Abstract
Trichoderma/Hypocrea is a genus of soil-borne or wood-decaying fungi containing members important to mankind as producers of industrial enzymes and biocontrol agents against plant pathogens, but also as opportunistic pathogens of immunocompromised humans. Species identification, while essential in view of the controversial properties of taxa of this genus, has been problematic by traditional methods. Here we will present a critical survey of the various identification methods in use. In addition, we will present an update on the taxonomy and phylogeny of the 88 taxa (which occur as 14 holomorphs, 49 teleomorphs and 25 anamorphs in nature) of Trichoderma/Hypocrea that have been confirmed by a combination of morphological, physiological and genetic approaches.
Keywords: Trichoderma/Hypocrea, Genus, Molecular taxonomy-phylogeny
WHY IS TRICHODERMA IMPORTANT?
The anamorphic fungal genus Trichoderma (Hypocreales, Ascomycota) contains cosmopolitan soil-borne fungi frequently also found on decaying wood (Samuels, 1996; Klein and Eveleigh, 1998), of which some are economically important producers of industrial enzymes (Trichoderma reesei=Hypocrea jecorina) (Kubicek and Penttilä, 1998) and antibiotics (Sivasithamparam and Ghisalberti, 1998), or have application as biocontrol agents against plant pathogens (i.e. T. harzianum=H. lixii; T. atroviride=H. atroviridis; T. asperellum) (Hjeljord and Tronsmo, 1998). More recently, T. longibrachiatum has also become known as opportunistic pathogen of immunocompromised mammals including humans (Kredics et al., 2003). Trichoderma has been recognized to comprise a significant amount of fungal biomass in soil (Nelson, 1982; Widden and Abitbol, 1980) and is frequently present as an indoor contaminant (Thrane et al., 2001). These diverse implications of Trichoderma/Hypocrea with human society render an accurate species and strain identification to be an important issue. However, classical approaches based on the use of morphological criteria are, as in several other fungi, difficult to apply to Trichoderma, due to the plasticity of characters. As a consequence, we shall review the current state of perception of taxa and other taxonomic ranks in Trichoderma and Hypocrea in this paper.
TOWARDS DEVELOPMENT OF A CONCEPT FOR THE GENUS TRICHODERMA
Although originally introduced by Persoon (1794), the taxonomy and identification of Trichoderma has remained problematic until relatively recently. Until 1969, nearly all strains of Trichoderma were identified in literature as “T. viride” (e.g. also all the cellulase-producing strains of T. reesei) owing to Bisby’s (1939) concept that Trichoderma consists of a single species. This led to the erroneous statement even in textbooks that “T. viride is an industrial cellulose producer”. As T. viride sensu stricto is a relatively rare species (1Kubicek, unpublished data; 2Samuels, personal communications), most of the taxa sampled and determined before 1969 are probably misidentified. Rifai (1969) adopted the concept of ‘aggregate’ species, and distinguished nine ‘species aggregates’, and admitted that some of them (particularly T. hamatum) likely contain two or more morphologically indistinguishable species. Bissett (1984;1991a;1991b;1991c;1992; Gams and Bissett, 1998) revised Rifai’s aggregate species by subtle recognition of continuities of morphological characters, expanded the morphological criteria to accommodate the wider range of morphological variation expressed by some anamorphs of Hypocrea and adopted some forms previously included in Gliocladium. While dissecting several of Rifai’s species aggregates into several defined taxa, Bissett (1991a) also established a subdivision of the genus into five sections: Longibrachiatum, Pachybasium, Trichoderma, Saturnisporum and Hypocreanum.
The advent of molecular tools for investigations in fungal taxonomy prompted research in the mid-nineties to re-assess the morphology-based taxonomy in Trichoderma. The laboratories of G.J. Samuels (Beltsville, MD, USA), T. Börner (Berlin, FRG) and C.P. Kubicek (Vienna, Aust.) collaboratively pioneered a revision of Bissett’s section Longibrachiatum. They combined the use of molecular markers (ITS1 and ITS2 sequence analysis, RAPD), physiological (isoenzyme analysis) and phenetic characters, and also for the first time included an analysis of potential teleomorphs of the Trichoderma spp. from this section (Kuhls et al., 1996; 1997; Samuels, 1996; Samuels et al., 1998; Turner et al., 1997). As a result, section Longibrachiatum was recognized to be monophyletic and to contain ten taxa, within which four pairs displayed teleomorph-anamorph relationships: H. schweinitzii/T. citrinoviride; H. pseudokoningii/T. pseudokoningii; H. jecorina/T. reesei and H. orientalis/T. longibrachiatum, p/p. They further merged section Saturnisporum, which included only two species, T. saturnisporum and T. ghanense (Doi et al., 1987), with section Longibrachiatum, and recognized the synonymy of T. ghanense with T. parceramosum. Yet, as a whole, the concept of section Longibrachiatum as defined by Bissett (1984) was largely confirmed, suggesting a degree of correlation between morphological and molecular approaches to taxonomy in Trichoderma.
Section Longibrachiatum is a comparably small section of Trichoderma, and phylogenetically most distant from the other sections. The relationship between morphological characters and molecular phylogeny became more complex, however, when larger sections of Trichoderma were investigated. Kindermann et al.(1998) attempted a first phylogenetic analysis of the whole genus. Using sequence analysis of the ITS1 region of rDNA they found that the largest section, section Pachybasium, is actually paraphyletic. Although the use of a single gene fragment alone is insufficient by today’s standards, this finding has been confirmed by phylogenetic analysis of several other genes (Kullnig-Gradinger et al., 2002; Chaverri et al., 2003b). In this context it is noteworthy that the nomenclatural type strain of sect. Pachybasium, Pachybasium hamatum (Bonord.)=T. hamatum, is not a member of the major one of the two Pachybasium clades (clade B) (Kindermann et al., 1998 ), but clusters together with T. pubescens and T. strigosum in a clade otherwise containing almost all taxa (i.e. with the exception of T. aureoviride) from section Trichoderma. Because of the lack of a distinctive morphological hiatus between clades A and B of Pachybasium (Kindermann et al., 1998), researchers have so far maintained the name Pachybasium for both clades, but it is clear that this is an unsatisfactory situation from a taxonomic point of view. Pachybasium B contains all taxa attributed to this section by Bissett (1991b) with the exception of the three mentioned above. In addition, Pachybasium B also poses the problem of strong genetic variability of several of its species (e.g. T. harzianum). Since its species (both ana- as well as teleomorphs) account for the majority of taxa found in field investigations (Kullnig et al., 2000; Kubicek et al., 2003; Wuczskowski et al., 2003; Gherbawy et al., 2004; Druzhinina et al., 2004b), this raises the question of how to identify species in this large heterogenous group.
WHAT IS A SPECIES IN TRICHODERMA?
