Abstract
Cyclic nucleotide–gated (CNG) ion channels are nonselective cation channels with a high permeability for Ca2+. Not surprisingly, they are blocked by a number of Ca2+ channel blockers including tetracaine, pimozide, and diltiazem. We studied the effects of dequalinium, an extracellular blocker of the small conductance Ca2+-activated K+ channel. We previously noted that dequalinium is a high-affinity blocker of CNGA1 channels from the intracellular side, with little or no state dependence at 0 mV. Here we examined block by dequalinium at a broad range of voltages in both CNGA1 and CNGA2 channels. We found that dequalinium block was mildly state dependent for both channels, with the affinity for closed channels 3–5 times higher than that for open channels. Mutations in the S4-S5 linker did not alter the affinity of open channels for dequalinium, but increased the affinity of closed channels by 10–20-fold. The state-specific effect of these mutations raises the question of whether/how the S4-S5 linker alters the binding of a blocker within the ion permeation pathway.
Keywords: CNG channels, dequalinium, voltage dependence, block, gating
INTRODUCTION
Much of what we know about ion channel function has been revealed through the use of molecules that block the ion-conducting pore (for review see Yellen, 1998). The identification of the P-loop as the region lining the pore, the hydrophobic nature of the pore lining, the location of a gate near the intracellular mouth, and the existence of a cavity behind the intracellular gate were all first discovered using pore blockers. Although blockers have been useful in elucidating pore structure, their true advantage comes from their use in studying rearrangements in the pore. In particular, examining the state dependence of block can yield information on the physical motions of channels during gating—something not accessible using structural approaches alone. State-dependent blockers are those for which the binding energy of the blocker is different in the closed and open states. If state-dependent block is observed, it can be inferred that the region to which the blocker binds moves during gating. Conversely, if block is not state dependent, it can be inferred that the blocker binding site does not undergo a significant rearrangement during gating.
For almost two decades, cyclic nucleotide–gated (CNG) channels have been known to be blocked by divalent cations such as Ca2+ and Mg2+ (Haynes et al., 1986; Colamartino et al., 1991; Zimmerman and Baylor, 1992; Root and MacKinnon, 1993; Eismann et al., 1994). Both Ca2+ and Mg2+ are permeant blockers: they traverse the channel but at a much slower rate than Na+ and K+ ions (Capovilla et al., 1983; Hodgkin et al., 1985; Torre et al., 1987; Nakatani and Yau, 1988). Another blocker of CNG channels is pseudechetoxin (PsTx), a peptide purified from the venom of the Australian king brown snake. When applied to the extracellular face of membrane patches containing homomeric CNGA2 channels, PsTx blocks the cGMP-dependent current. PsTx also blocks homomeric CNGA1 channels with high affinity, but is less effective on the native combination of CNGA1 plus CNGB1 found in rod photoreceptors (Brown et al., 1999).
Local anesthetics are also effective at blocking a wide variety of ion channels (Hille, 2001). Tetracaine has moderate affinity for native and expressed CNG channels (Schnetkamp, 1990; Ildefonse and Bennett, 1991; Quandt et al., 1991). Fodor et al. (1997a)(b) demonstrated that high affinity binding of tetracaine to the intracellular surface of CNGA1 and CNGA2 channels occurs preferentially when the pore is in the closed conformation. This state-dependent block was found to be due in part to the interaction of tetracaine with a negatively charged amino acid (E363) on the extracellular side of the selectivity filter (Fodor et al., 1997a). Neutralization of this negatively charged residue dramatically decreased tetracaine block of the closed state. The more favorable binding energy of tetracaine to closed channels compared with open channels implies that movement of tetracaine relative to E363 is coupled to channel gating. By piecing together information from state-dependent blockers that interact with different sites in and near the pore, a picture of the movements involved in activation can be constructed.
Here we examined the mechanism of CNG channel block by dequalinium. We show that the general mechanism of dequalinium block is conserved in CNGA1 and CNGA2 channels. In studies at 0 mV in which patches were equilibrated with dequalinium for many tens of minutes, block appeared to be state independent. In contrast, experiments designed to study block over a range of voltages revealed dequalinium to block closed channels with 3–5-fold higher affinity than open channels. This state dependence may have been masked by a slower, higher affinity block in the experiments at 0 mV. In addition, we found that mutations in the S4-S5 linker produced a 10–20-fold increase in the affinity of closed channels for dequalinium, without altering the affinity of open channels. This region of sequence is not thought to line the ion-permeation pathway, raising the question of whether the mutations altered dequalinium binding through a direct interaction, or indirectly through nonlocal perturbations of pore structure.
MATERIALS AND METHODS
Molecular Biology
Potential binding sites for dequalinium in the bovine rod CNGA1 channel were examined by generating channel chimeras. Chimeric channels were generated by replacing half of transmembrane segment S5 or S6 of CNGA1 with the corresponding segment of the olfactory CNGA2 channel. The constructs were designated to indicate the segment where the replacement was made. The constructs were generated by excising the segment for CNGA2 cDNA by using two different restriction enzymes to produce a cassette. This cassette was then ligated into CNGA1 cDNA, which was cut by the same two restriction enzymes. After transformation of bacteria with the ligated product, single colonies were selected for plasmid production. Identifications of chimera products were performed by restriction analysis, and verified by DNA sequencing
Because of difficulties using the above method for the S4/S5-CNGA2 chimera, we constructed it in two steps. We first used the QuickChange multisite-directed mutagenesis kit (Stratagene) to make CNGA2-S291N. The protocol provided with the QuickChange kit was used without any modifications. The entire coding region was sequenced and analyzed to confirm that the S291N mutation was created by this method, and to ensure against second-site mutations. We next used the QuickChange XL site-directed mutagenesis kit (Stratagene) to introduce the remaining three mutations of the S4/S5-CNGA2 chimera, as well a silent BsrGI site. The protocol provided with the kit was used without any modifications. Single colonies were picked for plasmid preparation. Primary screening for the mutation was executed by restriction analysis. Once identified, the entire coding region was sequenced to confirm the introduction of the mutations as well as to ensure against second-site mutations.
Similar methods were used to construct each of the other CNGA1/CNGA2 chimeras studied. Their chimeric regions were as follows (CNGA1 numbering): CNGA1-S5b, amino acids 315–327; CNGA1-P, amino acids 334–372; and CNGA1-S6a, amino acids 369–378.
RNA and Oocytes
DNA for CNGA1 was a gift from Dr. W.N. Zagotta and DNA for CNGA2 was a gift from Dr. R.R. Reed. cRNA was synthesized in vitro, using a standard in vitro transcription kit (mMessage mMachine; Ambion). Segments of ovary were removed from anesthetized Xenopus laevis. After gross mechanical isolation, individual oocytes were defolliculated by incubation with collagenase 1A (1 mg/ml) in Ca2+-free OR2 medium (82.5 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 5 mM HEPES, pH 7.6) for 0.75–1.5 h. The cells were then rinsed and stored in frog Ringer's solution (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5 mM HEPES, pH 7.6) at 14°C. Oocytes were injected with 50 nL cRNA solution within 2 d of harvest. Electrophysiological recordings were performed 2–10 d after injection.
