Abstract
Ribonucleotide reductase (RNR) catalyzes the conversion of nucleoside diphosphates to deoxynucleoside diphosphates. Crucial for rapidly dividing cells, RNR is a target for cancer therapy. In eukaryotes, RNR comprises a heterooligomer of α2 and β2 subunits. Rnr1, the α subunit, contains regulatory and catalytic sites; Rnr2, the β subunit (in yeast, a heterodimer of Rnr2 and Rnr4), houses the diferric-tyrosyl radical crucial for catalysis. Here, we present three x-ray structures of eukaryotic Rnr1 from Saccharomyces cerevisiae: one bound to gemcitabine diphosphate (GemdP), the active metabolite of the mechanism-based chemotherapeutic agent gemcitabine; one with an Rnr2-derived peptide, and one with an Rnr4-derived peptide. Our structures reveal that GemdP binds differently from its analogue, cytidine diphosphate; because of unusual interactions of the geminal fluorines, the ribose and base of GemdP shift substantially, and loop 2, which mediates substrate specificity, adopts different conformations when binding to GemdP and cytidine diphosphate. The Rnr2 and Rnr4 peptides, which block RNR assembly, bind differently from each other but have unique modes of binding not seen in prokaryotic RNR. The Rnr2 peptide adopts a conformation similar to that previously reported from an NMR study for a mouse Rnr2-based peptide. In yeast, the Rnr2 peptide binds at subsites consisting of residues that are highly conserved among yeast, mouse, and human Rnr1s, suggesting that the mode of Rnr1–Rnr2 binding is conserved among eukaryotes. These structures provide new insights into subunit assembly and a framework for structure-based drug design targeting RNR.
Keywords: allosteric regulation, crystallography, dNTP, chemotherapy, gemcitabine
Ribonucleotide reductases (RNRs) catalyze the reduction of ribonucleotides to deoxyribonucleotides, essential precursors of DNA synthesis. Crucial for rapidly proliferating cells, RNR is a target for anticancer (1, 2) and antiviral (2, 3) drugs. Gemcitabine, an analogue of deoxycytidine (2′-2′-difluorodeoxycytidine), is sequentially phosphorylated to the 5′-monophosphate form by deoxycytidine kinase and to difluorodeoxycytidine 5′-diphosphate (GemdP) by uridylate-cytidylate monophosphate kinase. In the presence of reductants, GemdP inactivates Rnr1. In the absence of reductants, with prereduced Rnr1 and Rnr2, inhibition occurs from the loss of the tyrosyl radical in Rnr2 (1). Recently, GemdP has been shown to inactivate both human R1 and R2 (JoAnne Stubbe, personal communication). Inhibition of RNR by GemdP leads to reduction of the pool of deoxyribonucleotide 5′-diphosphates available for DNA synthesis, presumably favoring incorporation of the gemcitabine triphosphate metabolite by DNA polymerase α, preventing chain elongation (4, 5).
RNRs require unusual metallocofactors to initiate radical-based nucleotide reduction and are divided into three classes based on their cofactor. Class I RNR, found in all eukaryotes, is a heterooligomer of α2 and β2 subunits (6). In eukaryotes, the α subunit, called Rnr1, contains the catalytic site, the substrate specificity site, and the activity site. The β2 subunit, usually a dimer of Rnr2, contains the diferric-tyrosyl (Y•) radical cofactor that initiates nucleotide reduction by a putative long-range proton-coupled electron transfer (7, 8). Recently, the intermolecular distance traveled by the free radical has been determined by EPR to be 33 Å (9). During catalysis, the radical transfer pathway in Escherichia coli is proposed to involve Y122, W48, and Y356 in Rnr2 and Y731, Y730, and C439 in Rnr1 (10).
In Saccharomyces cerevisiae, the active form of the β2 subunit is an Rnr2·Rnr4 heterodimer (11), in which Rnr4 stabilizes a helix in Rnr2 containing one of the iron ligands (12). The small subunit Rnr2 binds Rnr1 through its C-terminal residues (13). Hence, C-terminal Rnr2-based peptidomimetics (14–16) bind Rnr1, blocking RNR assembly and providing another mode of therapy for proliferative diseases such as cancer. Early reports on Rnr2 peptide-based inhibitors showed that they had in vivo efficacy against herpes simplex virus with nM dissociation constants (17–19), suggesting that similar potencies might be possible with anticancer peptidomimetics.
Although there are several structures reported for prokaryotic Rnr1s (20–25), until now, no such structure has been available for eukaryotes. Human and yeast Rnr1 share 66% sequence identity and 83% sequence similarity. In contrast, the sequence identity of human and E. coli Rnr1 is 27%, and the similarity is 43%. Potent renin inhibitors were designed before the availability of human and mouse structures by using homologous enzyme structures as templates (26–28), suggesting that, in the absence of a structure of Rnr1 from Homo sapiens or other higher eukaryote, the yeast structure can be used in structure-based drug design. We report the structures of Rnr1 (Fig. 1A) complexed with C-terminal nonapeptides of Rnr2 and Rnr4, with the effector–inhibitor pair AMPPNP (a nonhydrolysable analogue of ATP) and GemdP and the effector–substrate pair AMPPNP–cytidine diphosphate (CDP) to compare binding. These structures provide a molecular basis for understanding RNR assembly in eukaryotes and GemdP’s mode of binding to Rnr1.