Most of the taxa of Trichoderma have so far been defined on the basis of morphology, and gene sequence analysis was only used as a confirmative or distinctive complement. Thus, members of the genus are primarily defined by the application of the MSR (morphological species recognition) concept. Since strains of Trichoderma spp. apparently cannot be crossed, application of the BSR (biological species recognition) concept is impracticable. The genealogical concordance phylogenetic species recognition (GCPSR) (Taylor et al., 2000) is an attractive alternative or complement to the morphological species concept, but has not been widely applied to Trichoderma/Hypocrea. It requires the analysis of trees of several unlinked genes, and implies that the phylogenetic position of a true species will be concordant in at least some of them, and not be contradicted in the others. Kullnig-Gradinger et al.(2002) used four different loci (ITS1 and ITS2, mitssuDNA, short fifth tef1α intron, a fragment of ech42 large exon) to assess a global phylogeny of the genus. However, a stringent clade to clade concordance was not possible for most of the species because of insufficient phylogenetic resolution by the genes used. Chaverri et al.(2003a) analyzed ITS1 and ITS2, large tef1 intron, and short fragments of the actin (act1) and calmodulin (cal1) exon sequences in H. lixii/T. harzianum, and identified seven phylogenetic lineages which were concordant in most trees. However, since the isolates within the seven lineages could not be reliably distinguished morphologically, they were not given recognition as a species.
Taylor et al.(1999) proposed basing phylogenetic species concepts on the concordance between five or more gene trees. This requirement is not easily fulfilled in Trichoderma. In the past, most researchers made heavy use of ITS1 and/or ITS2 (Kuhls et al., 1997; Kindermann et al., 1998; Lieckfeldt et al., 1998; 2001; Dodd et al., 2000), because this gene cluster is present in >90 copies per genome and can thus be easily amplified. However, the use of ITS1 and ITS2 has meanwhile become discredited by the fact that some fungi, notably some sections in the closely related Fusarium, and plants have been shown to contain paralogous copies of the parts of the rDNA gene cluster (O’Donnell, 1992; Buckler et al., 1997; O’Donnell et al., 1998; Lieckfeldt and Seifert, 2000). Also, Chaverri et al.(2003b) reported unpublished data for the presence of divergent ITS1 and ITS2 sequences in T. strictipile and T. crassum. In contrast, we have so far not obtained evidence for the presence of paralogous ITS1 or ITS2 copies in most species of Trichoderma/Hypocrea, even though we specifically tested for it (1Mach et al., unpublished data). On the other hand, a serious drawback of the use of ITS1 and ITS2 is that it provides only poor phylogenetic resolution in some clades, particularly Pachybasium B (Kullnig-Gradinger et al., 2002; Chaverri et al., 2003a). Among 11 tested loci/fragments (Table 1) the most promising loci seem to be different fragments of translation elongation factor 1-alpha (EF-1α=tef1) different fragments of which were sequenced by different research groups (Fig.1). The gene has been cloned from H. jecorina and exceeds 2 kb in length, and consisted of several relatively large and variable introns and exons. Also, the coding portions of endochitinase 42 (ech42) and RNA-polymerase subunit 2 (rpb2) have displayed significant intra- and interspecific variability while other genes, such as the D1 and D2 regions of the 28S rDNA, or the small subunit of the mitochondrial ribosomal DNA (ssu-mDNA) have been used with limited success. Fragments of the calmodulin- and the actin-encoding genes (cal1, act1) had also been used in T. harzianum/H. lixii, but exhibited less powerful resolution than the large exon of tef1 (Table 1) (Chaverri et al., 2003a). It is important to mention that two different unalignable intron fragments of act1 are available in GenBank (Table 1).
Table 1.
Gene sequences used in molecular phylogeny of Trichoderma/Hypocrea
| Locus | Fragment | Length (kb) | N | Var. (%)* | Reference | |
| Internal transcribed spacer 1 and 2 | ITS1 and ITS2 | 0.4 | 50 | 25 | Kullnig-Gradinger et al., 2002 | |
| RNA coding genes | ||||||
| Small subunit of the mitochondrial ribosomal DNA | ssu-mDNA | 0.4 | 50 | 5 | Kullnig-Gradinger et al., 2002 | |
| 28S rDNA gene | 28S rDNA | 0.5 | 51 | 8 | Kullnig-Gradinger et al., 2002 | |
| Protein coding genes | ||||||
| Calmodulin | cal1 | Intron | 0.45 | 35** | 15 | Chaverri et al., 2003a |
| Actin | act1 | 1st intron | 0.35 | 35** | 5 | Chaverri et al., 2003a |
| 2nd intron | 0.8 | 18** | 6 | 1Samuels and Ismael, personal communication | ||
| RNA polymerase B subunit 2 | rpb2 | 0.4 | 97 | 40 | Chaverri et al., 2004 | |
| Endochitinase 42 | ech42 | Last large exon | 0.6 | 44*** | 33 | Kullnig-Gradinger et al., 2002 |
| Translation elongation factor 1-alpha | tef1 | 5th (small) intron | 0.1 | 47 | 30 | Kullnig-Gradinger et al., 2002 |
| 4th (large) intron | 0.35 | 125** | 29 | 2Druzhinina, unpublished data | ||
| Last large exon | 0.7 | 84 | 29 | Chaverri et al., 2004 | ||
Portion of parsimoni informative sites from the length of fragment
Only tested for closely related taxa
Data for section Longibrachiatum are not available
N indicates number of species investigated
Fig. 1.
A schematic structure of tef1 gene of H. jecorina (GeneBank accession number CAA80554), and location of primers used to amplify different parts of it for phylogenetic inferences
Notwithstanding the fact that both regions exhibit an extremely weak phylogenetic signal it is important to reach an agreement between workers regarding which fragment of this gene should be sequenced. We have also attempted to use histon 3A and β-tubulin gene sequences, which have proven worthwhile for phylogenetic analysis in Fusarium and other genera, but while the variation in the histon 3A gene was not high, several Trichoderma spp. contained multiple heterologous copies of the tub1 gene, thus rendering both genes not applicable for this purpose (2Kullnig-Gradinger and Kubicek, unpublished data). Unfortunately, none of these genes is optimal for phylogenetic resolution of the whole genus, or of large clades such as Pachybasium B: while the largest intron of tef1 provides excellent resolution and high clade support for closely related taxa such as the T. harzianum/H. lixii species clade (H. lixii, T. harzianum, T. aggressivum, T. tomentosum, T. cerinum, T. velutinum, H. tawa) or the group of H. rufa (T. viride, T. atroviride, T. koningii). However, the large introns in tef1 are less suited for resolving the phylogenetic elations of more distantly related species due to ambiguous alignments (2Druzhinina, unpublished data). On the other hand, the last large exon of tef1 contains only limited phylogenetic signals for analysis of diverse clades, and thus e.g. in a combined analysis of Pachybasium A and B resulted in lack of support for almost all basal branches, whereas the terminal branches had good support (Chaverri et al., 2003b; 2004). The same problem was even more apparent with rpb2. Therefore, the optimal combination of genes allowing the application of the GCPSR concept on the whole genus Trichoderma has not yet been applied. Detailed analysis of various loci available in GenBank (Table 1) may suggested that the simultaneous usage of (i) tefl arge intron and last large exon, (ii) rpb2 gene, (iii) ech42 last large exon and (iv) ITS1 and 2 as diagnostic regions may lead to the most reliable phylogeny. However a search for new phylogenetic markers is strongly recommended.
