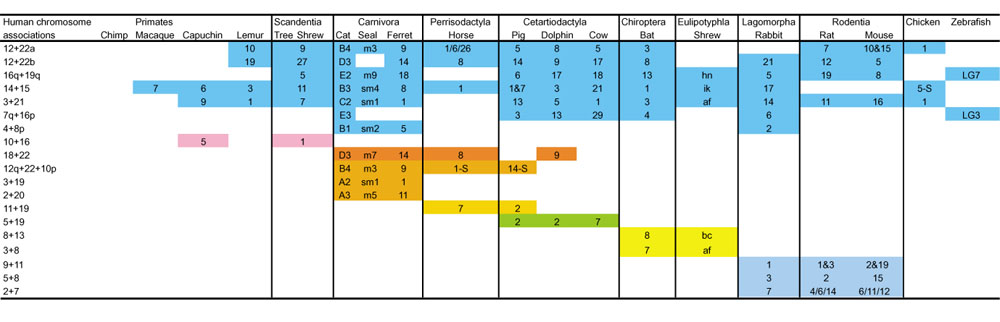
Human chromosome segment associations observed in other placental mammals by Zoo-FISH or gene mapping [2,22,32,37,41,48,49,68]. Numbers indicate the number of the chromosome in the indicated species that contains the human chromosome association shown in the left column. S, syntenic but uncertain if homologous in the absence of reciprocal chromosome painting. Note that multiple chromosome associations provide additional support for the cohort Glires (rodents plus lagomorphs) and Chiroptera plus Eulipotyphla; these clades are also supported by molecular data.
Recently, the phylogenomics approach has been applied to the genomes of living primates, allowing a reconstruction of the ancestral genome organization for this group of 279 living species [19,20,38,46]. The ancestral primate genome (Figure 3), which is thought to have existed some 60-70 million years ago, is remarkably similar to that of modern humans: 19 of the 26 ancestral chromosomes are unchanged - at least at the gross level revealed by chromosome painting - and the remaining chromosomes have only a single exchange that discriminates humans from the ancestral primate disposition. The average rate of genome exchanges among primates is remarkably slow (one exchange every 10 million years), as is the default rate across all mammals [1,2]. For most primate species, fewer than 20 rearrangements are sufficient to reassort the genome of a modern species into that of the primate ancestor (Figure 3) [38]. Similarly, the domestic cat genome can be derived by one fission and three fusions from the ancestral carnivore karyotype (Figure 3). Furthermore, as many as 18 of the ancestral primate chromosomes are found intact in the genomes of other placental mammals [2,47], suggesting that similar phylogenomic approaches can reach back further to the ancestors of all placental mammals (Figure 3).
Figure 3.
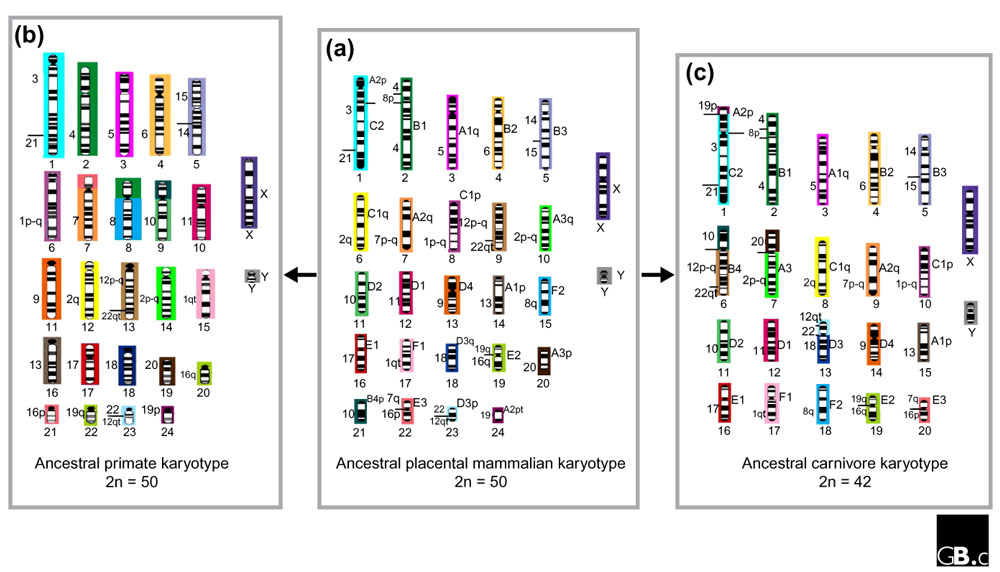
A hypothesis for the chromosome content of the ancestors of (a) all placental mammals, (b) all living primates, and (c) carnivores. To derive the ancestral syntenic karyotype of primates (b) from the ancestral placental mammalian karyotype, three fissions and three fusions are necessary, whereas only four fusions are needed to derive the ancestral karyotype of carnivores (c). Numbers to the left and right of each chromosome represent homologous regions of the human and cat genomes, respectively. Numbers below each chromosome are identifiers for the chromosome in each ancestral genome. The colors represent the chromosome in the ancestral mammalian genome from which each region originated. The G-banded karyotype depicted for the ancestral placental genome is speculative.