Fig. 1.
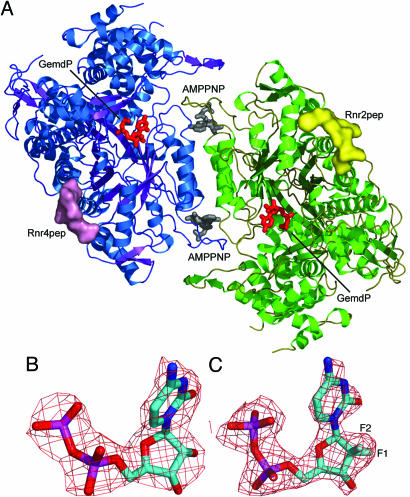
Rnr1 complexes. (A) The dimer of Rnr1 is displayed as a cartoon with the monomers colored blue and green. Surfaces for Rnr2pep and Rnr4pep are displayed in yellow and pink, respectively, and AMPPNP (gray) and GemdP (red) are shown binding to the specificity and catalytic sites. (B and C) The 2Fo–Fc electron density contoured at 1.0 σ for CDP (B) and GemdP (C).
Results and Discussion
GemdP Binding.
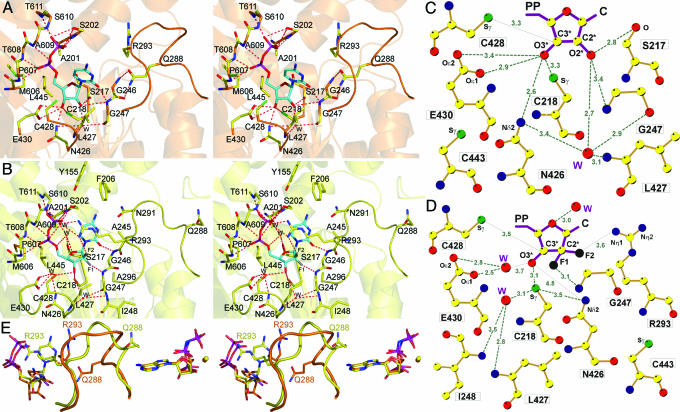
All our Rnr1 complexes, like those for most of the structures of Rnr1 reported in refs. 22, 24, 25, and 29, were obtained by the soaking method. The crystals remained isomorphous after soaking and contained well ordered ligands, as shown by the fact that our AMPPNP–CDP and AMPPNP–GemdP structures contained substrate that is fully visible in the 2Fo–Fc electron-density maps (Fig. 1B and C). GemdP differs from CDP only by substitution of two fluorines for the OH and the hydrogen atom bonded to 2′C of the ribose ring but adopts a different conformation when binding to Rnr1. In the AMPPNP–CDP structure, the 2′ and 3′ OH of the ribose are close to the catalytic N426 and E430 C428, where the thiyl radical is generated on Rnr1 by a series of coupled electron and proton transfers (7) and C218 of the reduced catalytic redox pair (C218 and C443) (Fig. 2A and B). In contrast, in the AMPPNP–GemdP structure, the ribose and, especially, the base of GemdP appear to bind higher in the pocket (toward the top of the page in Fig. 2A, C, and E), such that the 2′ carbon and the two fluorines of the GemdP ribose bind near the location of C2, N3, and O2 of CDP’s cytidine base in the AMPPNP–CDP structure (Fig. 2E). The GemdP ribose is displaced by an average of 2.3 Å and its cytidine base by an average of 3.8 Å compared with those of CDP. GemdP’s unique mode of binding places its ribose further away from N426 and E430 at the active site (Fig. 2A–D). Interestingly, a water molecule binds near the position left vacant by the displacement of the GemdP ribose 3′ OH, hydrogen-bonding the side chain of E430 and making van der Waals contact with the ribose 3′ OH in its new position (Fig. 2C and D). The conserved water molecule [see accompanying article (30)] that hydrogen bonds the 2′ OH in CDP is also observed in GemdP, and another water molecule is located 3.0 Å away from the O4′ atom and makes a second-sphere hydrogen bond to the side chain of S217 (Fig. 2C).
Fig. 2.
Catalytic-site interactions. (A) Stereoview of CDP (orange). Interacting atoms: oxygen, red; nitrogen, blue; phosphate, magenta; sulfur, green; substrate carbons, cyan; protein non-Cα carbons, yellow; Cα carbons, as secondary structure, orange. (B) Stereoview of GemdP. Interacting atoms are colored as in A, except that sulfur is orange; Cα carbons, as secondary structure, are yellow; and fluorines are gray. (C) Ligand plot of CDP ribose interactions. Colors are as in A, except that carbons are yellow. (D) Ligand plot of GemdP interactions. The van der Waals contact to L427 is omitted for clarity. (E) Stereoview of loop-2 superposition of AMPPNP–CDP (orange) and AMPPNP–GemdP (yellow). Substrate/inhibitor is seen on the left, and the effector is on the right. The color scheme is the same as in C, but fluorine is black.