A further important point in the use of the GCPSR concept is the justified use of the correct phylogenetic approach, so far, most workers employed the maximum parsimony method to analyze sequence data, which does not employ any modelling of evolution and therefore poses several problems when dealing with either very closely or very distantly related taxa (Salemi and Vandamme, 2003). Maximum likelihood methods would be adequate, but suffer from the long computation time they usually require. For this purpose, the Bayesian approach to phylogenetic inferences represents the most recent advance in phylogenetic analysis (Rannala and Yang, 1996; Huelsenbeck and Ronquist, 2001; Lutzoni et al., 2001). It shifts statistical interference away from an emphasis on hypothesis testing (P-values) and finding the single optimal tree, toward obtaining adequate estimates of uncertainty. Uncertainty is characterized through the use of the posterior distribution of a parameter, which is defined as the conditional probability of observing a particular parameter value given in the data. This involves the use of Markov chain Monte Carlo (MCMC) methods (Metropolis et al., 1953), which implements the simulation of a random walk through parameter space, which, if run long enough, will eventually converge on the stationary distribution of parameters (Lewis, 2001). Bayesian approaches have been introduced in the analysis of phylogenies of other genera, and had also been applied to Trichoderma/Hypocrea (Chaverri et al., 2003b; Druzhinina et al., 2004a). When combined with rigorous testing of evolutionary models, this approach yields excellent resolution even for difficult to resolve clades in Trichoderma (1Druzhinina et al., manuscript in preparation).
HOW CAN SPECIES OF TRICHODERMA BE IDENTIFIED?
While this review so far treated the concepts and advances in the recognition and definition of species of Trichoderma, nothing has been said about how an unknown isolate can be identified as one of these species. Morphological analysis is highly prone to error, and consequently roughly 50% of the Trichoderma spp. deposited in culture collections under names obtained by morphological analysis alone are wrong (2Kubicek, unpublished data)! Obviously, subjecting every new strain to a thorough GCPSR analysis would result in a safe identification, but most workers who study the occurrence of Trichoderma or Hypocrea in soil or other habitats will not invest such a massive use of money and time. In fact, several researchers are still mainly using morphological methods for identification of Trichoderma, although the use of ITS1 and ITS2 sequence analysis is becoming more and more popular. This is not in contradiction to the statement above that ITS1 and ITS2 do not provide sufficient phylogenetic resolution, because phylogeny and diagnosis underlie two different principles, i.e. even though the change in 2–3 nts may not provide sufficient phylogenetic information, it may be indicative of a given species if it is known that this species always contains these three nucleotides in the respective positions. Hence the intra- and interspecific variability must be fully known in order to confirm or refute species identity. Unfortunately, the preferred process used by some workers is to submit sequences to a BLAST, and then accept the best hit as species identity. This must be criticized for several reasons: first, the GenBank databases contain many sequences of Trichoderma isolates which had been incorrectly identified and thus occur under a false name (Table 2); second, many researchers without solid bioinformatic training are inexperienced with the principles of BLAST and take an E-value of 0.00 as identity without scrutinizing whether the sequence is identitical or only highly similar. Even if it is identical, it is imperative to know that no other species with the same sequence exists.
Table 2.
Example of a BLAST search with an ITS1 and ITS2 sequence from Trichoderma * which yielded ambiguous results
| Sequences producing significant alignments: | Score (bits) | E-value | |
| gi|19880152|gb|AF362109.1| | Trichoderma aureoviride strain T... | 1132 | 0.0 |
| gi|19880083|gb|AF359399.1| | Trichoderma aureoviride strain T... | 1116 | 0.0 |
| gi|32394933|gb|AY154947.1| | Trichoderma aggressivum Ir. 560 ... | 1112 | 0.0 |
| gi|19880067|gb|AF359267.1| | Trichoderma aureoviride strain T... | 1108 | 0.0 |
| gi|19032416|gb|AF345948.1| | Trichoderma harzianum isolate GJ... | 1100 | 0.0 |
| gi|19880081|gb|AF359397.1| | Trichoderma aureoviride strain T... | 1100 | 0.0 |
| gi|21239369|gb|AF501330.1| | Trichoderma aggressivum f. europ... | 1096 | 0.0 |
| gi|3095175|gb|AF055216.1|AF055216 | Trichoderma harzianum str... | 1082 | 0.0 |
| gi|27448757|gb|AF443912.1| | Trichoderma harzianum G.J.S. 00–... | 1045 | 0.0 |
| gi|9230639|gb|AF194019.1|AF194019 | Trichoderma aureoviride s... | 1045 | 0.0 |
| gi|9230631|gb|AF194011.1|AF194011 | Trichoderma harzianum str... | 1045 | 0.0 |
| gi|9230630|gb|AF194010.1|AF194010 | Trichoderma aureoviride s... | 1045 | 0.0 |
| gi|27448762|gb|AF443917.1| | Hypocrea lixii G.J.S. 91-138 int... | 1041 | 0.0 |
| gi|27448761|gb|AF443916.1| | Hypocrea lixii G.J.S. 94-53 inte... | 1041 | 0.0 |
| gi|27448758|gb|AF443913.1| | Trichoderma harzianum G.J.S. 00–... | 1041 | 0.0 |
| gi|27448771|gb|AF443926.1| | Hypocrea lixii G.J.S. 90-254 int... | 1039 | 0.0 |
| gi|27448769|gb|AF443924.1| | Hypocrea lixii G.J.S. 92-110 int... | 1037 | 0.0 |
| gi|27448764|gb|AF443919.1| | Hypocrea lixii G.J.S. 92-100 int... | 1037 | 0.0 |
| gi|27448760|gb|AF443915.1| | Hypocrea lixii G.J.S. 90-22 inte... | 1037 | 0.0 |
| gi|32394935|gb|AY154949.1| | Trichoderma harzianum Ir. 112 C ... | 1033 | 0.0 |
| gi|27448770|gb|AF443925.1| | Trichoderma harzianum G.J.S. 92–... | 1033 | 0.0 |
| gi|1813651|gb|U78881.1|THU78881 | Trichoderma harzianum isola... | 1033 | 0.0 |
| gi|32394941|gb|AY154955.1| | Trichoderma inhamatum Ir. 286 18... | 1029 | 0.0 |
Sequence AF 3362109 was used as query (T. harzianum, wrongly deposited as “T. aureoviride”), using BLASTN
As a solution to this problem, we have recently developed a DNA-barcode system for quick identification on the basis of defined nucleotide sequence differences in the ITS1 and ITS2 region (Druzhinina et al., 2004b). The method relies on the discriminatory power of ITS1 and ITS2, which could be shown to be capable of identifying 70 of a total of 77 investigated species of Trichoderma and Hypocrea. Some taxa could not be distinguished because of ITS1 and ITS2 sequence identity: T. crassum and T. longipile, which however, once delimitated by the barcode, can then easily be distinguished on the basis of morphology (Bissett, 1991b; Chaverri et al., 2003b); and T. tomentosum and T. cerinum. Although, the latter two are also difficult to distinguish morphologically, phylogenetic analysis has shown that the latter two taxa are the result of a recent allopatric speciation, T. cerinum found only in Eurasia whereas T. tomentosum only in the Americas (1Druzhinina et al., manscript in preparation). Therefore, knowledge of the geographic origin of an isolate with a T. tomentosum/T. cerinum sequence may be used to distinguish these two. Otherwise, the two must be distinguished by the aid of additional sequences or phenotype arrays (see below).