The original placental mammal genome organization
One of the primary goals now within reach of mammalian comparative mapping is to determine the arrangement of the ancestral placental mammalian genome. Once the ancestral genome is defined, it will be possible to determine the mode and tempo of evolutionary changes that have occurred in the phylogenetic lineages that gave rise to the genomes of members of the 18 extant placental orders. This would provide a framework for assessing lineage-specific rates of chromosomal evolution and would allow the explicit recognition of the different types of genomic exchange that characterize different mammalian taxa.
Figure 2 shows syntenic associations that are widespread in the eutherian orders. In several cases, these associations are also present in outgroup taxa, such as chicken and zebrafish [37,41]. The form and content of the ancestral genome of all placental mammals can be proposed from analyses of these data (Figure 3). On the basis of their widespread occurrence over several mammalian orders, it seems likely that the following associations homologous to human chromosomes or chromosome segments were present in the ancestral mammalian genome: 3+21, 4+8p+4, 7q+16p, 12p-q+22qt, 12qt+22q, 14+15, and 16q+19q (Figure 3). The number of hybridization signals found for each human chromosome and its distribution in extant orders provides additional data on the ancestral genome organization of placental mammals. Twelve human chromosomes (5, 6, 9, 11, 13-15, 17, 18, X, and Y) are found conserved in a block, either as single chromosomes or associated with other chromosome segments, in different species [2,19,41,48]. The remaining nine human homologs (1, 2, 7, 8, 10, 12, 16, 19, 22) are found in, at most, two fragments. For example, the regions homologous to human chromosome 2 are found in almost all mammals as two chromosomes. Their fusion in the human lineage is responsible for the origin of the human diploid number of 46; all great apes have 48 chromosomes. The ancestral genome of placental mammals probably had a diploid number (2n) of about 50 (Figure 3). The diploid numbers and chromosome associations present in the reconstructed ancestral genomemay be slightly modified when further mapping is performed in eutherian orders that have not yet been studied. Further chromosome painting with improved probes may help detect associations that have gone as yet unobserved. For instance, the 4+8p and 7q+16p associations were not observed in original painting studies on pig and cat but were first found in gene-mapping studies and then later confirmed using improved painting probes [2,22,49].
There remain major gaps in our understanding of genome organization in vertebrate groups nearer the transition between birds and placental mammals; data from these species would be useful to test these and similar hypotheses. First, gene-mapping data are too sparse in marsupials and monotremes to identify large regions of conserved synteny with eutherian autosomes [50]. The only successful chromosome-painting experiment to date between marsupials and placentals showed that a portion of the X chromosome (Xp11.2-Xqter) is conserved between the two lineages [17]. Second, all species of placental mammals examined to date by Zoo-FISH or gene mapping are limited to just two of four superordinal clades of placental mammals: 'Laurasiatheria' (Chiroptera (bats), Eulipotyphla (shrews, moles and others), Carnivora, Pholidota (pangolins), Cetartiodactyla (whales, ruminants and others), and Perissodactyla (horses and others)) and 'Euarchontoglires' (Primates, Scandentia (tree shrews), Dermoptera (flying lemurs), Rodentia and Lagomorpha (rabbits and others)) [44,45,51]. The two putative basal clades of mammals, Afrotheria (Proboscidea (elephants), Sirenia (including manatees), Tubulidentata (aardvarks), Hyracoidea (hyraxes), Afrosoricida (tenrecs and golden moles), and Macroscelidea (elephant shrews)) and Xenarthra (sloths, armadillos and anteaters), are unrepresented in gene mapping or Zoo-FISH experiments [2] (but see [52]). A comprehensive assessment of the history of placental mammalian genomes can be achieved only when these genomic 'missing link' species are examined in the context of outgroup species, such as marsupials, monotremes and other amniotes.