Current Recordings
Inside-out patch-clamp recordings were made using symmetrical low divalent NaCl/HEPES/EDTA solutions consisting of 130 mM NaCl, 3 mM HEPES and 200 μM EDTA (pH 7.2), with cGMP added to the intracellular solution only. The solution bathing the intracellular surface of the patch was changed using an RSC-200 rapid solution changer (Molecular Kinetics). Dequalinium chloride was prepared as follows: a 0.5 M dequalinium chloride stock was prepared in water and final dequalinium chloride dilutions were made using the low divalent solution described above in the presence of different cGMP concentrations. All chemicals were purchased from Sigma-Aldrich.
Currents were low pass-filtered at 2 kHz with an Axopatch 200B amplifier (Axon Instruments, Inc.), and sampled at a 10 kHz. Data were acquired and analyzed with the PULSE data acquisition software (Instrutech) and were plotted and fitted using Igor Pro (Wavemetrics, Inc.). For each experiment, currents in response to low and high concentrations of cGMP were monitored for several minutes and further experimentation did not proceed until these values had stabilized. This was necessary because of a “run up” in the fractional activation by cGMP that occurs after patch excision, and has been previously shown to represent dephosphorylation of the channels by endogenous patch-associated phosphatases (Gordon et al., 1992; Molokanova et al., 1997). All currents shown are difference currents in which the current in the patch in the absence of cyclic nucleotide has been subtracted. All dose–response curves were measured with a protocol in which the blocker and/or agonist were applied to the patch at a voltage of 0 mV. Since at this voltage the net current flux in our solutions is zero, the patches were held at 0 mV, and stepped to +20 mV for 20 ms at an interval of 5 s, unless otherwise stated (see Fig. 1 legend). Inside-out patches containing CNGA1, CNGA2, and CNGA1-S5a channels where held at 0 mV for 100 ms and stepped to +60 or to −60 mV (as indicated in the figure legends) for 20 s to ensure that the patches spent as much time as necessary to achieve steady-state of block at these voltages. All recordings were performed at room temperature. Smooth curves shown in dose–response relations to dequalinium chloride are fits with the Hill equation:
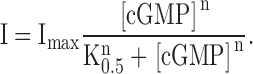 |
(1) |
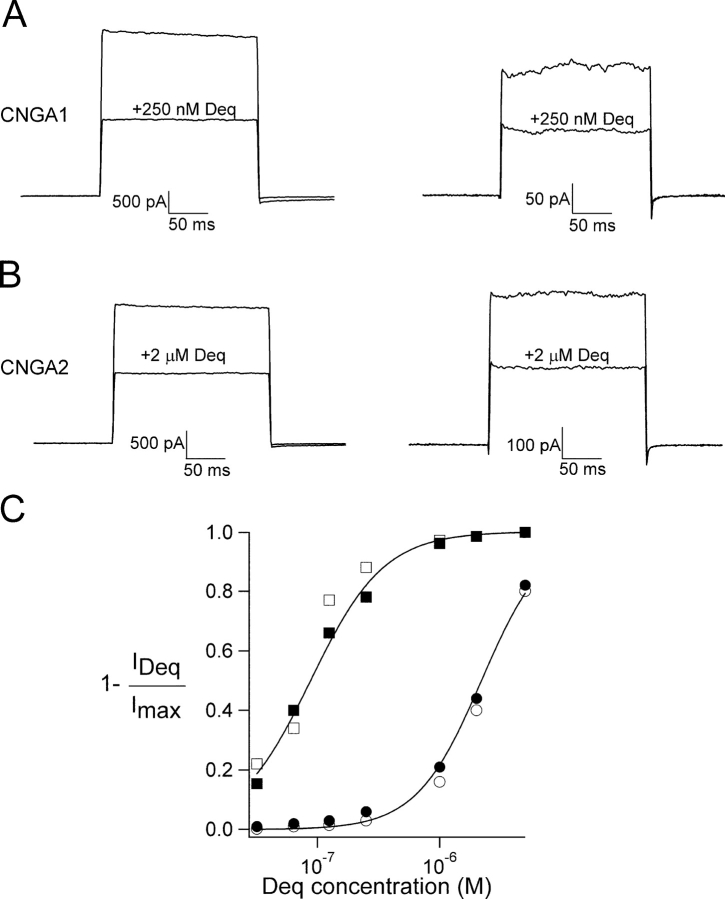
Figure 1.
State-independent block of CNGA1 and CNGA2 channels observed at 0 mV. (A and B) Current traces obtained when holding the patch at 0 mV with 20-ms steps to +20 mV at 5-s intervals. (A) CNGA1 channels in the absence of dequalinium and in the presence of 250 nM dequalinium with 2 mM (left) and 32 μM cGMP (right). Dequalinium blocked the current ∼50% under both conditions. (B) CNGA2 channels in the absence of dequalinium and in the presence of 2 μM dequalinium with 2 mM (left) and 1.8 μM cGMP (right). Dequalinium blocked the current ∼50% under both conditions. (C) Dose–response for block of CNGA1 channels (squares) and CNGA2 channels (circles) by dequalinium. Filled squares represent block in the presence of 2 mM cGMP and open squares represent block in the presence of 32 μM cGMP for CNGA1 channels. For CNGA2 channels, filled circles represent block in the presence of 2 mM cGMP and open circles represent block in the presence of 1.8 μM cGMP. The smooth curves are fits with Eq. 1 (see materials and methods) with IC50 = 189 nM and 192 nM for CNGA1 (high and low cGMP, respectively) and 2.4 μM (both for high and low cGMP) and the Hill coefficient = 1.4 in all cases. Error bars represent SEM, n = 5–7 patches.
To obtain voltage-current relations, the patches were stepped from a holding potential of 0 mV to between −60 and +80 mV in increments of 20 mV for 15 s to reach steady-state of block. Block versus voltage data were fit with the Boltzmann equation, unless otherwise indicated:
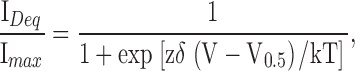 |
(2) |
where z is the valence of the blocker (two for dequalinium), δ is the electrical distance of the blocker in the pore, T is the absolute temperature, and k is Boltzmann's constant. Data are plotted, unless indicated otherwise, as the fractional current obtained by dividing the blocked current by the maximum unblocked current and reported as the mean ± SEM. Time courses of block could be fit with a single exponential.