ITS1 and ITS2 sequence differences were also unable to consistently distinguish between three taxa from the “H. rufa species complex” namely T. viride, T. koningii and T. atroviride. This is not because no eventually appropriate nt-differences would be detected, but because the species concept of this clade is currently under revision and several additional new taxa will be defined among them (2Samuels, personal communication). Sequence analysis of the large tef1 intron has already been shown to distinguish groups within T. viride and T. koningii, and will probably be the method of choice to complement the ITS based DNA-barcode for identification of these taxa in future.
While we think that DNA-barcode will enable many researchers to reliably identify their Trichoderma strains, it is obvious that some researchers will not have access or financial capability to use DNA-based methods, and therefore need alternatives. Samuels and colleagues, advocating the morphologically and physiologically based methods, proposed an interactive key for strain identification in Trichoderma, which, besides subtile differences in morphology (http://nt.ars-grin.gov/taxadescriptions/keys/TrichodermaIndex.cfm), makes use of differences in growth rates on PDA and SNA at 15, 20, 25, 30 and 35 °C (Chaverri et al., 2003b). This method is inexpensive, but is time consuming and requires a sufficient number of repetitions (n>5) for each sample to be reliable, thus becoming laborious with more than 50 samples being considered as the lowest limit for any ecological investigation. 1Bissett (unpublished) introduced a phenetic method, based on quantification of carbon assimilation patterns using BIOLOG MicroPlates™ (Biolog, Hayward, CA). Generally, phenotype arrays, commercialized by Biolog for identification of food and air-borne fungi, are not as suited for taxonomic purposes in Trichoderma because of the significant strain variation in some species (Kubicek et al., 2003). However, we have recently included phenotype arrays into an ‘integrated approach’ when describing the new taxon T. brevicompactum (Kraus et al., 2004). Phenotype array analysis of this new species and the phylogenetically closest neighbors method identified a number of carbon sources within the arrays, which were assimilated by T. brevicompactum at statistically different rates. This analysis is rapid and reliable, and even researchers for which phenotype arrays and/or the necessary reader are unavailable can use the published information to design simple agar plates with the respective carbon sources to verify an identification. We have recently used this system for distinguishing a number of species pairs which are morphologically very similar (e.g. T. tomentosum and T. cerinum; T. harzianum and T. aggressivum) (2Druzhinina and Bissett, unpublished data) and will continue to support by this method new species identified by GCPSR.
WHAT DO THE CURRENT DATA TELL US ABOUT THE PHYLOGENETICS OF TRICHODERMA?
Notwithstanding the limitations and caveats outlined above, the use of molecular tools enabled the taxonomy of Trichoderma to advance strongly over the last years, and we will therefore attempt to summarize the outcome. So far, 88 taxa have been recently redefined by combination of molecular and phenetic tools (Table 3). Four taxa introduced by Chaverri et al.(2004) (Table 4) still lack phylogenetic analysis. Among the 88, 14 anamorph-teleomorph relationships have been demonstrated and thus represent holomorphs, 49 have been described in Hypocrea, while the remaining 25 were described as Trichoderma. In the latter, two cases, the other (sexual/asexual) form has not been found in nature. It is possible (but unlikely) that many of these Hypocrea species occur naturally only in their teleomorph state and that many of the hitherto described Trichoderma species may lack a teleomorph state due to clonal evolution.
Table 3.
Current status of Trichoderma and Hypocrea taxa, and their attribution to phylogenetic sections and clades*
| Section | Clade1 | Anamorph2 | Teleomorph2 | Reference |
| Longibrachiatum | ||||
| T. longibrachiatum | H. orientalis | Samuels et al., 1998 | ||
| T. citrinoviride | H. schweinitzii | Samuels et al., 1998 | ||
| T. reesei | H. jecorina | Samuels et al., 1998 | ||
| T. ghanense | Samuels et al., 1998 | |||
| T. pseudokoningii | H. pseudokoningii | Samuels et al., 1998 | ||
| T. saturnisporum | Samuels et al., 1998 | |||
| T. konilangbra | Samuels et al., 1998 | |||
| T. effusum | Bissett et al., 2003 | |||
| T. sinensis | Bissett et al., 2003 | |||
| T. sp. MA | Wuczskowski et al., 2003 | |||
| H. andinensis | Samuels et al., 1998 | |||
| H. novazelandia | Samuels et al., 1998 | |||
| H. cerebriformis | 3Kubicek, unpublished data | |||
| H. poronoidea | 3Kubicek, unpublished data | |||
| H. peltata | Dodd et al., 2002 | |||
| Trichoderma | ||||
| H. pezizoides | Chaverri et al., 2004 | |||
| H. avellanea | Chaverri et al., 2004 | |||
| Rufa | T. viride | H. rufa | Lieckfeldt et al., 1999 | |
| T. atroviride | H. atroviridis | Dodd et al., 2003 | ||
| T. koningii | H. koningii | Lieckfeldt et al., 1998 | ||
| T. strigosum | Kullnig-Gradinger et al., 2002 | |||
| T. ovalisporum | Samuels, 2004 | |||
| T. erinaceum | Bissett et al., 2003 | |||
| H. stilbohypoxyli | Lu and Samuels, 2004 | |||
| Pachybasium A | T. hamatum | Kullnig-Gradinger et al., 2002 | ||
| T. pubescens | Kullnig-Gradinger et al., 2002 | |||
| T. asperellum | Kullnig-Gradinger et al., 2002 | |||
| H. neorufa | Dodd et al., 2002 | |||
| H. flavoconidia | Druzhinina et al., 2004a | |||
| Pachybasium B | ||||
| Pachybasioides | T. polysporum | H. pachybasioides | Lu et al., 2004 | |
| T. minutisporum | H. minutispora | Lu et al., 2004 | ||
| T. piluliferum | H. pilulifera | Lu et al., 2004 | ||
| H. parapilulifera | Lu et al., 2004 | |||
| H. stellata | Lu et al., 2004 | |||
| H. laciwombatensis | Lu et al., 2004 | |||
| Hypocreanum | H. citrina | Chaverri et al., 2004 | ||
| H. lactea | Chaverri et al., 2004 | |||
| H. sulphurea | Chaverri et al., 2004 | |||
| H. pulvinata | Chaverri et al., 2004 | |||
| Chlorospora | H. aureoviridis | Chaverri et al., 2004 | ||
| H. candida | Chaverri et al., 2004 | |||
| H. cremea | Chaverri et al., 2004 | |||
| H. surrotunda | Chaverri et al., 2004 | |||
| H. sinuosa | Chaverri et al., 2004 | |||
| H. chlorospora | Chaverri et al., 2004 | |||
| H. thelephoricola | Chaverri et al., 2004 | |||
| H. costaricensis | Chaverri et al., 2004 | |||