Comparative mapping data from the zebrafish genome [41] has offered some intriguing hypotheses addressing whether certain mammalian chromosomes may have been part of larger composite chromosomes that subsequently underwent fissions in different mammalian lineages. Postlethwait et al. [41] used the zebrafish gene map to infer the ancestral chromosomes that may have been present in the vertebrate ancestor of bony fishes and tetrapods. These authors propose that the widespread 2n = 48 karyotype characterizing most fish genomes - and the similarly high numbered karyotype defining many mammalian genomes - derives from excessive fissions during the tetrapod radiation and a single round of duplications in the fish lineage, and that tetrapods and fish both had a low-numbered, possibly 2n = 20-24, ancestral vertebrate karyotype [41]. Several large contiguous blocks of chromosome associations in the zebrafish are fragmented in human, cat and mouse genomes. Postlethwait et al. [41] hypothesize that there was an ancestral vertebrate chromosome consisting of homologs of human chromosomes 11+15+19q, which occurred in the ancestor of human and mouse but split independently in each lineage. Chromosome painting data clearly demonstrate, however, that the 19q+16p and 14+15 associations are ancestral for placental mammals (Figure 2) [19,38,52]. Apparently, both humans and muroid rodents have independently lost the ancestral association of 14+15. Thus, there is no evidence for the retention or occurrence of a 11+19+15 ancestral synteny in the modern primates or rodents, nor in other placental orders, especially given that 14+15 is also found in the chicken (Table 1). Convergence is the most likely reason for why this 11+19+15 association is found in both muroid rodents and zebrafish. Nevertheless, it cannot yet be ruled out that this synteny was found in the ancestor of all amniotes (birds, reptiles, and mammals) and was fragmented prior to the diversification of placental mammals.
Evolutionary breakpoints of genomes
As the resolution of comparative genome analysis increases, one might wonder what forces drive chromosomal rearrangements. Are certain regions of the mammalian genome, such as fragile sites, more prone to chromosome breakage than others? Are ancestral chromosome-exchange junction points randomly distributed? Do they correlate with common chromosomal polymorphisms or translocations that cause neoplastic transformation? Would analysis of sequence motifs at the junction of evolutionary breakpoints reveal structures that promote breakage?
Recent large-scale comparative sequencing efforts in the mouse genome have begun to provide some insight into chromosome breakpoints, through comparison with homologous human chromosomal regions. Two studies of the mouse genome observed a preponderance (60-70%) of highly repetitive elements at the junctions of human conserved segments, particularly simple-sequence repeats such as TCTG tandem repeats [42,53]. Transposable elements, Alu-mediated recombination and minisatellites have also been found associated with chromosome rearrangements [54,55,56]. Not all analyses of chromosomal breakpoints between human and mouse have revealed a markedly increased percentage of sequence repeats, however [57]. It may be necessary to take into account the evolutionary age of breakpoints, in order to discriminate more recent rearrangements from older events that may have been overprinted by genetic change [58]. This will be best accomplished by more precise alignment of ordered gene maps [22,28,59,60] from multiple mammalian genomes and by interpretation of these maps with regard to phylogeny.
In conclusion, comparative genome analysis in mammals has revealed complex differences in the rates and patterns of change in different mammalian species. Development and refinement of comparative maps, combining both chromosome-painting and gene-mapping approaches in a broader number of species, will allow us to estimate lineage-specific rates of chromosomal change and to make more accurate reconstructions of ancestral genomes. This will be further enhanced by the full genome sequences of rodents, which are anticipated in the near future. Undoubtedly, a large amount of fascinating insight and function will finally be gained concerning the organization and distribution of genes within mammalian chromosomes, distributions and composition of breakpoints, and the forces driving genome evolution.