Although structures produced by soaking sometimes differ from structures produced by cocrystallization, the CDP- and GemdP-bound structures were both obtained by soaking under similar conditions. Thus, we attribute the differences observed between CDP and GemdP binding to the unique chemical properties of the geminal fluorine atoms, which are more hydrophobic and can form hydrogen bonds (31). According to this study, geminal fluorines can form hydrogen bonds to donor nitrogen atoms with hydrogen-bond lengths ranging from 3.0 to 3.6 Å and C-F.N angles ranging from 60° to 180°. In our AMPPNP–GemdP structure, the F2 fluorine that replaces the hydrogen atom makes a weak hydrogen bond (3.6 Å) with the guanidinium group of R293 from loop 2 with a C-F.N angle of 142°. This F2 fluorine has been shown to hydrogen-bond an arginine in deoxycytidine kinase, which phosphorylates gemcitabine (32). The F1 geminal fluorine forms a hydrogen bond (3.1 Å) with the amide nitrogen of G247, with a C-F.N angle of 141°. In contrast, in the AMPPNP–CDP complex, CDP does not interact with R293, whereas the 2′ OH forms a longer (3.5 Å) hydrogen bond to the amide nitrogen of G247. The hydrogen bond between the 2′ OH of CDP and the CO of S217 is missing in the AMPPNP–GemdP structure. Moreover, GemdP’s F1 makes a close van der Waals contact (3.3 Å) with CD2 of L427, possibly because of the more hydrophobic nature of fluorine; the corresponding distance in the AMPPNP–CDP complex is 4.2 Å. As regards the catalytic residues, the 3′ carbon of GemdP is within 3.5 Å of C428, and the 3′ OH of GemdP is 3.1 Å from the C218. Moreover, as in the AMPPNP–CDP structure, the OH of Y741 that is in the free-radical relay pathway is within 3.5 Å of the Sγ of C428 in the AMPPNP–GemdP structure. These distances should still permit mechanism-based inhibition, which requires abstraction of the GemdP’s 3′ hydrogen atom by a thiyl radical generated at C428 by a series of coupled proton and electron transfers from Y183• of Rnr2 (33).
Interactions with Substrate Bases.
The different position of GemdP’s base occurs in conjunction with significant changes in ligand–protein interactions with respect to CDP. In the AMPPNP–GemdP structure, the guanidinium group of R293 from loop 2 makes van der Waals contact with the cytidine base and forms a second-sphere hydrogen bond to the phosphates via a water molecule, adopting a conformation it cannot make in CDP because of steric clashes with the base of CDP itself (Fig. 2 C and E). Although this interaction is not found in any of our effector–substrate complexes (30), a similar interaction has been reported for the dATP–CDP structure from Thermotoga maritima, where the guanidinium group of the equivalent R207 forms a salt bridge with the phosphate (22). R293 in the AMPPNP–CDP complex does not interact with the base, and, instead, Q288 makes van der Waals contact with it (Fig. 2 A and E). Loop-2 conformation differs significantly between the two structures, so that Q288 in GemdP is barred from the position it occupies in CDP because of clashes with K292 (data not shown) and points toward the effector. In contrast, in the T. maritima dATP–CDP Rnr1 (22), the CDP base interacts with the residue corresponding to Q288. The unusual mode of binding of GemdP also results in Y155, F206, and N291, contacting the cytidine base from above (Fig. 2C). These interactions are missing in the AMPNP–CDP structure.
Additionally, GemdP’s base makes van der Waals contact with S202, and its O2 atom hydrogen-bonds the amide nitrogen of G246. In the CDP structure, the O2 atom hydrogen-bonds the amide nitrogen of G247 and side chains of L427 and C428 contact the bottom of the base. The phosphates of CDP and GemdP bind similarly: the α phosphate to main-chain amines from P607 to A609 and the β phosphate to main-chain amines and side-chain hydroxyls of S610, T611, and S202 (Fig. 2 A and B).
Interactions with Effector.
In both the AMPPNP–CDP and AMPPNP–GemdP complexes, a single Mg2+ ion coordinates the β and γ phosphates of AMPPNP. The position of Q288 in the AMPPNP–GemdP structure, near the effector, contrasts sharply with its position in the AMPPNP–CDP structure, pointing toward the substrate (Fig. 2E). This difference is reflected in the different pattern of hydrogen bonds from loop 2 to the effector. In the AMPPNP–CDP complex, the adenine ring makes two hydrogen bonds with D287 (see Fig. 4A, which is published as supporting information on the PNAS web site). In the AMPPNP–GemdP structure, AMPPNP makes one hydrogen bond to the backbone amide of D287 via N1, which also makes a hydrogen bond with the backbone amide of Q288 (see Fig. 4B). This substantial change in loop-2 conformation in the presence of a different ligand at the catalytic site may be due to the unusual mode of binding of the inhibitor GemdP. The effector is complexed with magnesium in both complexes, and loop 1 interacts with effector phosphates via main-chain amides and the side chain of R256, folding over the effector-binding site.