| H. thailandica | Chaverri et al., 2004 | |||
| H. virecentiflava | Chaverri et al., 2004 | |||
| Lixii/catoptron | T. harzianum | H. lixii | Chaverri et al., 2004 | |
| T. aggressivum | Samuels et al., 2002 | |||
| T. tomentosum | Chaverri et al., 2004 | |||
| T. cerinum | Bissett et al., 2003 | |||
| T. velutinum | Bissett et al., 2003 | |||
| T. sp. DAOM 175928 | 4Druzhinina, unpublished data | |||
| H. tawa | 4Druzhinina, unpublished data | |||
| H. atrogelatinosa | Chaverri et al., 2003b | |||
| H. ceracea | Chaverri et al., 2004 | |||
| H. cinnamomea | Chaverri et al., 2004 | |||
| H. straminea | Chaverri et al., 2004 | |||
| H. catoptron | Chaverri et al., 2004 | |||
| Virens | T. virens | H. virens | Kullnig-Gradinger et al., 2002 | |
| T. crassum | H. crassa | Chaverri et al., 2004 | ||
| Semiorbis | H. semiorbis | Chaverri et al., 2004 | ||
| H. hunua | Kullnig-Gradinger et al., 2002 | |||
| T. fertile | Kullnig-Gradinger et al., 2002 | |||
| T. oblongisporum | Kullnig-Gradinger et al., 2002 | |||
| Strictipilis | T. strictipilis | H. strictipilosa | Chaverri et al., 2004 | |
| T. longipile | Chaverri et al., 2004 | |||
| H. cuneispora | Chaverri et al., 2004 | |||
| H. aureoviridis var. macrospora | 4Druzhinina, unpublished data | |||
| Stromatica | T. stromaticum | Chaverri et al., 2004 | ||
| T. rossicum | Bissett et al., 2003 | |||
| T. sp. PPRI 3559 | Kullnig-Gradinger et al., 2002 | |||
| Ceramica | H. ceramica | Chaverri et al., 2004 | ||
| H. estonica | Chaverri et al., 2004 | |||
| Lutea | H. lutea | Chaverri et al., 2004 | ||
| H. megalomagna | Chaverri et al., 2004 | |||
| T. brevicompactum | Kraus et al., 2004 | |||
| Psychrophila | H. psychrophila | Chaverri et al., 2004 | ||
| H. megacitrina | Chaverri et al., 2004 | |||
| “Lone lineages” | T. spirale | Kullnig-Gradinger et al., 2002 | ||
| T. helicum | Bissett et al., 2003 | |||
| H. gelatinosa | Chaverri et al., 2004 | |||
| H. chromosperma | Chaverri et al., 2004 | |||
| H. sulawensis | Chaverri et al., 2004 | |||
| H. nigrovirens | Chaverri et al., 2004 | |||
| H. phyllostachidis | Chaverri et al., 2004 |
Only taxa, which have been verified by molecular analyses are included in the table. The following species were not included, as they have recently been abandoned: T. inhamatum (=T. harzianum); T. fasciculatum (=T. strictipilis), T. flavofuscum (=T. virens) and T. croceum (=T. polysporum)
Clades follow the nomenclature of Chaverri et al.(2004) and define phylogenetic groups of species, which received high statistic support in all investigations performed so far
Species names listed in the same line indicate anamorph-teleomorph relationships, and are only given for cases where both forms have been found in nature
Kubicek, C.P., 2003
Druzhinina, I., 2004
Table 4.
Hypocrea spp. with Trichoderma anamorphs, whose phylogenetic position is unknown
Phylogenetic studies of these 88 species showed that Trichoderma and Hypocrea form a single holomorph genus, within which two major clades can be distinguished (Fig.2, Table 3): one, which contains all the taxa described in section Longibrachiatum (Samuels et al., 1998), T. effusum and T. sinensis (Bissett et al., 2003), Trichoderma sp. MA 3239 (Wuczskowski et al., 2003), H. cerebriformis and H. poronoidea (3Kubicek, unpublished data). H. patella forms a stable basal branch to the Longibrachiatum clade. At the moment it is unclear whether H. peltata should be included in section Longibrachiatum or forms a sister clade. For the sake of simplicity we include it in this section. The second clade forms several well supported subclades: one, leading to members of section Trichoderma including “Pachybasium A” (T. viride/H. rufa, T. koningii, T. atroviride, T. ovalipsorum, H. neorufa, H. stilbohypoxyli, T. erinaceum, T. asperellum, T. hamatum, T. pubescens and T. strigosum), and which also contains H. pezizoides, H. avellanea (Chaverri et al., 2003b), and T. flavoconidia (Druzhinina et al., 2004a). The branch leading to this clade is frequently accompanied, but usually not strongly supported, by two sister clades: the Pachybasioides clade and the Hypocreanum clade. The former contains H. pachybasioides/T. polysporum (=T. croceum), H. pilulifera, H. parapilulifera, H. stellata, H. minutispora/T. minutisporum and H. laciwombatensis (Lu and Samuels, 2004), whereas the latter includes H. citrina, H. lactea, H. sulphurea and H. pulvinata (Chaverri et al., 2004; 4Overton, personal communication).
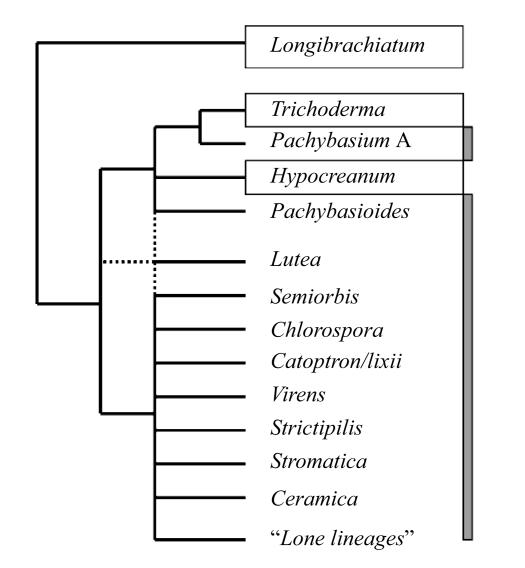
Fig. 2.
Schematic representation of the phylogenetic relationships of the currently recognized sections and clades in the genus Trichoderma. Clades representing sections as defined by Bissett (1991a) are boxed. The two clades of Pachybasium (A and B) are given by a vertical grey bar. The dotted line indicates the ambiguous phylogenetic placement of the H. lutea clade. Note that this is not a phylogenetic tree, but only a scheme based on several different phylogenetic trees published by Kullnig-Gradinger et al.(2002) and Chaverri et al.(2004). Branches are only given if they were strongly supported in all trees obtained until now
The other, large clade contains one weakly supported branch, but otherwise remains essentially unresolved by all genes or gene fragments used so far (Kullnig-Gradinger et al., 2002; Chaverri et al., 2003a; 2003b; 2004). This weakly supported branch leads to H. aureoviridis/T. aureoviride and a group of Hypocrea spp. described recently (H. aureoviridis var. macrospora, H. candida, H. cremea, H. surrotunda, H. sinuosa, H. chlorospora, H. thelephoricola, H. costaricensis, H. thailandica and H. virecentiflava) with very similar morphological and genetic characters. Apart from this clade, several terminal subclades can be identifed which contain closely related species, but whose phylogenetic relationship remains unclear: H. lixii/catoptron (containing H. lixii/T. harzianum, T. aggressivum, T. tomentosum, T. cerinum, T. velutinum, T. sp. DAOM 172928, H. tawa, H. atrogelatinosa, H. ceracea, H. cinnamomea, H. straminea and H. catoptron); H. virens (H. virens/T. virens, T. flavofuscum, T. crassum); H. semiorbis (H. semiorbis, T. oblongisporum, T. fertile); H. strictipilosa (H. strictipilosa/T. strictipile, H. aureoviridis var. macrospora, H. cuneispora, T. longipile, T. fasciculatum (synonymized with T. strictipile); H. ceramica (H. ceramica, H. estonica); and H. stromatica (H. stromatica/T. stromaticum, T. rossicum, T. sp. PPRI 3559).