References
- O'Brien SJ, Menotti-Raymond M, Murphy WJ, Nash WG, Wienberg J, Stanyon R, Copeland NG, Jenkins NA, Womack JE, Marshall Graves JA. The promise of comparative genomics in mammals. Science. 1999;286:458–481. doi: 10.1126/science.286.5439.458. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Eisenberg JF, Miyamoto M, Hedges SB, Kumar S, Wilson DE, Menotti-Raymond M, Murphy WJ, Nash WG, Lyons LA, et al. Comparative genomics mammalian radiations. Science. 1999;286:Wall Chart. [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Sawyer JR, Dunham K. The striking resemblance of high resolution G-banded chromosomes of man and chimpanzee. Science. 1980;208:1145–1148. doi: 10.1126/science.7375922. [DOI] [PubMed] [Google Scholar]
- Nash WG, O'Brien SJ. Conserved regions of homologous G-banded chromosomes between orders in mammalian evolution: carnivores and primates. Proc Natl Acad Sci USA. 1982;79:6631–6635. doi: 10.1073/pnas.79.21.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutrillaux B, Couturier J. The ancestral karyotype of carnivora - comparison with that of Platyrrhine monkeys. Cytogenet Cell Genet. 1983;35:200–208. doi: 10.1159/000131867. [DOI] [PubMed] [Google Scholar]
- Lalley PA, Minna JD, Francke U. Conservation of autosomal gene synteny groups in mouse and man. Nature. 1978;274:160–163. doi: 10.1038/274160a0. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Nash WG. Genetic mapping in mammals: chromosome map of the domestic cat. Science. 1982;216:257–265. doi: 10.1126/science.7063884. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Cevario SJ, Martenson JS, Thompson MA, Nash WG, Chang E, Graves JA, Spencer JA, Cho KW, Tsujimoto H, Lyons LA. Comparative gene mapping in the domestic cat (Felis catus). J Hered. 1997;88:408–414. doi: 10.1093/oxfordjournals.jhered.a023127. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Wienberg J, Lyons LA. Comparative genomics: lessons from cats. Trends Genet. 1997;13:393–399. doi: 10.1016/s0168-9525(97)01297-3. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ. Mammalian genome mapping: lessons and prospects. Curr Opin Genet Dev. 1991;1:105–111. doi: 10.1016/0959-437x(91)80050-v. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Womack JE, Lyons LA, Moore KJ, Jenkins NA, Copeland NG. Anchored reference loci for comparative genome mapping in mammals. Nat Genet. 1993;3:103–112. doi: 10.1038/ng0293-103. [DOI] [PubMed] [Google Scholar]
- Wienberg J, Jauch A, Stanyon R, Cremer T. Molecular cytotaxonomy of primates by chromosomal in situ suppression hybridization. Genomics. 1990;8:347–350. doi: 10.1016/0888-7543(90)90292-3. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Cremer T, Arnason U, Weier H-U, Lima-de-Faria A, Frönicke L. Comparative chromosome painting discloses homologous segments in distantly related mammals. Nat Genet. 1994;6:342–347. doi: 10.1038/ng0494-342. [DOI] [PubMed] [Google Scholar]
- Wienberg J, Stanyon R. Chromosome painting in mammals as an approach to comparative genomics. Curr Opin Genet Dev. 1995;39:792–797. doi: 10.1016/0959-437x(95)80013-u. [DOI] [PubMed] [Google Scholar]
- Glas R, Marshall-Graves JA, Toder R, Ferguson-Smith M, O'Brien PC. Cross-species chromosome painting between human and marsupial directly demonstrates the ancient region of the mammalian X. Mamm Genome. 1999;10:1115–1116. doi: 10.1007/s003359901174. [DOI] [PubMed] [Google Scholar]
- Jauch A, Wienberg J, Stanyon R, Arnold N, Tofanelli S, Ishida T, Cremer T. Reconstructing genomic arrangements in great apes and gibbons by chromosome painting. Proc Natl Acad Sci USA. 1992;89:8611–8615. doi: 10.1073/pnas.89.18.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary BP, Raudsepp T, Fronicke L, Scherthan H. Emerging patterns of comparative genome organization in some mammalian species as revealed by Zoo-FISH. Genome Res. 1998;8:577–589. doi: 10.1101/gr.8.6.577. [DOI] [PubMed] [Google Scholar]
