Abstract
An ancient genome duplication (PPP1) that predates divergence of the cereals has recently been recognized. We report here another potentially older large-scale duplication (PPP2) event that predates monocot-dicot divergence in the genome of rice (Oryza sativa L.), as inferred from the age distribution of pairs of duplicate genes based on recent genome data for rice. Our results suggest that paleopolyploidy was widespread and played an important role in the evolution of rice.
Keywords: Oryza sativa, Polyploidy, Genome evolution, Age distribution of duplicate genes, Monocot-dicot divergence
INTRODUCTION
Genome duplication or polyploidy is common in flowering plants (Wendel, 2000). An ancient polyploidy (paleopolyploidy) is difficult to detect because extensive gene loss or inactivation and chromosomal rearrangements can occur after a large-scale duplication. Genomic sequence analyses performed on yeast (S. cerevisiae), Arabidopsis and rice (Oryza sativa L.) provided strong evidence for ancient polyploidies (Wolfe and Shields, 1997; Arabidopsis Genome Initiative, 2000; Paterson et al., 2004; Vision et al., 2000; Goff et al., 2002; Simillion et al., 2002; Blanc and Wolfe, 2004). In these analyses, many non-overlapped duplicate regions with conserved gene order and orientation, or duplicate blocks have been detected intro-genome, and this has been considered to be evidence of polyploidy (Wolfe and Shields, 1997). The age distributions duplicate genes have also been widely used to infer potential polyploidy events in plant species (Vision et al., 2000; Goff et al., 2002; Blanc and Wolfe, 2004). Large-scale duplication events lead to a dramatic increase in the number of duplicate genes. Pairs of paralogs of a particular age that correspond to a large-scale duplication are expected to give rise to an independent peak in the age distribution. An initial peak that accounts for the most recently duplicated genes is also expected. Based on this method and data regarding unigenes, widespread paleopolyploidy has been found in many plant species, such as Arabidopsis, Zea mays etc. (Blanc and Wolfe, 2004).
A paleopolyploidy (PPP1) or “whole-genome duplication” in rice was first suggested by Goff et al.(2002), who based their suggestion on the age distribution of paralogous protein pairs on a genome scale and dated the event at around 40–50 million years ago. Based on their own early assembly of the unfinished genome sequence, Vandepoele et al.(2003) and Paterson et al.(2003) reported the duplication of rice chromatin. Paterson et al.(2004) found many clear non-overlapped duplicate blocks in the rice genome and suggested that a whole-genome duplication event occurred ~70 million years ago, which predates the divergence of the grass family based on the current genome sequence of rice. About 10 years ago, chromosomal duplications (such as chromosome 11–12 and 1–5 duplication) were reported in rice genome based on genetic maps using DNA markers (Kishimoto et al., 1994; Nagamura et al., 1995; Wang et al., 2000).
In this study based on the age distribution of duplicate genes and using an effective approach to control background noise, another potentially older paleopolyploidy (PPP2) that predates monocot-dicot divergence was detected in the rice genome.
MATERIALS AND METHODS
The rice (osa1, version 2.0) and Arabidopsis thaliana (ath1, version 5.0) genome annotation databases were downloaded from The Institute for Genomic Research (TIGR) (http://www.tigr.org).
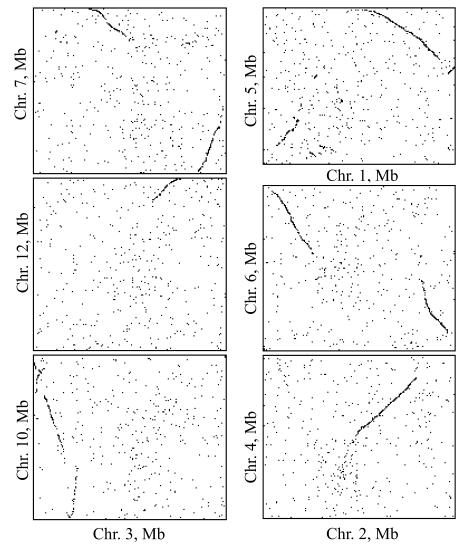
A total of 59712 annotated coding sequences of rice (version 2.0) and 26207 of Arabidopsis (version 5.0) encoded by their chromosomal order were compared by reciprocal BLASTN searching (E<10−14) for any two chromosomes. Two sequences were defined as one-to-one paralogous or pairs of a duplicate gene when each was the best hit of the other. Coding sequences that show a BLASTN match (E<10−10) with members of the rice and Arabidopsis repeat databases by TIGR should first be removed. A pair of duplicate genes identified by this method is presented as a single dot in Fig.1. Based on dot-plots of pairs of duplicate genes among chromosomes, 13 duplicate blocks corresponding to the latest ancient large-scale duplication (PPP1) and a recent duplication between chromosomes 11 and 12 were identified and duplicate blocks of PPP1 were used in the next analysis of distribution age.
Fig. 1.