Modeling
Predictions of the voltage dependence from Scheme I were calculated numerically using the analytical expression for the open probability as a function of nucleotide concentration. The voltage dependence of block was calculated at 2 mM for open channels (both CNGA1 and CNGA2 channels) and at 32 μM (for CNGA1) and 1.8 μM (for CNGA2) for closed channels. The voltage dependence of the binding constants for dequalinium was assumed to be of the form:
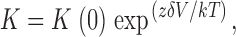 |
(3) |
where K(0) is the binding constant at 0 mV, zδ is the product of the dequalinium valence and “electrical distance”, V is the voltage, k the Boltzmann constant and T the temperature. All simulations were performed using Igor Pro (Wavemetrics).
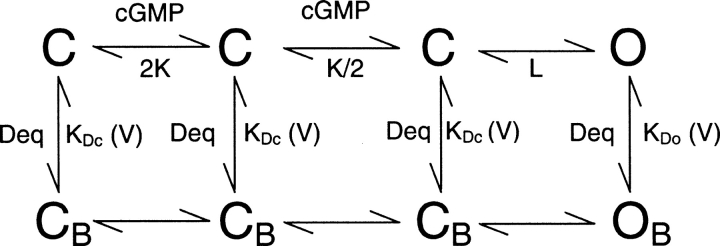
SCHEME I.
RESULTS
We explored the mechanism of block of CNG channels by the organic divalent cation dequalinium. We have previously shown that dequalinium blocks CNGA1 channels from the intracellular side with high apparent affinity (190 nM), in a voltage-dependent manner (zδ = 1), and can be electrostatically repelled by ions in the permeation pathway, suggesting that its mechanism involves a physical occlusion of the pore. In our studies performed at 0 mV, dequalinium appeared to bind to open and closed channels with equal affinity.
As an additional tool in probing the interactions between the channels and dequalinium, we used a related CNG channel, CNGA2. We compared block by dequalinium at 0 mV in CNGA1 channels to that of CNGA2 channels. Fig. 1 shows that block by dequalinium at 0 mV in both types of channels occurs with equal affinity for open channels (in the presence of saturating cGMP concentrations) and for closed channels (in the presence of subsaturating cGMP concentrations). In Fig. 1, A and B, currents activated by a saturating cGMP concentration are shown on the left, and currents activated by concentrations of cGMP below the K1/2 are shown on the right. A given concentration of dequalinium blocked the same fraction of current in both cases. To further study the apparent affinity of the channels for dequalinium, we measured dose–response relations for block at the cGMP concentrations examined above. For these experiments, we held the patches at 0 mV, a voltage in which no current was observed in our symmetrical solutions. At intervals of 5 s we then briefly (20 ms) jumped the voltage to +20 mV to drive current through the open channels. Using this pulse protocol we found that dequalinium inhibited CNGA2 channels with an IC50 of 2.4 μM ± 0.01 (Fig. 1 C, circles), regardless of the cGMP concentration used to activate the channels. Although the affinity for CNGA2 channels was ∼10-fold lower than for CNGA1 channels (190 nM, Fig. 1 C, squares) (Rosenbaum et al., 2002), for both types of channels block occurred in a state-independent manner when measured at 0 mV.
To further understand the molecular mechanism of block by dequalinium, we expanded our studies to a broader range of membrane potentials. Examples of typical currents measured at two potentials are shown in Fig. 2, A and B. Instead of holding at the potential of interest for many minutes, we instead held the patches at 0 mV and stepped to various potentials for sufficient time for the blocker to reach steady-state. A pulse to +60 mV is shown in Fig. 2 A and a pulse to −60 mV is shown in Fig. 2 B. The time course of block could be fit with a single exponential (gray line) and was faster at +60 mV (τ = 2.9 s) than at −60 mV (τ = 17 s). This relatively fast block is quite different from that observed when holding the membrane potential at 0 mV, and then stepping the voltage to +20 mV for 20 ms every 5 s. As shown in Fig. 2 C, this experimental protocol produced a very slow block (τ = 59 min) that reached steady-state only after many tens of minutes.
Figure 2.
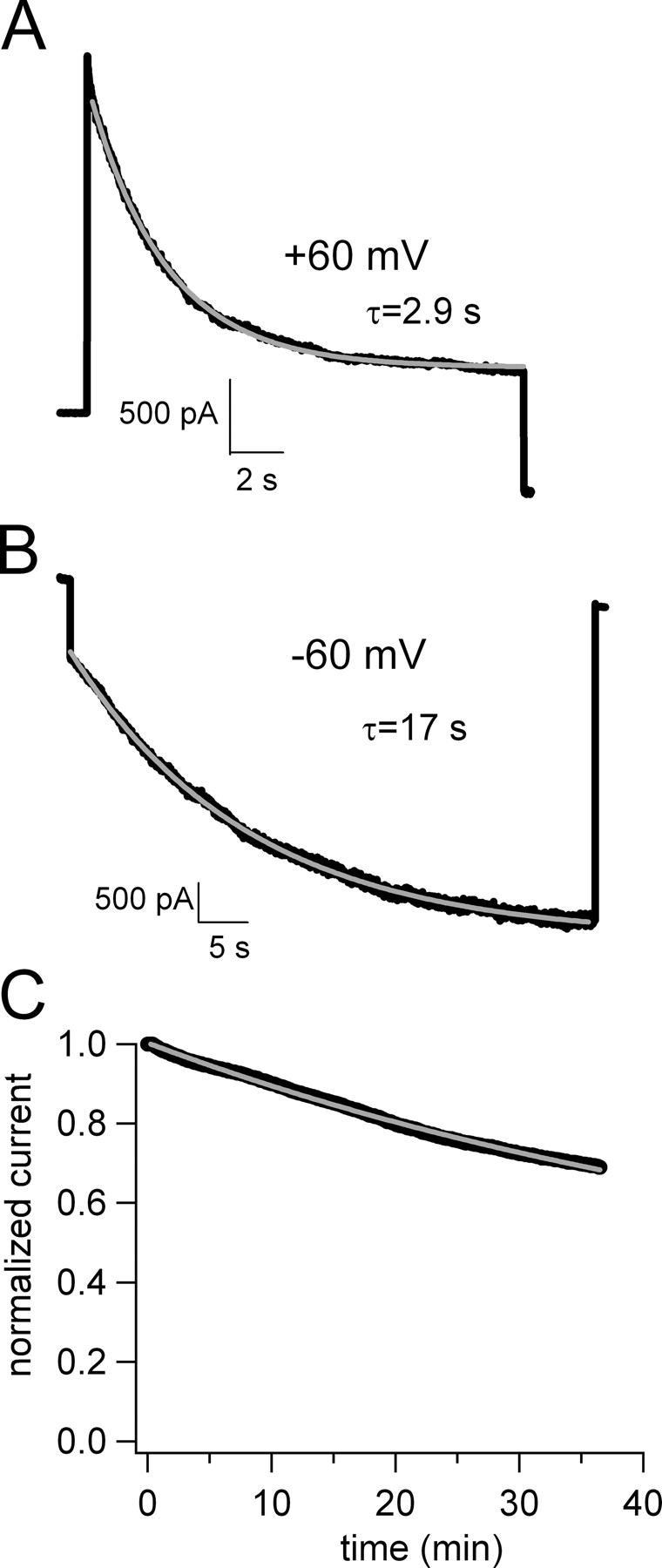
Time course of block of CNGA1 channels during depolarizing and hyperpolarizing voltage steps. (A) Block by 50 nM dequalinium during a voltage step to +60 mV. The gray curve is a fit with a single exponential, with a time constant τ = 2.9 s. (B) Block by 50 nM dequalinium during a voltage step to −60 mV. The gray curve is a fit with a single exponential, with a time constant τ = 17 s. (C) Time course of block obtained by using a protocol in which the holding potential was held at 0 mV and pulsed for 20 ms to +20 mV every 5 s. The gray line is a fit with a single exponential, yielding τ = 59 min.