- Müller S, Stanyon R, O'Brien PCM, Ferguson-Smith MA, Plesker R, Wienberg J. Defining the ancestral karyotype of all primates by multidirectional chromosome painting between tree shrews, lemurs and humans. Chromosoma. 1999;108:393–400. doi: 10.1007/s004120050391. [DOI] [PubMed] [Google Scholar]
- Müller S, Stanyon R, Finelli P, Archidiancono N, Wienberg J. Molecular cytogenetic dissection of human chromosomes 3 and 21 evolution. Proc Natl Acad Sci USA. 2000;97:206–211. doi: 10.1073/pnas.97.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Sun S, Chen ZQ, Yuhki N, Hirschmann D, Menotti-Raymond M, O'Brien SJ. A radiation hybrid map of the cat genome: implications for comparative mapping. Genome Res. 2000;10:691–702. doi: 10.1101/gr.10.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Gilbert DJ, Eppig JT, Maltais LJ, Miller JC, Dietrich WF, Weaver A, Lincoln SE, Steen RG, et al. A genetic linkage map of the mouse: current applications and future prospects. Science. 1993;262:57–66. doi: 10.1126/science.8211130. [DOI] [PubMed] [Google Scholar]
- Cox DR, Burmeister M, Price ER, Kim S, Myers RM. Radiation hybrid mapping: a somatic cell genetic method for constructing high-resolution maps of mammalian chromosomes. Science. 1990;250:245–250. doi: 10.1126/science.2218528. [DOI] [PubMed] [Google Scholar]
- Walter MA, Spillett DJ, Thomas P, Weissenbach J, Goodfellow PN. A method for constructing radiation hybrid maps of whole genomes. Nat Genet. 1994;7:22–28. doi: 10.1038/ng0594-22. [DOI] [PubMed] [Google Scholar]
- Watanabe TK, Bihoreau MT, McCarthy LC, Kiguwa SL, Hishigaki H, Tsuji A, Browne J, Yamasaki Y, Mizoguchi-Miyakita A, Oga K, et al. A radiation hybrid map of the rat genome containing 5,255 markers. Nat Genet. 1999;22:27–36. doi: 10.1038/8737. [DOI] [PubMed] [Google Scholar]
- Mellersh CS, Hitte C, Richman M, Vignaux F, Priat C, Jouquand S, Werner P, Andre C, DeRose S, Patterson DF, et al. An integrated linkage-radiation hybrid map of the canine genome. Mamm Genome. 2000;11:120–130. doi: 10.1007/s003350010024. [DOI] [PubMed] [Google Scholar]
- Band MR, Larson JH, Rebeiz M, Green CA, Heyen DW, Donovan J, Windish R, Steining C, Mahyuddin P, Womack JE, Lewin HA. An ordered comparative map of the cattle and human genomes. Genome Res. 2000;10:1359–1368. doi: 10.1101/gr.145900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawken RJ, Murtaugh J, Flickinger GH, Yerle M, Robic A, Milan D, Gellin J, Beattie CW, Schook LB, Alexander LJ. A first-generation porcine whole-genome radiation hybrid map. Mamm Genome. 1999;10:824–830. doi: 10.1007/s003359901097. [DOI] [PubMed] [Google Scholar]
- Geisler R, Rauch GJ, Baier H, van Bebber F, Brobeta L, Dekens MP, Finger K, Fricke C, Gates MA, Geiger H, et al. A radiation hybrid map of the zebrafish genome. Nat Genet. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- Hukriede NA, Joly L, Tsang M, Miles J, Tellis P, Epstein JA, Barbazuk WB, Li FN, Paw B, Postlethwait JH, et al. Radiation hybrid mapping of the zebrafish genome. Proc Natl Acad Sci USA. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk WB, Korf I, Kadavi C, Heyen J, Tate S, Wun E, Bedell JA, McPherson JD, Johnson SL. The syntenic relationship of the zebrafish and human genomes. Genome Res. 2000;10:1351–1358. doi: 10.1101/gr.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavagna P, Menotti A, Stanyon R. Genomic homology of the domestic ferret with cats and humans. Mamm Genome. 2000;11:866–870. doi: 10.1007/s003350010172. [DOI] [PubMed] [Google Scholar]