Selected dot-plots of pairs of duplicate genes in the rice genome. Syntenic lines of duplicate blocks corresponding to the latest ancient large-scale duplication (PPP1) are discernible. Pairs of duplicate genes identified as one-to-one paralogs are presented as single dots. The annotation of the TIGR rice assembly (osa1, version 2.0) was used
Amino acid substitution rates (d A) were estimated by the method of Goff et al.(2002): protein sequences of one-to-one paralogs were used to estimate their d A values using the aaml program in the Phylogenetic Analysis by Maximum Likelihood (PAML) package (Yang, 1999) with the Dayhoff matrix. The divergence time was calculated based on a molecular clock rate of 9×10−10 nonsynonymous substitutions per site per lineage per year and 2.25 nonsynonymous substitutions per amino acid change.
RESULTS AND DISCUSSION
Many non-overlapping duplicate blocks, which cover almost all regions of rice chromosomes, have been reported using the first assembly (osa1, version 1.0) of the rice genome by TIGR (Paterson et al., 2004). Here, based on the current version (version 2.0), almost the same duplicate blocks were detected (Fig.1). An ancient large-scale duplication, or a whole-genome duplication (PPP1) and a recent segmental duplication between chromosomes 11 and 12 (11–12 duplication, data not shown) are involved in the formation of the duplicate blocks in Fig.1. The ancient whole-genome duplication was believed to have occurred about 70 million years ago, and thus predates the divergence of the cereals and also the latest detectable ancient polyploidy (PPP1) event in the rice genome.
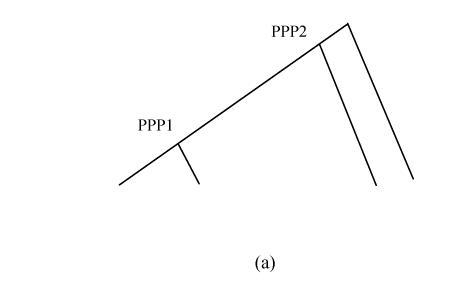
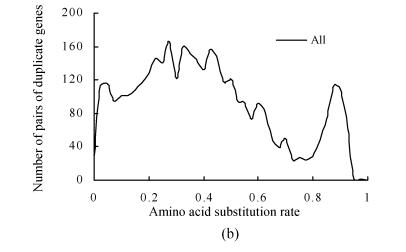
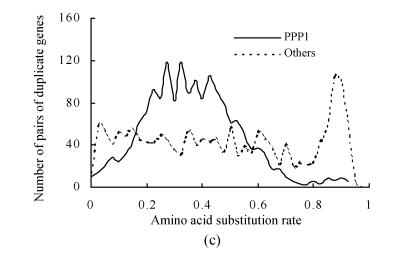
The amino acid substitution rates (d A) of pairs of duplicate genes on duplicate blocks of PPP1 were calculated. Three clear peaks were observed in the frequency distribution of their d A values (Fig.2b). The duplicate gene pairs on syntenic lines of duplicate blocks that corresponded to PPP1 were further separated from the overall distribution. The results showed that PPP1 gave rise to the first peaks (Fig.2c). The second peak was still retained and strongly suggested another older large-scale duplication, which may have been a whole-genome duplication (PPP2) for the rice genome (Fig.2a). The peak value of the distribution of PPP2 was ~0.875, which suggests that the ancient polyploidy event occurred about 220 million years ago, which predates monocot-dicot divergence, with an assumption of a molecular clock.
Fig. 2.
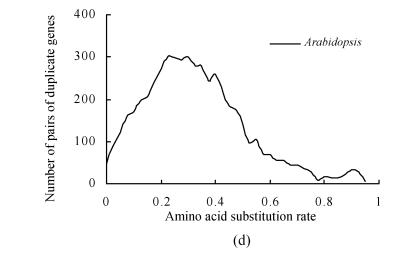
Frequency distributions of amino acid substitution rates (d A) obtained from pairs of duplicate genes on duplicate blocks of the latest large-scale duplication (PPP1) in the rice genome. These distributions suggest another potential older large-scale duplication (PPP2). (a) A phylogenic tree with suspected two large-scale duplication events: PPP1 that predates the divergence of the cereals (Paterson et al., 2004) and PPP2 that predates monocot-dicot divergence (this study); (b) Frequency distributions of d A values obtained from pairs of duplicate genes on duplicate blocks of PPP1; (c) Compositional analysis of (b). Pairs of duplicate genes of PPP1 were divided. In addition to the peaks in age distribution corresponding to PPP1, another obvious peak that suggests an older large-scale duplication (PPP2) was detected; (d) Frequency distributions of amino acid substitution rates obtained from pairs of duplicate genes among chromosomes (inter-chromosome) in the Arabidopsis genome. Pairs of duplicate genes were identified using the same method as in rice
A relatively small initial peak was observed in our study. Only inter-chromosome pairs of duplicate genes were used in our analysis and mass intro-chromosome pairs of new duplicate genes were excluded. Therefore, the initial peak in our figures will be small.