Binding of Rnr2 and Rnr4 Peptides.
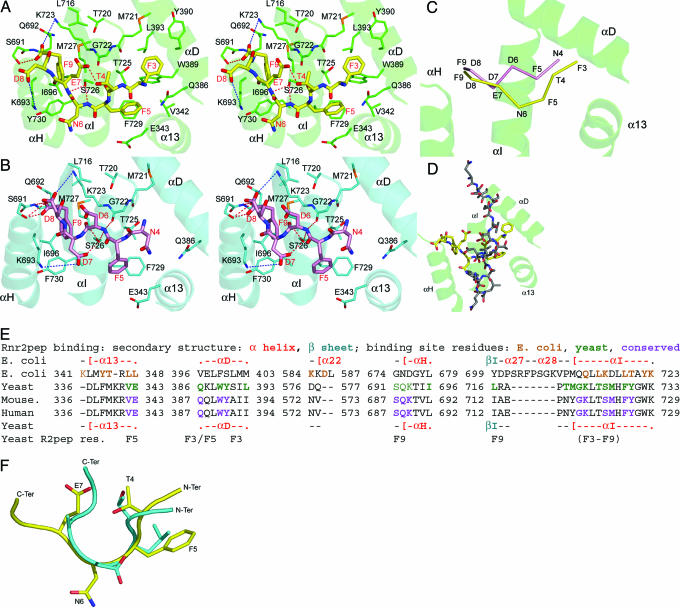
Peptides comprising the nine C-terminal residues of Rnr2 (Rnr2pep, sequence GAFTFNEDF) and Rnr4 (Rnr4pep, sequence KEINFDDDF) compete with their respective intact proteins for binding to Rnr1 (13). On this basis, a class of anticancer and antiviral inhibitors that disrupt RNR assembly are being developed (14–16, 34). In our Rnr1–Rnr2pep and Rnr1–Rnr4pep complex structures, the last seven amino acids of Rnr2pep (3FTFNEDF9) and six amino acids of Rnr4pep (4NFDDDF9) were ordered. Rnr4pep binds Rnr1 in a partially extended conformation, whereas Rnr2pep binds similarly at its C terminus but bulges between residues 4–7 so that the N termini of the peptides are out of register by one residue (Fig. 3A–C), resulting in different binding subsites for Rnr2pep and Rnr4pep, such that only F9 is fully shared. This finding indicates that one monomer of Rnr1 can bind either Rnr2 or Rnr4 but not both.
Fig. 3.
Binding of Rnr2 and Rnr4 peptides to Rnr1. The structures of Rnr1–Rnr2pep complex in stereo (A), Rnr1–Rnr4pep complex in stereo (B), and Rnr4pep superimposed on the Rnr2pep complex (C). Cα backbones are shown for Rnr2pep (yellow) and Rnr4pep (pink), and nearby helices are drawn from Rnr1 from the Rnr2pep complex (green). (D) E. coli Rnr2pep (gray) superimposed on yeast Rnr2p (yellow) and Rnr1 (green). (E) Sequence alignment through Rnr2pep-binding site. Secondary structure is shown for E. coli (above) and yeast (below). Residues that interact with the peptide are colored. The Rnr2pep residues binding a given subsite are listed below the corresponding sequence. (F) Mouse Rnr2pep (cyan) superimposed on yeast Rnr2pep (yellow); some side chains are omitted for clarity.
Both our peptide structures suggest that most of the binding energy appears to result from hydrophobic interactions, hydrogen bonds, and some ion-pair interactions. For instance, Rnr2pep makes three hydrogen bonds to Rnr1 and forms two long-range ion-pair interactions with K693 and K723, whereas F9, F5, and F3 are tucked into hydrophobic pockets (Fig. 3A). The Rnr4pep makes seven hydrogen bonds and two long-range ion pairs with K693 and K723, whereas the aromatic side chain of F9 occupies its hydrophobic pocket, and F5 stacks edge-to-face with F729 of αI (Fig. 3B). These extra interactions are consistent with Rnr1-binding data for similar C-terminal nonapeptides, which show that Rnr4pep binds Rnr1 slightly more strongly than Rnr2pep (13).
In the E. coli Rnr1 structure, an Rnr2-derived peptide binds between α13 and αI, roughly parallel to αI. However, our peptides bind Rnr1 almost orthogonal to αI, with their N termini near α13 and αD and their C termini near αH (Fig. 3D). These differences could be significant in structure-based drug design of anticancer peptidomimetics (15), which, until now, were designed based on the E. coli Rnr1 structure. Moreover, these results indicate that prokaryotic and eukaryotic Rnr1 bind Rnr2 differently.