The presence of species of section Hypocreanum as a subclade in clade “Pachybasium B” deserves some comments: Samuels (1996) hypothesized that the anamorphs of species of Hypocrea with effused stromata having colourless conidia on irregularly verticillate conidiophores, which Bissett (1991a) placed in Trichoderma sect. Hypocreanum may be synanamorphs or spermatial states, and therefore inappropriately placed in Trichoderma. Because the anamorphs had not been found in nature, Gams and Bisset (1998) omitted sect. Hypocreanum from their treatment of Trichoderma. However, phylogenetic data proved that the respective taxa are clearly members of the genus. Kullnig-Gradinger et al.(2002) postulated that the ability to form the Trichoderma-like anamorph may have been lost during evolution of these species. A phylogeny based on 28S-rDNA sequence analysis showed that genera having Verticillium-like anamorphs (Aphysiostroma, Podocrea, Arachnocrea) occur in a stable basal position to the genus Trichoderma implying that Trichoderma evolved from them. We thus favour the interpretation that the ability to form the Trichoderma-like anamorph morphology, which may have supported the switch from fungicolous to saprophytic habitats, was subsequently lost in some evolutionary lines. Other Hypocrea species with Verticillium-anamorph morphology such as H. tawa or H. hunua occur nested in other clades (Table 1) (Kullnig-Gradinger et al., 2002), indicating that the loss of genes (or their expression) required form the Trichoderma-like anamorph occured several times during the evolution of the genus.
Phylogenetic analysis revealed that three other species (H. lutea/H. melanomagna/T. brevicompactum) also cluster together, but their association with any of the large clades is unclear and dependent on the genes and species used (1Kubicek, unpublished data).
Besides these, there are still a number of species for which no close neighbor is known, and for which the phylogenetic position within Pachybasium B could so far not be identified: H. psychrophila/H. megacitrina, which are found in the vicinity of the Hypocreanum clade; T. spirale, which has phylogenetic affinity to T. virens and T. harzianum; T. helcum, which has a slight affinity to T. spirale; and H. gelatinosa, H. chromosperma, H. sulawensis, H. nigrovirens and H. phyllostachidis, which all occur as Lone Lineages of unresolved Pachybasium B.
WHAT NEXT?
The current information, as summarized above, places Trichoderma among the fungal genera most thoroughly investigated taxonomically today, in view of the fact that at least two but on average more gene sequences are known for every recognized species. While the 88 species recently characterized by molecular methods are phylogenetically well supported, their evolution and phylogenetic relationship has so far been more difficult to resolve. This may either reflect the lack of known ancestors (as with most other fungi), or the occurrence of high selection pressure during the evolution of these clades, consequently leading to dichotomous trees. However, we must emphasize that these 88 species are still probably only a minor fraction of the existing number of taxa. The CABI online database of fungal names (http://www.indexfungorum.org) lists 401 Hypocrea spp., and while several of them may be redundant (i.e. synonyms of other described species) or not correctly placed in the genus (e.g. H. pallida), this may be compensated by others yet to be described. One should bear in mind that many of the currently known Hypocrea spp. had been isolated and described by Yoshimichi Doi (see Samuels, 1996) from sampling in Japan, the Western Pacific and South America. In our lab, Walter Jaklitsch has recently initiated a study on the biodiversity of Hypocrea spp. in Central Europe, and his preliminary data indicate the presence of at least 10–15 new undescribed taxa in the samples from the first 18 months. Given the comparatively small area investigated, and the fact that huge areas like Africa or Central Asia have not been studied at all, we expect that 400 will be even too low.
The number of Trichoderma spp. may remain lower, but also here a further rise can be anticipated. From our own study on the biodiversity of Trichoderma, at least 10 putative new ‘phylogenetic spp.’ are currently in the pipeline, and will be described in the near future. In addition, screening of so far neglected geographic areas will likely show new taxa (Kullnig et al., 2000; Kubicek et al., 2003; Wuczskowski et al., 2003), and several such investigations have recently been completed or are being undertaken (e.g. New Zealand, S.L. Dodd and coworkers; Iran, D. Zafari; Sardinia, Q. Migheli and I. Druzhinina; China, T. Xu; Rwanda, J. Bissett and I. Druzhinina). Still, areas like most of the African continent and the Pacific have not been investigated.
As emphasized above, most of the phylogenetic analyses are incomplete, due to the limited suitability of the gene sequences used so far (Table 1). To improve this situation, novel gene sequences are needed. Researchers need to be aware that there is no single “universal, all-purpose” gene for phylogenetic analysis of this genus, and the suitability of a given gene should always be tested first before applying it to a given phylogenetic problem.
Finally, a safe species concept may also aid in identification and safe comparison of biochemical and physiological properties of Trichoderma strains used in biocontrol. While these strains have been uniformly been called “T. harzianum” in the past, leading to the situation that the name T. harzianum is synonymized with biocontrol agent, there is now increasing evidence that actually several, genetically diverse species are used in biocontrol (Hermosa et al., 2000; 2004; Kullnig et al., 2001). The species identification tools now in hand will help to answer the question whether particular taxa are to be preferred on particular hosts or plants.
Acknowledgments
This article is dedicated to the memory of the late Dr. Elke Lieckfeldt, and her pioneering contributions to the molecular taxonomy of Trichoderma. The authors are grateful to Walter Jaklitsch from their laboratory and to various colleagues for sharing unpublished information.