We examined the voltage dependence of block by dequalinium at both saturating and subsaturating concentrations of cGMP. The relative current present at each voltage is shown in Fig. 3 A for CNGA1 channels, and in Fig. 3 B for CNGA2 channels. In each case, the block of open channels (saturating cGMP) is shown by the filled symbols and the block of closed channels (subsaturating cGMP) is shown by the open symbols. Fits with the Boltzmann equation yielded V1/2 values of 107 and 86 mV for open and closed states, respectively, in CNGA1 channels and V1/2 values of 95 and 60 mV for open and closed states, respectively, in CNGA2 channels. From these experiments it is evident that block was more efficient at depolarized potentials, and that it occurred with a higher affinity when the channels were closed.
Figure 3.
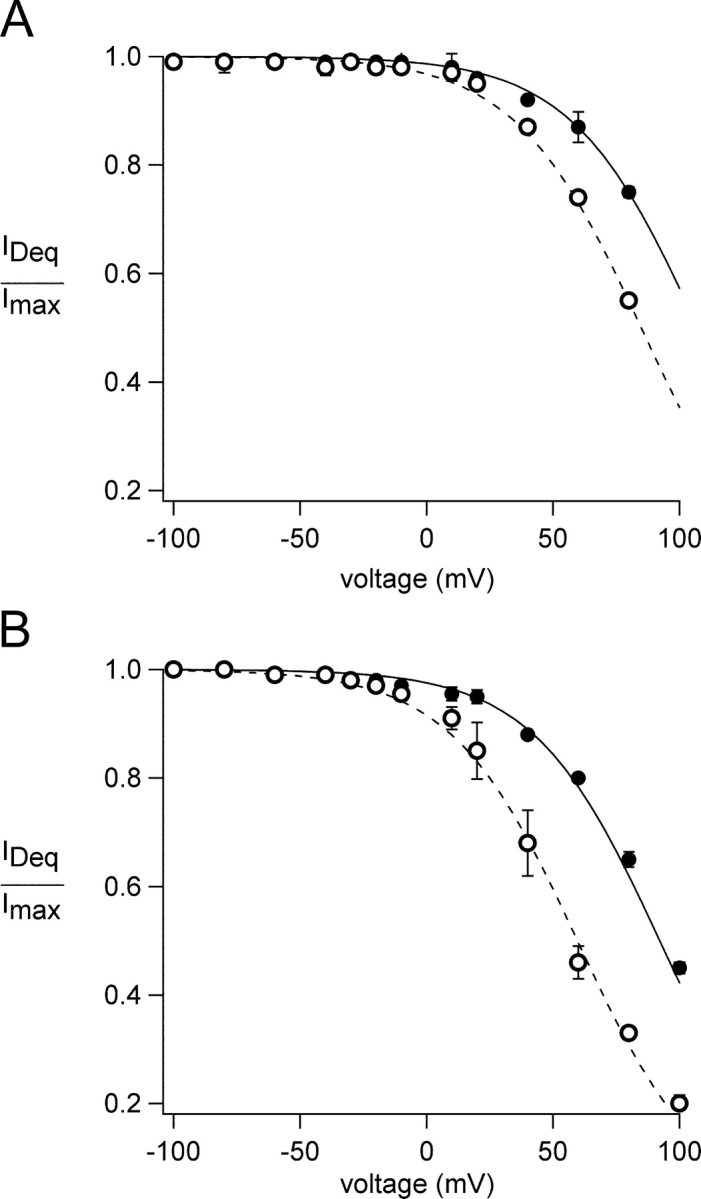
Voltage dependence of block in CNGA1 and CNGA2 channels. (A) Block of CNGA1 channels by 50 nM dequalinium in the presence of 2 mM cGMP (filled circles) or 32 μM cGMP (open circles). The curves represent fits with Scheme I, with KDo = 4 μM and KDc = 1.3 μM. The value of zδ was 1 in both cases. (B) Block of CNGA2 channels by 500 nM dequalinium in the presence of 2 mM cGMP (filled circles) or 1.8 μM cGMP (open circles). The curves represent fits with Scheme I, with KDo = 20 μM, and KDc = 3.6 μM The value of zδ was 1 in both cases. Dequalinium concentrations below the IC50 of block were used since otherwise block at depolarized potentials becomes rapidly saturated and the voltage-dependence of block cannot be correctly determined.
We used the following simple model to describe how state-dependent block by dequalinium could occur in CNAG1 and CNGA2 channels:
The model assumes that two ligand binding events precede a concerted opening transition; this approach has been shown to be sufficient to explain the steady-state activation by cGMP (Karpen et al., 1988; Gordon and Zagotta, 1995a). In this model, the channels are blocked by dequalinium with different affinities in the closed and in the open states, but with no change in the voltage dependence of block (i.e., zδ = 1 for both closed and open states). The solid curves in Fig. 3 represent the predictions of the model for channels in the presence of saturating cGMP concentrations (when most of the channels are open) and the dotted curves represent the predictions for channels in the presence of subsaturating cGMP concentrations (when most of the channels are closed). These results show that KDc is 3–5 times more favorable than KDo (see legend for Fig. 3), giving rise to the observed state dependence.
Organic cations that block CNG channels in a voltage-dependent manner have been described previously. One of these, tetracaine, is like dequalinium in that it has a higher apparent affinity for CNGA1 channels compared with CNGA2 channels. The basis of this difference was found to lie not in the tetracaine binding site but rather in the different energetics of activation of CNGA1 and CNGA2 (Fodor et al., 1997b). CNGA1 channels have an intrinsically less favorable allosteric conformational change than do CNGA2 channels (Goulding et al., 1994; Gordon and Zagotta, 1995a). This difference has several functional consequences, including: a lower apparent affinity for activation by cGMP in CNGA1; cAMP is a partial agonist for CNGA1 channels but a full agonist for CNGA2 channels; and closed-channel blockers such as tetracaine bind with higher apparent affinity to CNGA1 channels than to CNGA2 channels.