As in mouse RNR, the C-terminal Rnr2 heptapeptide in yeast may constitute the minimal peptide length required for binding Rnr1 (35). Many residues interacting with Rnr2pep in yeast are conserved in mouse and human Rnr1 (Fig. 3E). In an NMR study, the N-acetylated mouse Rnr2 C-terminal heptapeptide (AcFTLDADF), P7 peptide, formed a nonstandard reverse turn between residues (2TLDA5) when it bound mouse Rnr1 (16). The first five residues of Rnr2pep (3FTFNE7) and the mouse peptide (1FTLDA5) superimpose with a Cα RMSD of 0.98 Å (Fig. 3F). The last two residues of the mouse peptide show less order in the NMR structure and superimpose poorly with Rnr2pep. Residues of mouse Rnr2 corresponding to F3 and F9 of Rnr2pep are considered critical for Rnr2’s binding to Rnr1 (35). This finding is not surprising, because the subsite that binds F3 in our structure comprises aromatic residues W389 and Y390, hydrophobic residues M721 and L393, and polar residues Q386 and T725. All but M721 and L393 are conserved in mouse, and these are substituted by Y and I, respectively. F9 binds at a subsite comprising Q692, K693, I696, K723, S726, M727, and Y730. I696 becomes L in mouse Rnr1; all others are conserved. Modeling an F-to-W substitution in the peptide at F9 introduces steric hindrance at the subsite, consistent with observed weaker binding of a similarly modified mouse peptide (15). The negatively charged C terminus of Rnr2pep is stabilized by an ion-pair interaction with K723 and a hydrogen bond to S691; these residues are conserved in mouse and may explain why removing the C-terminal negative charge decreases affinity (35). Moreover, a photoincorporation study using an azidophenyl derivative of the C-terminal seven residues of mouse Rnr2 showed that the peptide interacts with α helix 724–738 of mouse Rnr1 (36), equivalent to αI in yeast Rnr1. This helix makes the bulk of Rnr2pep interactions (Fig. 3E). Recently, to test the feasibility of using yeast Rnr1 for knowledge-based drug design of peptidomimetics that disrupt mammalian RNR assembly, we soaked the mouse P7 peptide (35) into our orthorhombic crystals and collected an x-ray data set. Initial difference Fourier electron-density maps reveal that the P7 peptide binds almost identically to the yeast Rnr2pep (data not shown, C.D., H.X., and B. S. Cooperman, unpublished results). Taken together, these results suggest evolutionary conservation of the peptide-binding site on eukaryotic Rnr1.
Conclusions
This work provides a molecular basis for understanding how eukaryotic Rnr1 binds GemdP, the diphosphate metabolite of the anticancer agent gemcitabine, compared with CDP, the analogous substrate. The unusual chemistry of the geminal fluorines is responsible for GemdP’s unique mode of binding. Our peptide complexes show that the Rnr2pep and Rnr4pep have different modes of binding to Rnr1, with the exception of F9 in both peptides, which bind at the same subsite. The binding-site overlap will only permit either Rnr2 or Rnr4 to bind a given Rnr1 monomer, resulting in asymmetric binding. The Rnr1–Rnr2pep complex structure provides an excellent model for designing Rnr2-based peptidomimetics and small molecules that disrupt RNR assembly, a potential class of anticancer and antiviral inhibitors (14, 15, 34).
Materials and Methods
Protein Purification and Crystallization.
The yeast Rnr1 expression plasmid as described in ref. 37 was used throughout this study. Yeast Rnr1 was overexpressed in E. coli BL21(DE3) pLysS strains as described in ref. 38. The cells were lysed by using the freeze–thaw method, and the protein was purified by using peptide-affinity chromatography as described in ref. 39.
Yeast Rnr1 was crystallized in the space group P21212 by using the hanging-drop method at 298 K. The crystals grew with a well solution containing 0.1 M sodium acetate, pH 6.5, 20–25% PEG 3350, and 0.2 M ammonium sulfate. One microliter of the well solution was mixed with 1 μl of protein at a concentration of 20 mg/ml. The AMPPNP–CDP and AMPPNP–GemdP complexes were obtained by soaking the orthorhombic crystals for 3 h in mother liquor containing 20 mM DTT and 10 mM MgCl2. Additionally, the soaking buffer of AMPPNP–GemdP contained 20 mM AMPPNP and GemdP, whereas the AMPPNP–CDP soaking buffer contained 20 mM AMPPNP and CDP. The Rnr2 peptide (GAFTFNEDF) and the Rnr4 peptide (KEINFDDDF) were synthesized at the Keck facility at Yale University (New Haven, CT) and soaked similarly. Although longer soaking times were explored, the ligands bound similarly. However, as the resolution limit of diffraction was lowered by longer soak times because of crystal deterioration, the structures reported were obtained with 3-h soaks.
X-Ray Data Collection, Structure Determination, and Refinement.