- Breen M, Thomas R, Binns MM, Carter NP, Langford CF. Reciprocal chromosome painting reveals detailed regions of conserved synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics. 1999;61:145–155. doi: 10.1006/geno.1999.5947. [DOI] [PubMed] [Google Scholar]
- Stanyon R, Consigliere S, Muller S, Morescalchi A, Neusser M, Wien-berg J. Fluorescence in situ hybridization (FISH) maps chromosomal homologies between the dusky titi and squirrel monkey. Am J Primatol. 2000;50:95–107. doi: 10.1002/(SICI)1098-2345(200002)50:2<95::AID-AJP1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Stanyon R, Yang F, Cavagnau P, O'Brien PC, Bagga M, Ferguson Smith MA, Wienberg J. Reciprocal chromosome painting shows that genomic rearrangement between rat and mouse proceeds ten times faster than between humans and cats. Cytogenet Cell Genet. 1999;84:150–155. doi: 10.1159/000015244. [DOI] [PubMed] [Google Scholar]
- Burt DW, Bruley C, Dunn IC, Jones CT, Ramage A, Law AS, Morrice DR, Paton IR, Smith J, Windsor D, et al. The dynamics of chromosome evolution in birds and mammals. Nature. 1999;402:411–413. doi: 10.1038/46555. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Stanyon R. Phylogenomics. Ancestral primate viewed. Nature. 1999;402:365–366. doi: 10.1038/46450. [DOI] [PubMed] [Google Scholar]
- Schibler L, Vaiman D, Oustry A, Giraud-Delville C, Cribiu EP. Comparative gene mapping: a fine-scale survey of chromosome rearrangements between ruminants and humans. Genome Res. 1998;8:901–915. doi: 10.1101/gr.8.9.901. [DOI] [PubMed] [Google Scholar]
- Erlich J, Sankoff D, Nadeau JH. Synteny conservation and chromosome rearrangements during mammalian evolution. Genetics. 1997;147:289–296. doi: 10.1093/genetics/147.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Puttagunta R, Gordon LA, Meyer GE, Kapfhamer D, Lamerdin E, Kantheti P, Portman K, Chung WK, Jenne DE, Olsen AS, et al. Comparative maps of human 19p13.3 and mouse chromosome 10 allow identification of sequences at evolutionary breakpoints. Genome Res. 2000;10:1369–1380. doi: 10.1101/gr.145200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash WG, Wienberg J, Ferguson-Smith MA, Menninger JC, O'Brien SJ. Comparative genomics: tracking chromosome evolution in the family Ursidae using reciprocal chromosome painting. Cytogenet Cell Genet. 1998;83:182–192. doi: 10.1159/000015176. [DOI] [PubMed] [Google Scholar]
- Madsen O, Scally M, Douady CJ, Kao DJ, DeBry R, Adkins R, Amrine HM, Stanhope MJ, deJong WW, Springer MS. Parallel adaptive radiations in two major clades of placental mammals. Nature. 2001;409:610–614. doi: 10.1038/35054544. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O'Brien SJ. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- Haig D. A brief history of human autosomes. Philos Trans R Soc Lond B Biol Sci. 1999;354:1447–1470. doi: 10.1098/rstb.1999.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM, Vandeberg JL. Comparative gene mapping: introduction. ILAR J. 1998;39:47–260. [Google Scholar]
- Glas R, Wakefield MJ, Toder R, Graves JAM. Comparative chromosome painting. ILAR J. 1998;39:Wall Chart. [PubMed] [Google Scholar]
- Fronicke L, Wienberg J. Comparative chromosome painting defines the high rate of karyotype changes between pigs and bovids. Mamm Genome. 2001;12:442–449. doi: 10.1007/s003350010288. [DOI] [PubMed] [Google Scholar]
- Toder R, Waugh O'Neill R, Graves JAM. Chromosome painting in marsupials. ILAR J. 1998;39:92–95. doi: 10.1093/ilar.39.2-3.92. [DOI] [PubMed] [Google Scholar]
- Springer MS, DeJong WW. Which mammalian supertree to bark up? Science. 2001;291:1709–1711. doi: 10.1126/science.1059434. [DOI] [PubMed] [Google Scholar]