Does the differential affinity of dequalinium for CNGA1 compared with CNGA2 represent the same kind of closed-channel block mechanism observed with tetracaine? Scheme I can be used to address this question. For the fits in Fig. 3, the equilibrium constant between the closed and open state, L, was taken as 17 for CNGA1 and 10,000 for CNGA2 (Fodor et al., 1997a,b; Rosenbaum et al., 2003). The affinities of the open and closed states that come out of the fits thus take into account the more favorable energetics of activation of CNGA2 channels compared with CNGA1 channels. The dissociation constants for block of closed and open CNGA1 channels were 20–30 times more favorable than those for block of CNGA2 channels. Thus, intrinsic differences in gating of CNGA1 and CNGA2 channels were not sufficient to explain the different affinity of each for dequalinium.
The experiments above indicate that the mechanism for dequalinium block is conserved in CNGA1 and CNGA2 channels, but that the apparent affinity for dequalinium block of CNGA1 channels is greater than that for CNGA2 channels. The sequences of CNGA1 and CNGA2 are 59% identical and 70% conserved overall, with higher identity in the core region and cyclic nucleotide binding domain. We reasoned that differences in their sequence might underlie the difference in their apparent affinities for dequalinium. We therefore tested a series of chimeras, using the CNGA1 sequence as a background with regions of sequence from CNGA2 substituted in.
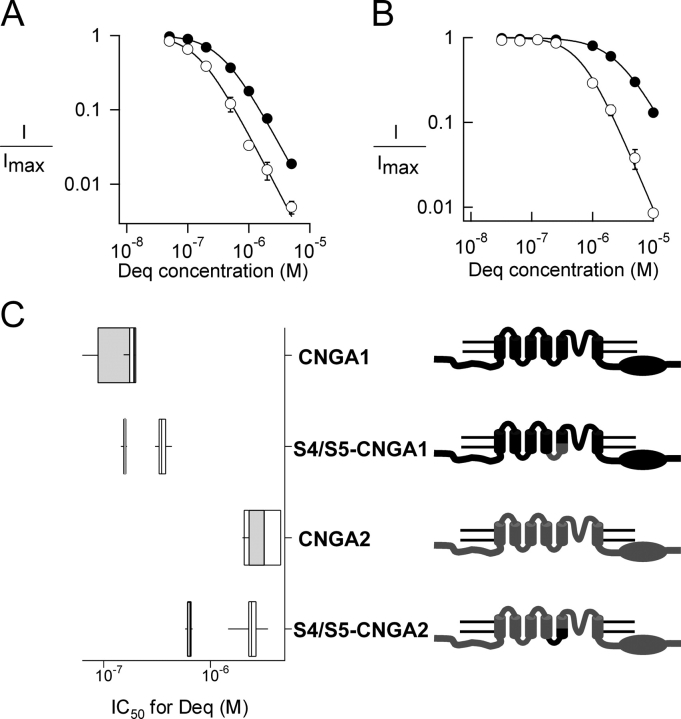
We examined the apparent affinity for dequalinium of several chimeras at 0 mV. Surprisingly, chimeras of the P-loop (CNGA1-P) and S6 (CNGA1-S6a), both regions known to line the ion conducting pore, hardly altered the apparent affinity for dequalinium (Table I). Mutations in the S5 region also did not affect the affinity for the blocker (CNGA1-S5b). However, when we examined chimeras in which the S4-S5 linker had been swapped, state dependence was apparent at 0 mV. For the S4/S5-CNGA1 chimera, the IC50 for block was 360 nM at 2 mM cGMP and 156 nM at 32 μM cGMP (filled and open symbols, respectively, Fig. 4 A). For the S4/S5-CNGA2 chimera, the IC50 for block was 2.7 μM at 2 mM cGMP and 650 nM at 1.8 μM cGMP (filled and open symbols, respectively, Fig. 4 B).
TABLE I.
Affinity of Chimeric Channels for Dequalinium
| Construct | IC50 for Deq block |
|---|---|
| CNGA1 | 189 ± 4.19 nM |
| CNGA1-R342Y | 91 ± 0.1 nM |
| CNGA1-R342D | 123 ± 0.2 nM |
| CNGA1-S5b | 260 ± 0.1 nM |
| CNGA1-P | 80 ± 0.1 nM |
| CNGA1-S6a | 218 ± 8.46 nM |
| S4/S5-CNGA1 | 360 ± 8.7 nM |
| S4/S5-CNGA2 | 2.7 ± 0.33 μM |
| CNGA2 | 2.4 ± 0.29 μM |
Regions that affect binding of dequalinium to CNG channels. A CNGA1 channel background (denoted as the CNGA1 prefix in the names of the constructs) was used to construct chimeras where specific regions where replaced by the corresponding regions of CNGA2, except for S4/S5-CNGA2 chimera where the background was that of CNGA2. The mean IC50 values for each of the constructs shown were obtained in the presence of 2 mM cGMP.
Figure 4.
State-dependent block of the S4/S5-linker chimeras by dequalinium at 0 mV. Block by different dequalinium concentrations of (A) S4/S5-CNGA1 and (B) S4/S5-CNGA2 channels. Filled symbols represent block in the presence of 2 mM cGMP and open symbols represent block in the presence of subsaturating cGMP concentrations (32 μM for S4/S5-CNGA1 and 1.8 μM for S4/S5-CNGA2). Fits to the Hill equation yielded mean IC50 values for S4/S5-CNGA1 channels of 360 nM at 2 mM cGMP and 156 nM with 32 μM cGMP, and for S4/S5-CNGA2 channels of 2.7 μM at 2 mM cGMP and 650 nM at 1.8 μM cGMP. (C) Block of wild-type and mutant channels. A CNGA1 channel background (depicted in black) was used to construct chimeras where specific regions where replaced by the corresponding regions of CNGA2 (depicted in gray). The IC50 values for each of the constructs obtained in the presence of saturating cGMP (2 mM, white boxes) and subsaturating cGMP (32 μM or 1.8 μM, gray boxes) are shown for each construct. For the box and whisker plot the line represent the median of the data, the box surrounds the 25th through 75th percentile, and the whiskers extend to the 5th and 95th percentiles.
The data from the experiments described above are summarized in Fig. 4 C. The box plots shown in this figure represent the IC50 for dequalinium in the presence of saturating cGMP concentrations (white boxes) and in the presence of subsaturating cGMP concentrations (gray boxes), measured at 0 mV. As evidenced from these data, the S4/S5 chimeras display state-dependent block even at 0 mV, a phenotype distinct from the state-independent block at 0 mV observed in wild-type channels (Fig. 1).