One concern in our study is the reality of the ancient peak in the age distribution detected in the rice genome. It is located at a very marginal position, which raises the question of whether this is a ghost peak. Three steps were used to prevent such mistakes in this study: (1) masking of repeat sequences (genes); (2) a rigid reciprocal BLASTN best-hit search method was used to identify pairs of duplicate genes, so-called one-to-one paralogs, in a genome; and (3) a stricter standard was used to identify one-to-one paralogs in this study: a successful pair must first have less than an 10−14 BLASTN match. These three steps basically prevent the misidentification of un-related genes as pairs of one-to-one paralogs. Meanwhile, the age distribution of pairs of duplicate genes from Arabidopsis was estimated using the same method that we used for the rice genome. No similar ancient peak was observed in the Arabidopsis genome (Fig.1d). The results suggest that the ancient peak in the age distribution observed in the rice genome is not an artifact of our method, but rather is an actual peak that is due to a potential ancient polyploidy.
Our analysis of the Arabidopsis genome failed to show clear evidence of a polyploidy that predated monocot-dicot divergence (Fig.1d), although a large-scale duplication near or prior to the divergence of monocots from the dicot had been reported (Vision et al., 2000; Simillion et al., 2002; Chapman et al., 2004). It is possible that rapid evolution in Arabidopsis has made it difficult to detect the ancient polyploidy. In contrast, rice has been suggested to have an evolutionarily stable genome (Ilic et al., 2003).
Controlling background noise is an important part of large-scale genome analysis. Long evolutionary processes make it difficult to identify the signals of ancient events in a genome. Two key steps were used to control background noise in this study: (1) Only inter-chromosome pairs of duplicate genes were used: both small-scale (gene level) and large-scale duplication played significant roles in genome evolution (Gu et al., 2002). Many small-scale duplications occur intro-chromosome and can significantly affect the detection of large-scale duplication events; (2) Only pairs on duplicate blocks of PPP1 were used: clear, long and intact duplicate blocks of the latest ancient polyploidy were identified in the rice genome. No similar long and intact duplicate blocks have yet been observed in other species. Such distinct blocks provide a sound basis for further searching for older evolutionary events in the rice genome. The hidden footprints of some ancient evolutionary events were uncovered and therefore became detectable.
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 30270810, 90208022 and 30471067) and IBM Shared University Research (Life Science), China
References
- 1.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 2.Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman BA, Bowers JE, Schulze SR, Paterson AH. A comparative phylogenetic approach for dating whole genome duplication events. Bioinformatics. 2004;20:180–185. doi: 10.1093/bioinformatics/bth022. [DOI] [PubMed] [Google Scholar]
- 4.Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Quail PH. A draft sequence of the rice genome (Oryza sativa L. ssp japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 5.Gu X, Wang Y, Gu J. Age distribution of human gene families shows significant roles of both large- and small-scale duplications in vertebrate evolution. Nature Genet. 2002;31:205–209. doi: 10.1038/ng902. [DOI] [PubMed] [Google Scholar]
- 6.Ilic K, SanMiguel PJ, Bennetzen JL. A complex history of rearrangement in an orthologous region of the maize, sorghum, and rice genomes. Proc. Natl. Acad. Sci. USA. 2003;100:12265–12270. doi: 10.1073/pnas.1434476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kishimoto N, Higo H, Abe K, Arai S, Saito A, Higo K. Identification of the duplicated segments in rice chromosomes 1 and 5 by linkage analysis of cDNA markers of known functions. Theor. Appl. Genet. 1994;88:722–726. doi: 10.1007/BF01253976. [DOI] [PubMed] [Google Scholar]
- 8.Nagamura Y, Inoue T, Antonio B, Shimano T, Kajiya H, Shomura A, Lin S, Kuboki Y, Harushima Y, Kurata N, et al. Conservation of duplicated segments between rice chromosomes 11 and 12. Breed Sci. 1995;45:373–376. [Google Scholar]
- 9.Paterson AH, Bowers JE, Peterson DG, Estill JC, Chapman BA. Structure and evolution of cereal genomes. Curr. Opin. Genet. Dev. 2003;13:644–650. doi: 10.1016/j.gde.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simillion C, Vandepoele K, Van Montagu MCE, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vandepoele K, Simillion C, Van de Peer Y. Evidence that rice and other cereals are ancient aneuploids. Plant Cell. 2003;15:2192–2202. doi: 10.1105/tpc.014019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis . Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Liu K, Zhang Q. Segmental duplications are common in rice genome. Acta. Bot. Sin. 2000;42:1150–1155. [Google Scholar]
- 15.Wendel JF. Genome evolution in polyploids. Plant. Mol. Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- 16.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z. Phylogenetic Analysis by Maximum Likelihood (PAML) Version 2. London, UK: University College; 1999. [Google Scholar]