The native P21212 data were collected at BioCARS, at the Advanced Photon Source (APS). Although data were collected for the dATP–CDP complex at Northeastern Collaborative Access Team, the AMPPNP–CDP complex crystals diffracted to a higher resolution. Data for the AMPPNP–GemdP, AMPPNP–CDP, and peptide complexes were collected at our in-house x-ray facility by using an R-AXIS IV++ imaging plate mounted on a Rigaku rotating anode with X-stream cooling. Cryogenic data collection was performed at 100 K by transferring crystals into reservoir solution containing an additional 15% glycerol, then flash-freezing in liquid nitrogen. The data were integrated and scaled by using hkl2000 (40) (see Table 1).
Table 1.
Data collection and refinement statistics for ligand and peptide complex structures
| AMPPNP–CDP | AMPPNP–GemdP | R2 peptide | R4 peptide | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21212 | P21212 | P21212 | P21212 |
| Cell dimensions a, b, c, Å | 107.8, 117.5, 64.6 | 107.6, 117.2, 64.0 | 108.1, 117.6, 64.2 | 108.3, 117.7, 64.5 |
| Wavelength, Å | 1.54180 | 1.54180 | 1.54180 | 1.54180 |
| Resolution, Å | 50–2.9 | 50–2.3 | 50–2.4 | 50–2.8 |
| Unique reflections | 18,752 | 35,816 | 32,238 | 21,005 |
| Rsym, %* | 12.7 (40.8) | 9.1 (36.8) | 7.0 (47.2) | 10.3 (46.7) |
| I/σ(I) | 7.6 (3.4) | 9.9 (2.7) | 11.4 (3.2) | 8.8 (4.3) |
| Completeness, %* | 99.7 (100.0) | 97.5 (87.4) | 98.0 (97.0) | 99.8 (100.0) |
| Redundancy | 4.6 | 4.5 | 5.3 | 6.9 |
| Refinement | ||||
| Resolution, Å | 50–2.9 | 50–2.3 | 50–2.4 | 50–2.8 |
| No. of reflections | 16,829 | 36,701 | 28,071 | 18,789 |
| Rwork/Rfree† | 0.183/0.236 | 0.204/0.240 | 0.222/0.263 | 0.199/0.258† |
| No. of atoms | ||||
| Protein | 5,209 | 5,182 | 5,129 | 5,129 |
| Ligand/ion | 57‡ | 59§ | 97¶ | 55∥ |
| Water | 133 | 207 | 74 | 58 |
| B-factors | ||||
| Protein | 37.6 | 35.9 | 47.6 | 45.2 |
| Ligand/ion | 38.1 | 42.3 | 65.9 | 74.2 |
| Water | 31.4 | 33.4 | 38.2 | 35.1 |
| rms deviations | ||||
| Bond lengths, Å | 0.013 | 0.0065 | 0.014 | 0.014 |
| Bond angles, ° | 1.59 | 1.34 | 1.53 | 1.62 |
*Highest resolution shell is shown in parentheses.
†Rwork and Rfree = Σ∥Fo|−|Fc∥/Σ|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes. For the calculation of Rfree, 10% of the reflection data were selected and omitted from refinement.
‡The ligand/ion is CDP and AMPPNP.
§The ligand/ion is GemdP and AMPPNP.
¶The ligand/ion is R2 peptide.
∥The ligand/ion is R4 peptide.
The structure of yeast Rnr1 was determined by the multiwavelength anomalous dispersion method (41) by using a HgBr2-derivatized crystal of the P21212 form (30). The complex crystals are all isomorphous to the native P21212 form, and the structures were directly determined by the difference Fourier technique (Table 1). The graphics program o (42) was used for model building into omit maps, interspersed with refinement using both cns (43) and refmac (44). During the course of refinement, simulated annealing omit maps were computed by using the program cns and examined. The final models were all evaluated with the program procheck (45), and 99% of all residues were in the allowed region of the Ramachandran plot, with >85% in the most favored region.
Figures were prepared with the programs pymol (46) and ligplot (47).
Supplementary Material
Acknowledgments
We thank Dr. JoAnne Stubbe for critical review of the manuscript; Dr. Rodney Rothstein (Columbia University, New York) for providing expression plasmids; Drs. Anna Gardberg, Brad Bennett, and Joseph Brunzelle for help during data collection; Sanath Wijerathna and James Fairman for useful discussion; and the members of the BMC beamline at BioCARS and the members of the Northeastern Collaborative Access Team at Advanced Photon Source for data collection. The GemdP was a gift from the Eli Lilly Corporation, Indianapolis, synthesized by Dr. Don Saba. This work was supported by National Institutes of Health Grant 2 R01 CA1000827-03 from the National Cancer Institute.
Abbreviations
- CDP
cytidine diphosphate
- GemdP
difluorodeoxycytidine 5′-diphosphate
- RNR
ribonucleotide reductase.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2CVU (complex with AMPPNP and CDP), 2EUD (complex with AMPPNP and gemcitabine diphosphates), 2CVY (complex with R2 peptide), and 1ZZD (complex with R4 peptide)].