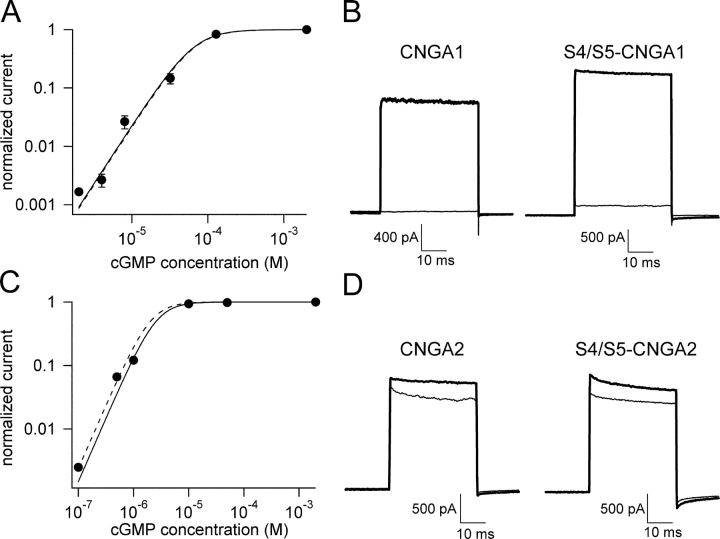
To determine whether this phenomenon was due to general changes in the behavior of these channels, we compared the dose–response relations for activation by cGMP and the fractional activation by cAMP of the chimeras to that of their corresponding wild-type homologue. The data indicate that the overall behavior of the chimeras was very similar to that of the corresponding wild type channels, with a K1/2 for activation by cGMP of 66 μM (Fig. 5 A) and a fractional activation by cAMP of 6% (Fig. 5 B) for S4/S5-CNGA1 , and a K1/2 for activation by cGMP of 2.6 μM (Fig. 5 C) and a fractional activation by cAMP of 86% (Fig. 5 D) in S4/S5-CNGA2.
Figure 5.
cGMP activation curves for S4/S5-chimeras. (A and C) Activation of the S4/S5-CNGA1 and S4/S5-CNGA2 chimeras by cGMP. Data were fit with Eq. 1 (see materials and methods). The dashed lines represents the activation curve for wild-type CNGA1 (A) and CNGA2 channels (C). The K1/2 for activation by cGMP was 66 μM for S4/S5-CNGA1 and 2.6 μM for S4/S5-CNGA2. (B and D) Fractional activation by cAMP (thin traces) was of ∼6% of the maximal current activated by cGMP (thick traces) for both wild-type CNGA1 (B, left) and S4/S5-CNGA1 channels (B, right), and 86% for wild-type CNGA2 (D, left) and S4/S5-CNGA1 (D, right)
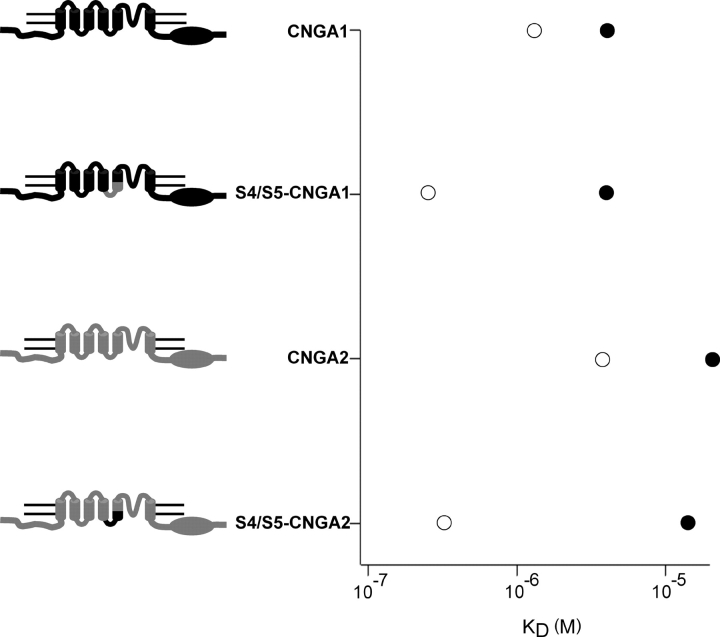
We next examined the voltage dependence of block in the two S4/S5 chimeras. As shown in Fig. 6, block of both chimeras was state dependent, with closed channels (Fig. 6, open symbols) being blocked at more hyperpolarized potentials than open channels (Fig. 6, filled symbols). Fitting these data with the Boltzmann equation, gave V1/2 values for S4/S5-CNGA1 of 61 mV for closed channels and 120 mV for open channels. For S4/S5-CNGA2, V1/2 values of −2 and 80 mV were obtained for closed and open channels, respectively. Values for zδ were 1 in all cases. Compared with their wild-type homologues, the voltage dependence of block was thus comparable for open channels, but shifted toward hyperpolarized potentials for closed channels. Fits to these data with Scheme I gave the equilibrium constants for block shown in Fig. 7. Compared with wild-type channels, the affinity for block in the closed state (KDc; open circles) was greatly increased (5–10-fold) for the chimeras, with little or no change in the affinity of block of the open state (KDo, filled circles).
Figure 6.

Voltage dependence of block in S4/S5-CNGA1 and S4/S5-CNGA2 channels. (A) Block of S4/S5-CNGA1 channels by 50 nM dequalinium in the presence of 2 mM cGMP (filled circles) and 32 μM cGMP (open circles). The curves represent fits with Scheme I, with KDo = 4 μM and KDc = 250 nM. (B) Block of S4/S5-CNGA2 channels by 500 nM dequalinium in the presence of 2 mM cGMP (filled circles) and 1.8 μM cGMP (open circles). The curves represent fits with Scheme I, with KDo = 14 μM and KDc = 300 nM.
Figure 7.
Summary of the effects of mutations in the S4/S5 region on affinity of open and closed channels for dequalinium. Regions of sequence from CNGA1 are shown in black and regions of sequence from CNGA2 are shown in gray.
We also studied the effect of mutating a positively charged amino acid in the turret region, which faces the external side of the channel (R342Y and R342D). This site is likely near to E363, which is known to interact with the blocker tetracaine (Fodor et al., 1997a). We found that these mutations had a small effect on the IC50 for block by dequalinium (see Table I). The R342 mutations, like E363 mutations, produced decreases in the favorability of opening, observed as increases in K1/2 for cGMP and decreases in the maximal activation by cAMP. Thus, the increased affinity of dequalinium block of R342Y and R342D channels is likely explained by the better binding of this closed-state selective blocker to channels with lower open probability.