Footnotes
Project supported by FWF-grants P-12748-MOB and P-16601 to C.P.K., Austria
Kubicek, C.P., 2003
Samuels, G.J., 2003
Mach, R.L., et al., 1999
Kullnig-Gradinger, C.M., Kubicek, C.P., 2004
Samuels, G.J., Ismael, E., 2004
Druzhinina, I., 2004
Druzhinina, I., Bissett, J., Kubicek, C.P., 2004. Bayesian inferences towards the phylogenetic species recognition of biocontrol fungi Hypocrea lixii/Trichoderma harzianum and closely related species. Manuscript in preparation
Kubicek, C.P., 2000
Druzhinina, I., Bissett, J., Kubicek, C.P., 2004. Bayesian inferences towards the phylogenetic species recognition of biocontrol fungi Hypocrea lixii/Trichoderma harzianum and closely related species. Manuscript in preparation
Samuels, G.J., 2004
Bissett, J., 2004
Druzhinina, I., Bissett, J., 2004
Kubicek, C.P., 2003
Overton, B., 2004
Kubicek, C.P., 2003
References
- 1.Bisby GR. Trichoderma viride Pers. ex Fries, and notes on Hypocrea . Trans Brit Mycol Soc. 1939;23:149–168. [Google Scholar]
- 2.Bissett J. A revision of the genus Trichoderma. I. Sect. Longibrachiatum sect. nov. Can J Bot. 1984;62:924–931. [Google Scholar]
- 3.Bissett J. A revision of the genus Trichoderma. II. Infrageneric classification. Can J Bot. 1991;69:2357–2372. [Google Scholar]
- 4.Bissett J. A revision of the genus Trichoderma. III. Sect. Pachybasium . Can J Bot. 1991;69:2373–2417. [Google Scholar]
- 5.Bissett J. A revision of the genus Trichoderma. IV. Additional notes on section Longibrachiatum . Can J Bot. 1991;69:2418–2420. [Google Scholar]
- 6.Bissett J. Trichoderma atroviride . Can J Bot. 1992;70:639–641. [Google Scholar]
- 7.Bissett J, Szakacs G, Nolan CA, Druzhinina I, Kullnig-Gradinger CM, Kubicek CP. Seven new taxa of Trichoderma from Asia. Can J Bot. 2003;81:570–586. [Google Scholar]
- 8.Buckler ES, Ippolito A, Holtsfort TE. The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics. 1997;145:821–832. doi: 10.1093/genetics/145.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaverri P, Castlebury LA, Samuels GJ, Geiser D. Multilocus phylogenetic structure within the Trichoderma harzianum/Hypocrea lixii complex. Mol. Phylogenet. Evol. 2003;27:302–313. doi: 10.1016/s1055-7903(02)00400-1. [DOI] [PubMed] [Google Scholar]
- 10.Chaverri P, Castlebury LA, Overton BE, Samuels GJ. Hypocrea/Trichoderma: species with conidiophore elongations and green conidia. Mycologia. 2003;95:1100–1140. [PubMed] [Google Scholar]
- 11.Chaverri P, Candoussau F, Samuels GJ. Hypocrea phyllostachydis and its Trichoderma anamorph, a new bambusicolous species from France. Mycol Progr. 2004;3:29–36. [Google Scholar]
- 12.Dodd SL, Crowhurst RN, Rodrigo AC, Samuels GJ, Hill RA, Stewart A. Examination of Trichoderma phylogenies derived from ribosomal DNA sequence data. Mycol Res. 2000;104:23–34. [Google Scholar]
- 13.Dodd SL, Lieckfeldt E, Chaverri P, Overton BE, Samuels GJ. Taxonomy and phylogenetic relationships of two species of Hypocrea with Trichoderma anamorphs. Mycol Progr. 2002;1:409–428. [Google Scholar]
- 14.Dodd SL, Lieckfeldt E, Samuels GJ. Hypocrea atroviridis sp. nov., the teleomorph of Trichoderma atroviride . Mycologia. 2003;95:27–40. doi: 10.1080/15572536.2004.11833129. [DOI] [PubMed] [Google Scholar]
- 15.Doi Y. Revision of the Hypocreales with cultural observations. IV. The genus Hypocrea and its allies in Japan. (2) Enumeration of the species. Bull Natl Sci Museum Tokyo. 1972;15:649–751. [Google Scholar]
- 16.Doi Y, Abe Y, Sugyama J. Trichoderma sect. Saturnisporum, sect. nov. and Trichoderma ghanense, sp. nov. Bull Nat Sci Museum, Ser B (Botany) 1987;12:1–9. [Google Scholar]
- 17.Druzhinina I, Chaverri P, Fallah P, Kubicek CP, Samuels GJ. Hypocrea flavoconidia, a new species with yellow conidia from Costa Rica. Stud Mycol. 2004 in press. [Google Scholar]
- 18.Druzhinina I, Koptchinski A, Komon M, Bissett J, Szakacs G, Kubicek CP. A DNA-barcode for strain identification in Trichoderma . 2004 Manuscript submitted. [Google Scholar]
- 19.Gams W, Bissett J. Morphology and Identification of Trichoderma . In: Kubicek CP, Harman GE, editors. Trichoderma and Gliocladium. Vol. 1. Basic Biology, Taxonomy and Genetics. London: Taylor and Francis Ltd.; 1998. pp. 3–34. [Google Scholar]
- 20.Gherbawy Y, Druzhinina I, Shaban GM, Wuczkowsky M, Yaser M, El-Naghy MA, Prillinger HJ, Kubicek CP. Trichoderma populations from alkaline agricultural soil in the Nile valley, Egypt, consist of only two species. Mycol Prog. 2004;3:211–218. [Google Scholar]
- 21.Hermosa MR, Grondona I, Iturriaga EA, Diaz-Minguez JM, Castro C, Monte E, Garcia-Acha I. Molecular characterization and identification of biocontrol isolates of Trichoderma spp. Appl Environ Microbiol. 2000;66:1890–1898. doi: 10.1128/aem.66.5.1890-1898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hermosa MR, Keck E, Chamorro I, Rubio B, Sanz L, Vizcaino JA, Grondona I, Monte E. Genetic diversity shown in Trichoderma biocontrol isolates. Mycol Res. 2004;108:897–906. doi: 10.1017/s0953756204000358. [DOI] [PubMed] [Google Scholar]
- 23.Hjeljord L, Tronsmo A. Trichoderma and Gliocladium in Biological Control: An Overview. In: Harman GE, Kubicek CP, editors. Trichoderma and Gliocladium. Vol. 2. Enzymes, Biological Control and Commercial Applications. London: Taylor and Francis Ltd.; 1998. pp. 131–151. [Google Scholar]
- 24.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 25.Kindermann J, El-Ayouti Y, Samuels GJ, Kubicek CP. Phylogeny of the genus Trichoderma based on sequence analysis of the internal transcribed spacer region 1 of the rDNA clade. Fungal Genet Biol. 1998;24:298–309. doi: 10.1006/fgbi.1998.1049. [DOI] [PubMed] [Google Scholar]
- 26.Klein D, Eveleigh DE. Ecology of Trichoderma . In: Kubicek CP, Harman GE, editors. Trichoderma and Gliocladium. Vol. 1. Basic Biology, Taxonomy and Genetics. London: Taylor and Francis Ltd.; 1998. pp. 57–74. [Google Scholar]
- 27.Kraus G, Druzhinina I, Bissett J, Zafari D, Prillinger HJ, Szakacs G, Zare R, Gams W, Kubicek CP. Trichoderma brevicompactum sp. nov. Mycologia. 2004;96:1057–1071. [PubMed] [Google Scholar]
- 28.Kredics L, Antal Z, Doczi I, Manczinger L, Kevei F, Nagy E. Clinical importance of the genus Trichoderma. A review. Acta Microbiol Immunol Hung. 2003;50:105–117. doi: 10.1556/AMicr.50.2003.2-3.1. [DOI] [PubMed] [Google Scholar]
- 29.Kubicek CP, Penttilä ME. Regulation of Production of Plant Polysaccharide Degrading Enzymes by Trichoderma . In: Harman GE, Kubicek CP, editors. Trichoderma and Gliocladium. Vol. 2. Enzymes, Biological Control and Commercial Applications. London: Taylor and Francis Ltd.; 1998. pp. 49–71. [Google Scholar]