References
- 1.van der Donk W. A., Yu G., Silva D. J., Stubbe J., McCarthy J. R., Jarvi E. T., Matthews D. P., Resvick R. J., Wagner E. Biochemistry. 1996;35:8381–8391. doi: 10.1021/bi960190j. [DOI] [PubMed] [Google Scholar]
- 2.Szekeres T., Fritzer-Szekeres M., Elford H. L. Crit. Rev. Clin. Lab. Sci. 1997;34:503–528. doi: 10.3109/10408369709006424. [DOI] [PubMed] [Google Scholar]
- 3.Mayhew C. N., Phillips J. D., Greenberg R. N., Birch N. J., Elford H. L., Gallicchio V. S. Stem Cells. 1999;17:345–356. doi: 10.1002/stem.170345. [DOI] [PubMed] [Google Scholar]
- 4.Rosell R., Danenberg K. D., Alberola V., Bepler G., Sanchez J. J., Camps C., Provencio M., Isla D., Taron M., Diz P., Artal A. Clin. Cancer Res. 2004;10:1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- 5.Huang P., Chubb S., Hertel L. W., Grindey G. B., Plunkett W. Cancer Res. 1991;51:6110–6117. [PubMed] [Google Scholar]
- 6.Stubbe J. Curr. Opin. Struct. Biol. 2000;10:731–736. doi: 10.1016/s0959-440x(00)00153-6. [DOI] [PubMed] [Google Scholar]
- 7.Mao S. S., Holler T. P., Yu G. X., Bollinger J. M., Jr, Booker S., Johnston M. I., Stubbe J. Biochemistry. 1992;31:9733–9743. doi: 10.1021/bi00155a029. [DOI] [PubMed] [Google Scholar]
- 8.Stubbe J. Curr. Opin. Chem. Biol. 2003;7:183–188. doi: 10.1016/s1367-5931(03)00025-5. [DOI] [PubMed] [Google Scholar]
- 9.Bennati M., Robblee J. H., Mugnaini V., Stubbe J., Freed J. H., Borbat P. J. Am. Chem. Soc. 2005;127:15014–15015. doi: 10.1021/ja054991y. [DOI] [PubMed] [Google Scholar]
- 10.Yee C. S., Seyedsayamdost M. R., Chang M. C., Nocera D. G., Stubbe J. Biochemistry. 2003;42:14541–14552. doi: 10.1021/bi0352365. [DOI] [PubMed] [Google Scholar]
- 11.Perlstein D. L., Ge J., Ortigosa A. D., Robblee J. H., Zhang Z., Huang M., Stubbe J. Biochemistry. 2005;44:15366–15377. doi: 10.1021/bi051616+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sommerhalter M., Voegtli W. C., Perlstein D. L., Ge J., Stubbe J., Rosenzweig A. C. Biochemistry. 2004;43:7736–7742. doi: 10.1021/bi049510m. [DOI] [PubMed] [Google Scholar]
- 13.Chabes A., Domkin V., Thelander L. J. Biol. Chem. 1999;274:36679–36683. doi: 10.1074/jbc.274.51.36679. [DOI] [PubMed] [Google Scholar]
- 14.Pellegrini M., Liehr S., Fisher A. L., Laub P. B., Cooperman B. S., Mierke D. F. Biochemistry. 2000;39:12210–12215. doi: 10.1021/bi001323a. [DOI] [PubMed] [Google Scholar]
- 15.Pender B. A., Wu X., Axelsen P. H., Cooperman B. S. J. Med. Chem. 2001;44:36–46. doi: 10.1021/jm000335r. [DOI] [PubMed] [Google Scholar]
- 16.Fisher A., Laub P. B., Cooperman B. S. Nat. Struct. Biol. 1995;2:951–955. doi: 10.1038/nsb1195-951. [DOI] [PubMed] [Google Scholar]
- 17.Moss N., Beaulieu P., Duceppe J. S., Ferland J. M., Gauthier J., Ghiro E., Goulet S., Grenier L., Llinas-Brunet M., Plante R., et al. J. Med. Chem. 1995;38:3617–3623. doi: 10.1021/jm00018a022. [DOI] [PubMed] [Google Scholar]
- 18.Paradis H., Gaudreau P., Brazeau P., Langelier Y. J. Biol. Chem. 1988;263:16045–16050. [PubMed] [Google Scholar]
- 19.Moss N., Beaulieu P., Duceppe J. S., Ferland J. M., Garneau M., Gauthier J., Ghiro E., Goulet S., Guse I., Jaramillo J., et al. J. Med. Chem. 1996;39:4173–4180. doi: 10.1021/jm960324r. [DOI] [PubMed] [Google Scholar]
- 20.Uhlin U., Eklund H. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 21.Logan D. T., Andersson J., Sjoberg B. M., Nordlund P. Science. 1999;283:1499–1504. doi: 10.1126/science.283.5407.1499. [DOI] [PubMed] [Google Scholar]
- 22.Larsson K. M., Jordan A., Eliasson R., Reichard P., Logan D. T., Nordlund P. Nat. Struct. Mol. Biol. 2004;11:1142–1149. doi: 10.1038/nsmb838. [DOI] [PubMed] [Google Scholar]