DISCUSSION
Here, we have continued our study of dequalinium block of CNG channels. We have previously shown that dequalinium blocks CNGA1 channels in a state-independent manner at 0 mV. However, our experiments examining the voltage dependence of closed and open channels clearly show a state dependence at all potentials (Fig. 3). Although we cannot definitively reconcile these findings, we believe that differences in the experimental protocols used for the two types of experiments may be responsible. For the experiments at 0 mV, we held at this potential for 5 s and stepped to +20 mV every 5 s for 20 ms. In many patches, we observed that steady-state block could be reached only after waiting for 45 min (Fig. 2 C). In contrast, when examining voltage dependence, we held at 0 mV for 10 ms and stepped to various potentials for up to 20 s, until steady-state of block was achieved (Fig. 2, A and B). It is possible that using the latter experimental paradigm we missed a second, much slower, state-independent component of block. This hypothesis is supported by the steady-state level of block shown in the traces of Fig. 1 and the time course for block by dequalinium shown in Fig. 2 C. From the values of KD calculated from Scheme I (Fig. 7), we expect that, at saturating cGMP concentrations, 4 μM dequalinium should have produced a 50% block of the current. In fact, 250 nM produced a 50% block of the current at 0 mV (Fig. 1 A, left). These data indicate that the block observed at 0 mV likely contained an additional, very slow, state-independent component.
For the S4-S5 linker chimeras, state dependence was observed even at 0 mV. However, the efficacy of block observed was still greater than expected from the open- and closed-channel affinities calculated from the fits to the voltage dependence with Scheme I (Figs. 6 and 7). Whenever two affinities for a blocker are observed, the higher affinity site will always dominate at equilibrium. It may be that the increase in closed-state affinity of the chimeras allowed the state-dependent component to dominate over the slower, state-independent component of block. The slower component is clearly still contributing, as evidenced from the lesser amount of state dependence and the greater than expected efficacy of block observed at 0 mV.
State-dependent block by dequalinium resembles block of CNG channels by other compounds, such as the local anesthetic tetracaine. Tetracaine blocks CNG channels in a closed state-dependent fashion (Fodor et al., 1997a). In our previous study we showed that at the single-channel level, a low concentration of dequalinium (5 nM) reduced the open probability of CNGA1 channels by ∼50% at +60 mV. If block were equally effective in the closed and open states, we would expect the dwell times for the open and closed states to be reduced equally. Instead, we found a pronounced increase in closed dwell time with little or no change in the open dwell time (Rosenbaum et al., 2003). This much higher change in closed dwell time compared with open dwell time is predicted by the preferential block of closed channels compared with open channels outlined in Scheme I.
The current-voltage relation of block by dequalinium in both CNGA1 and CNGA2 channels shows that zδ = 1. The valence (z) for dequalinium is 2, consistent with block of dequalinium half-way through the pore (Hille, 2001). As discussed in our previous study (Rosenbaum et al., 2003), this explanation holds only if the membrane potential decays linearly across the pore and the dequalinium positive charges are close together, assumptions that are almost certainly untrue. A second possibility is that one of the charges of the molecule traverses the whole length of the channel's pore. It is more likely, however, that the zδ arises as a combination of an “effective valence” of dequalinium, z′, such that 1 < z′ < z interacting at some distance δ. The voltage dependence of dequalinium block in CNGA1 and CNGA2 thus suggests that the blocker interacts either within the pore or with an alternative region in the protein within the membrane's electric field.
Our experiments using the S4/S5-CNGA1 and S4/S5-CNGA2 chimeras indicate that mutations in the S4/S5-linker selectively increased the affinity of closed channels for dequalinium, without altering the affinity of open channels.
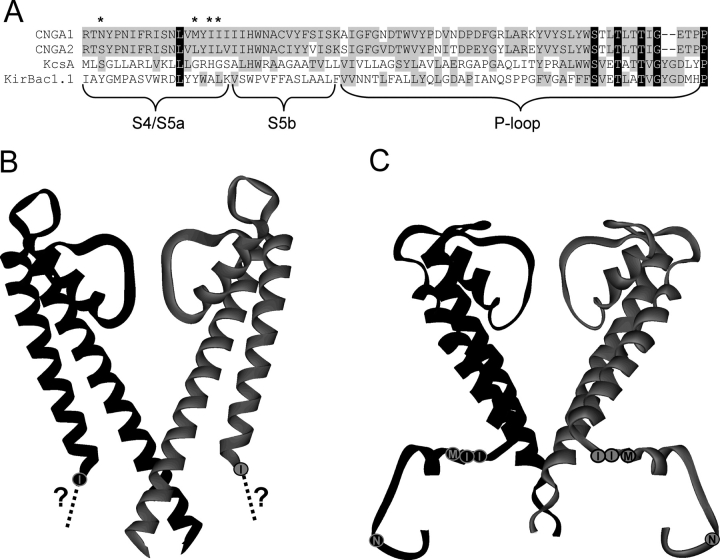
To determine where the mutations for the chimeras are located in the channels' structures, we compared the sequences of CNGA1 and CNGA2 channels to those of two bacterial potassium channels in which the crystal structure for the closed state has been resolved, KcsA (Doyle et al., 1998) and KirBac 1.1 (Kuo et al., 2003) in Fig. 8 A. As seen in Fig. 8 B, the most COOH-terminal of our chimeric mutations corresponds to the first amino acid in the crystal structure of KcsA, with the remaining chimeric mutations positioned before the region where the structure begins. In the KirBac 1.1 channel shown in Fig. 8 C, the amino acids corresponding to those we have mutated in the S4/S5 chimeras are clearly located in the intracellular side of the structure.
Figure 8.
Localization of amino acids of the S4/S5 chimeras in the channel structure. (A) Sequence alignment of four types of ion channels, CNGA1, CNGA2, KcsA, and KirBac 1.1. Asterisks denote amino acids mutated in the S4-S5 linker chimeras. (B) Structure of the KcsA K+ channel (Doyle et al., 1998). The circle with the “I” represents the position of one of the amino acids we mutated in the S4/S5 chimeras of CNG channels. The dotted line and the question mark indicated that the region of sequence containing the other S4-S5 linker mutations is not resolved. (C) Structure of the KirBac 1.1. K+ channel (Kuo et al., 2003). The positions analogous to the four mutations of the S4/S5 chimeras we produced are shown as circles.
Although sequence similarity in the region containing our mutations is low, the alignment of the sequences coding for the CNGA1, CNGA2, KcsA and KirBac 1.1 channels suggests that the region we have mutated is unlikely to line the ion-conducting pore. Given that the mutated residues map to intracellular regions in KcsA and KirBac 1.1 (Fig. 8), at least two mechanisms for the effects of the chimeras on dequalinium block must be considered. One is that the structure of CNG channels does not resemble that of the bacterial K+ channels in this region. The poor conservation of primary sequence in this region (Fig. 8 A) could mean that in CNG channels the mutated residues do indeed line the ion-conducting pore, at least in the closed state. Another possibility is that the structures of CNG and K+ channels are conserved in this region, and that the chimeric mutations lie outside the pathway for permeant ions. If this were the case, the mutated residues could alter dequalinium affinity either if they were part of the dequalinium binding site, or indirectly by altering the structure of other residues in the pore. Although we cannot explain the specifics of why the CNGA1 to CNGA2 and CNGA2 to CNGA1 mutations in this region both shifted the affinity of the blocker for the closed state in the same direction (more favorable), it may be that generic perturbations in this region favor the interaction of the blocker in the closed state.