- Richard F, Lombard M, Dutrillaux B. Phylogenetic origin of human chromosomes 7, 16, and 19 and their homologs in placental mammals. Genome Res. 2000;10:644–651. doi: 10.1101/gr.10.5.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher MT, Roe BA, Chen F, Do T, Do A, Malaj E, Reeves RH. Chromosome evolution: the junction of mammalian chromosomes in the formation of mouse chromosome 10. Genome Res. 2000;10:1463–1467. doi: 10.1101/gr.146600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray YH. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16:461–468. doi: 10.1016/s0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- Vergnaud G, Denoeud F. Minisatellites: mutability and genome architecture. Genome Res. 2000;10:899–907. doi: 10.1101/gr.10.7.899. [DOI] [PubMed] [Google Scholar]
- Prak ET, Kazazian HH., Jr Mobile elements and the human genome. Nat Rev Genet. 2000;1:134–144. doi: 10.1038/35038572. [DOI] [PubMed] [Google Scholar]
- Lund J, Chen F, Hua A, Roe B, Budarf M, Emanuel BS, Reeves RH. Comparative sequence analysis of 634 kb of the mouse chromosome 16 region of conserved synteny with the human velacardiofacial syndrome region on chromosome 22q11.2. Genomics. 2000;63:374–383. doi: 10.1006/geno.1999.6044. [DOI] [PubMed] [Google Scholar]
- Ji Y, Eichler EE, Schwartz S, Nicholls RD. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 2000;10:597–610. doi: 10.1101/gr.10.5.597. [DOI] [PubMed] [Google Scholar]
- Yang YP, Womack JE. Parallel radiation hybrid mapping: a powerful tool for high-resolution genomic comparison. Genome Res. 1998;8:731–736. doi: 10.1101/gr.8.7.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Shan S, Chen ZQ, Pecon-Slattery J, O'Brien SJ. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res. 1999;9:1223–1230. doi: 10.1101/gr.9.12.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienberg J, Froenicke L, Stanyon R. Insights into mammalian genome organization and evolution by molecular cytogenetics. In Comparative Genomics Edited by Clark MS Dordrecht: Kluver Academic Publishers, 2000. pp. 207–244.
- Rogers J, Mahaney MC, Witte SM, Nair S, Newman D, Wedel S, Rodriguez LA, Rice KS, Slifer SH, Perelygin A, et al. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics. 2000;67:237–247. doi: 10.1006/geno.2000.6245. [DOI] [PubMed] [Google Scholar]
- Korstanje R, O'Brien PC, Yang F, Rens W, Bosma AA, van Lith HA, van Zutphen LF, Ferguson-Smith MA. Complete homology maps of the rabbit (Oryctolagus cuniculus) and human by reciprocal chromosome painting. Cytogenet Cell Genet. 1999;86:317–322. doi: 10.1159/000015325. [DOI] [PubMed] [Google Scholar]
- Scheetz TE, Raymond MR, Nishimura DY, McClain A, Roberts C, Birkett C, Gardiner J, Zhang J, Butters N, Sun C, et al. Generation of a high-density rat EST map. Genome Res. 2001;11:497–502. doi: 10.1101/gr.151601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielec PE, Gallagher DS, Womack JE, Busbee DL. Homologies between human and dolphin chromosomes detected by heterologous chromosome painting. Cytogenet Cell Genet. 1998;81:18–25. doi: 10.1159/000015002. [DOI] [PubMed] [Google Scholar]
- Volleth M, Klett C, Kollak A, Dixkens C, Winter Y, Just W, Vogel W, Hameister H. Zoo-FISH analysis in a species of the order Chiroptera: Glossophaga soricina (Phyllostomidae). Chromosome Res. 1999;7:57–64. doi: 10.1023/a:1009227428727. [DOI] [PubMed] [Google Scholar]
- Nadeau JH, Taylor BA. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci USA. 1984;81:814–818. doi: 10.1073/pnas.81.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Reboul J, Lutfalla G, Burt DW. Human chromosomes 3 and 21 are the products of an ancestral gene arrangement that is at least 300 million years old. Mamm Genome. 2000;11:806–807. doi: 10.1007/s003350010156. [DOI] [PubMed] [Google Scholar]