- 23.Sintchak M. D., Arjara G., Kellogg B. A., Stubbe J., Drennan C. L. Nat. Struct. Biol. 2002;9:293–300. doi: 10.1038/nsb774. [DOI] [PubMed] [Google Scholar]
- 24.Uppsten M., Farnegardh M., Jordan A., Eliasson R., Eklund H., Uhlin U. J. Mol. Biol. 2003;330:87–97. doi: 10.1016/s0022-2836(03)00538-2. [DOI] [PubMed] [Google Scholar]
- 25.Larsson K. M., Andersson J., Sjoberg B. M., Nordlund P., Logan D. T. Structure (London) 2001;9:739–750. doi: 10.1016/s0969-2126(01)00627-x. [DOI] [PubMed] [Google Scholar]
- 26.Blundell T. L., Cooper J., Foundling S. I., Jones D. M., Atrash B., Szelke M. Biochemistry. 1987;26:5585–5590. doi: 10.1021/bi00392a001. [DOI] [PubMed] [Google Scholar]
- 27.Lunney E. A., Hamilton H. W., Hodges J. C., Kaltenbronn J. S., Repine J. T., Badasso M., Cooper J. B., Dealwis C., Wallace B. A., Lowther W. T., et al. J. Med. Chem. 1993;36:3809–3820. doi: 10.1021/jm00076a008. [DOI] [PubMed] [Google Scholar]
- 28.Dealwis C. G., Frazao C., Badasso M., Cooper J. B., Tickle I. J., Driessen H., Blundell T. L., Murakami K., Miyazaki H., Sueiras-Diaz J., et al. J. Mol. Biol. 1994;236:342–360. doi: 10.1006/jmbi.1994.1139. [DOI] [PubMed] [Google Scholar]
- 29.Eriksson M., Uhlin U., Ramaswamy S., Ekberg M., Regnstrom K., Sjoberg B. M., Eklund H. Structure (London) 1997;5:1077–1092. doi: 10.1016/s0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 30.Xu H., Faber C., Uchiki T., Fairman J. W., Racca J., Dealwis C. Proc. Natl. Acad. Sci. USA. 2006;103:4022–4027. doi: 10.1073/pnas.0600443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carosati E., Sciabola S., Cruciani G. J. Med. Chem. 2004;47:5114–5125. doi: 10.1021/jm0498349. [DOI] [PubMed] [Google Scholar]
- 32.Sabini E., Ort S., Monnerjahn C., Konrad M., Lavie A. Nat. Struct. Biol. 2003;10:513–519. doi: 10.1038/nsb942. [DOI] [PubMed] [Google Scholar]
- 33.Stubbe J., van der Donk W. A. Chem. Biol. 1995;2:793–801. doi: 10.1016/1074-5521(95)90084-5. [DOI] [PubMed] [Google Scholar]
- 34.Gao Y., Kashlan O. B., Kaur J., Tan C., Cooperman B. S. Biopolymers. 2005;80:9–17. doi: 10.1002/bip.20184. [DOI] [PubMed] [Google Scholar]
- 35.Fisher A., Yang F. D., Rubin H., Cooperman B. S. J. Med. Chem. 1993;36:3859–3862. doi: 10.1021/jm00076a015. [DOI] [PubMed] [Google Scholar]
- 36.Davis R., Thelander M., Mann G. J., Behravan G., Soucy F., Beaulieu P., Lavallee P., Graslund A., Thelander L. J. Biol. Chem. 1994;269:23171–23176. [PubMed] [Google Scholar]
- 37.Zhao X., Muller E. G., Rothstein R. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen H. H., Ge J., Perlstein D. L., Stubbe J. Proc. Natl. Acad. Sci. USA. 1999;96:12339–12344. doi: 10.1073/pnas.96.22.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang F. D., Spanevello R. A., Celiker I., Hirschmann R., Rubin H., Cooperman B. S. FEBS Lett. 1990;272:61–64. doi: 10.1016/0014-5793(90)80449-s. [DOI] [PubMed] [Google Scholar]
- 40.Minor W., Tomchick D., Otwinowski Z. Struct. Fold. Des. 2000;8:R105–R110. doi: 10.1016/s0969-2126(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 41.Hendrickson W. A., Horton J. R., LeMaster D. M. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 43.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Collaborative Computational Program Number 4 (CCP4) Science and Engineering Research Council. Warrington, U.K.: Daresbury Laboratory; 1985. [Google Scholar]
- 45.Laskowski R. A., Rullmann J. A., MacArthur M. W., Kaptein R., Thornton J. M. J. Biol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 46.DeLano W. L. pymol. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 47.Wallace A. C., Laskowski R. A., Thornton J. M. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.