- 30.Kubicek CP, Bissett J, Druzhinina I, Kullnig-Gradinger CM, Szakacs G. Genetic and metabolic diversity of Trichoderma: a case study on South East Asian isolates. Fungal Genet Biol. 2003;38:310–319. doi: 10.1016/s1087-1845(02)00583-2. [DOI] [PubMed] [Google Scholar]
- 31.Kuhls K, Lieckfeldt E, Samuels GJ, Kovacs W, Meyer W, Petrini O, Gams W, Börner T, Kubicek CP. Molecular evidence that the asexual industrial fungus Trichoderma reesei is a clonal derivative of the ascomycete Hypocrea jecorina . Proc. Natl Acad Sci USA. 1996;93:7755–7760. doi: 10.1073/pnas.93.15.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhls K, Lieckfeldt E, Samuels GJ, Meyer W, Kubicek CP, Börner T. Revision of Trichoderma section Longibrachiatum including related teleomorphs based on an analysis of ribosomal DNA internal transcribed spacer sequences. Mycologia. 1997;89:442–460. [Google Scholar]
- 33.Kullnig CM, Szakacs G, Kubicek CP. Molecular identification of Trichoderma species from Russia, Siberia and the Himalaya. Mycol Res. 2000;104:1117–1125. [Google Scholar]
- 34.Kullnig CM, Krupica T, Woo SL, Mach RL, Rey M, Benitez T, Lorito M, Kubicek CP. Confusion abounds over identity of Trichoderma biocontrol isolates. Mycol Res. 2001;105:769–772. [Google Scholar]
- 35.Kullnig-Gradinger CM, Szakacs G, Kubicek CP. Phylogeny and evolution of the fungal genus Trichoderma–a multigene approach. Mycol Res. 2002;106:757–767. [Google Scholar]
- 36.Lewis PO. Phylogenetic systematics turns over a new leaf. Trends Ecol Evol. 2001;16:30–37. doi: 10.1016/s0169-5347(00)02025-5. [DOI] [PubMed] [Google Scholar]
- 37.Lieckfeldt E, Seifert KA. An evaluation of the use of ITS sequences in the taxonomy of the Hypocreales . Stud Mycol. 2000;45:35–44. [Google Scholar]
- 38.Lieckfeldt E, Samuels GJ, Börner T, Gams W. Trichoderma koningii: neotypification and Hypocrea teleomorph. Can J Bot. 1998;76:1507–1522. [Google Scholar]
- 39.Lieckfeldt E, Samuels GJ, Nirenberg HI, Petrini O. A morphological and molecular perspective of Trichoderma viride: is it one or two species. Appl Environ Microbiol. 1999;65:2418–2428. doi: 10.1128/aem.65.6.2418-2428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieckfeldt E, Kullnig CM, Kubicek CP, Samuels GJ, Börner T. Trichoderma aureoviride: phylogenetic position and characterization. Mycol Res. 2001;105:313–322. [Google Scholar]
- 41.Lu BS, Samuels GJ. Hypocrea stilbohypoxyli and its Trichoderma koningii-like anamorph from Puerto Rico on Stilbohypoxylon Muelleri. Sydowia. 2004 manuscript submitted. [Google Scholar]
- 42.Lu BS, Druzhinina I, Chaverri P, Fallah P, Gradinger C, Kubicek CP, Samuels GJ. Hypocrea/Trichoderma species with pachybasium-like conidiophores: teleomorphs for T. minutisporum and T. polysporum, and their newly discovered relatives. Mycologia. 2004;96:310–342. [PubMed] [Google Scholar]
- 43.Lutzoni F, Pagel M, Reeb V. Major fungal lineages are derived from lichen symbiotic ancestors. Nature. 2001;411:937–940. doi: 10.1038/35082053. [DOI] [PubMed] [Google Scholar]
- 44.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. Equations of state calculations for fast computing machines. J Chem Phys. 1953;21:1087–1091. [Google Scholar]
- 45.Nelson EE. Occurrence of Trichoderma in a Douglas-fir soil. Mycologia. 1982;74:280–284. [Google Scholar]
- 46.O’Donnell K. Ribosomal DNA internal transcribed spacers are highly divergent in the phytopathogenic ascomycete Fusarium sambucinum (Gibberella pulicaris) Curr Genet. 1992;22:213–220. doi: 10.1007/BF00351728. [DOI] [PubMed] [Google Scholar]
- 47.O’Donnell K, Cigelnik E, Nirenberg HI. Molecular systematics and phylogeography of the Gibberella fuji-kuroi species complex. Mycologia. 1998;90:465–493. [Google Scholar]
- 48.Persoon CH. Disposito Methodica Fungorum in Classes, Ordines, Familias et Genera. In: Römer JJ, editor. Neues Magazin für Botanik. Zürich: Ziegler und Söhne; 1794. pp. 63–128. (Ger). [Google Scholar]
- 49.Rannala B, Yang Z. Probability distribution of molecular evolutionary trees: a new method of phylogenetic interference. J Mol Evol. 1996;43:304–311. doi: 10.1007/BF02338839. [DOI] [PubMed] [Google Scholar]
- 50.Rifai MA. A revision of the genus Trichoderma . Mycol Pap. 1969;116:1–116. [Google Scholar]
- 51.Salemi M, Vandamme A. The Phylogenetic Handbook. A Practical Approach to DNA and Protein Phylogeny. UK: Cambidge University Press; 2003. [Google Scholar]
- 52.Samuels GJ. Trichoderma: A review of biology and systematics of the genus. Mycol Res. 1996;100:923–935. [Google Scholar]
- 53.Samuels GJ. Trichoderma ovalisporum: A new endophytic species with potential to control frosty pod rot of cocoa. Mycol. Prog. 2004 in press. [Google Scholar]
- 54.Samuels GJ, Petrini O, Kuhls K, Lieckfeldt E, Kubicek CP. The Hypocrea schweinitzii complex and Trichoderma sect. Longibrachiatum . Stud Mycol. 1998;41:1–54. [Google Scholar]
- 55.Samuels GJ, Dodd SL, Gams W, Castlebury LA, Petrini O. Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus . Mycologia. 2002;94:146–170. [PubMed] [Google Scholar]
- 56.Sivasithamparam K, Ghisalberti EL. Secondary Metabolism in Trichoderma and Gliocladium . In: Kubicek CP, Harman GE, editors. Trichoderma and Gliocladium. Vol. 1. Basic Biology, Taxonomy and Genetics. London: Taylor and Francis Ltd.; 1998. pp. 139–191. [Google Scholar]
- 57.Taylor JW, Jacobson DJ, Fisher M. The evolution of asexual fungi: speciation and classification. Annu Rev Phytopathol. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- 58.Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- 59.Thrane U, Poulsen SB, Nirenberg HI, Lieckfeldt E. Identification of Trichoderma strains by image analysis of HPLC chromatograms. FEMS Microbiol Lett. 2001;203:249–255. doi: 10.1111/j.1574-6968.2001.tb10849.x. [DOI] [PubMed] [Google Scholar]
- 60.Turner D, Kovacs W, Kuhls K, Lieckfeldt E, Peter B, Arisan-Atac I, Strauss J, Samuels GJ, Börner T, Kubicek CP. Biogeography and phenotypic variation in Trichoderma sect. Longibrachiatum and associated Hypocrea species. Mycol Res. 1997;101:449–459. [Google Scholar]
- 61.Widden P, Abitbol JJ. Seasonality of Trichoderma species in a spruce-forest soil. Mycologia. 1980;72:775–784. [Google Scholar]
- 62.Wuczskowski M, Druzhinina I, Gherbawy Y, Klug B, Prillinger HJ, Kubicek CP. Taxon pattern and genetic diversity of Trichoderma in a mid-European, primeval floodplain-forest. Microbiol Res. 2003;158:125–134. doi: 10.1078/0944-5013-00193. [DOI] [PubMed] [Google Scholar]