Acknowledgments
We would like to thank: Professors W.N. Zagotta and R.R. Reed for their kind gifts of the CNGA1 and CNGA2 cDNA's, respectively; Li Hua for making some of the CNGA1-CNGA2 chimeras; and Professor Bertil Hille, Margorie Anderson, and Trisha Davis for creating a supportive research environment. We also thank Drs. F. Frank, M. Rosenbaum, W.N. Zagotta, and A. Stein for helpful discussion.
This work was funded by the National Eye Institute (R01EYI3007 to S.E. Gordon).
Olaf S. Andersen served as editor.
Abbreviations used in this paper: CNG, cyclic nucleotide–gated; PsTx, pseudechetoxin.
References
- Brown, R.L., T.L. Haley, K.A. West, and J.W. Crabb. 1999. Pseudechetoxin—a peptide blocker of cyclic nucleotide-gated ion channels. Biophys. J. 76:A7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capovilla, M., A. Caretta, L. Cervetto, and V. Torre. 1983. Ionic movements through light-sensitive channels of toad rods. J. Physiol. 343:295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colamartino, G., A. Menini, and V. Torre. 1991. Blockage and permeation of divalent cations through the cyclic GMP-activated channel from tiger salamander retinal rods. J. Physiol. 440:189–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, D.A. J. Morais Cabral, R.A. Pfuetzner, A. Kuo, J.M. Gulbis, S.L. Cohen, B.T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- Eismann, E., F. Muller, S.H. Heinemann, and U.B. Kaupp. 1994. A single negative charge within the pore region of a cGMP-gated channel controls rectification, Ca2+ blockage, and ionic selectivity. Proc. Natl. Acad. Sci. USA. 91:1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor, A.A., K.D. Black, and W.N. Zagotta. 1997. a. Tetracaine reports a conformational change in the pore of cyclic nucleotide-gated channels. J. Gen. Physiol. 110:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor, A.A., S.E. Gordon, and W.N. Zagotta. 1997. b. Mechanism of tetracaine block of cyclic nucleotide-gated channels. J. Gen. Physiol. 109:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, S.E., D.L. Brautigan, and A.L. Zimmerman. 1992. Protein phosphatases modulate the apparent agonist affinity of the light-regulated ion channel in retinal rods. Neuron. 9:739–748. [DOI] [PubMed] [Google Scholar]
- Gordon, S.E., and W.N. Zagotta. 1995. a. Localization of regions affecting an allosteric transition in cyclic nucleotide-activated channels. Neuron. 14:857–864. [DOI] [PubMed] [Google Scholar]
- Goulding, E.H., G.R. Tibbs, and S.A. Siegelbaum. 1994. Molecular mechanism of cyclic-nucleotide-gated channel activation. Nature. 372:369–374. [DOI] [PubMed] [Google Scholar]
- Haynes, L.W., A.R. Kay, and K.W. Yau. 1986. Single cyclic GMP-activated channel activity in excised patches of rod outer segment membrane. Nature. 321:66–70. [DOI] [PubMed] [Google Scholar]
- Hille, B. 2001. Ion Channels of Excitable Membranes. 3rd edition. Sinauer, Sunderland, MA. xviii, 814, [818] of plates pp.
- Hodgkin, A.L., P.A. McNaughton, and B.J. Nunn. 1985. The ionic selectivity and calcium dependence of the light-sensitive pathway in toad rods. J. Physiol. 358:447–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ildefonse, M., and N. Bennett. 1991. Single-channel study of the cGMP-dependent conductance of retinal rods from incorporation of native vesicles into planar lipid bilayers. J. Membr. Biol. 123:133–147. [DOI] [PubMed] [Google Scholar]
- Karpen, J.W., A.L. Zimmerman, L. Stryer, and D.A. Baylor. 1988. Gating kinetics of the cyclic-GMP-activated channel of retinal rods: flash photolysis and voltage-jump studies. Proc. Natl. Acad. Sci. USA. 85:1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, A., J.M. Gulbis, J.F. Antcliff, T. Rahman, E.D. Lowe, J. Zimmer, J. Cuthbertson, F.M. Ashcroft, T. Ezaki, and D.A. Doyle. 2003. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 300:1922–1926. [DOI] [PubMed] [Google Scholar]
- Molokanova, E., B. Trivedi, A. Savchenko, and R.H. KrAm. 1997. Modulation of rod photoreceptor cyclic nucleotide-gated channels by tyrosine phosphorylation. J. Neurosci. 17:9068–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani, K., and K.W. Yau. 1988. Calcium and light adaptation in retinal rods and cones. Nature. 334:69–71. [DOI] [PubMed] [Google Scholar]
- Quandt, F.N., G.D. Nicol, and P.P. Schnetkamp. 1991. Voltage-dependent gating and block of the cyclic-GMP-dependent current in bovine rod outer segments. Neuroscience. 42:629–638. [DOI] [PubMed] [Google Scholar]
- Root, M.J., and R. MacKinnon. 1993. Identification of an external divalent cation-binding site in the pore of a cGMP-activated channel. Neuron. 11:459–466. [DOI] [PubMed] [Google Scholar]
- Rosenbaum, T., M. Awaya, and S.E. Gordon. 2002. Subunit modification and association in VR1 ion channels. BMC Neurosci. 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, T., L.D. Islas, A.E. Carlson, and S.E. Gordon. 2003. Dequalinium: A novel, high-affinity blocker of CNGA1 channels. J. Gen. Physiol. 121:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetkamp, P.P. 1990. Cation selectivity of and cation binding to the cGMP-dependent channel in bovine rod outer segment membranes. J. Gen. Physiol. 96:517–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre, V., G. Rispoli, A. Menini, and L. Cervetto. 1987. Ionic selectivity, blockage and control of light-sensitive channels. Neurosci. Res. Suppl. 6:S25–S44. [DOI] [PubMed] [Google Scholar]
- Yellen, G. 1998. The moving parts of voltage-gated ion channels. Q. Rev. Biophys. 31:239–295. [DOI] [PubMed] [Google Scholar]
- Zimmerman, A.L., and D.A. Baylor. 1992. Cation interactions within the cyclic GMP-activated channel of retinal rods from the tiger salamander. J. Physiol. 449:759–783. [DOI] [PMC free article] [PubMed] [Google Scholar]