Abstract
Seashore landfill aquifers are environments of special physicochemical conditions (high organic load and high salinity), and microbes in leachate-polluted aquifers play a significant role for intrinsic bioremediation. In order to characterize microbial diversity and look for clues on the relationship between microbial community structure and hydrochemistry, a culture-independent examination of a typical groundwater sample obtained from a seashore landfill was conducted by sequence analysis of 16S rDNA clone library. Two sets of universal 16S rDNA primers were used to amplify DNA extracted from the groundwater so that problems arising from primer efficiency and specificity could be reduced. Of 74 clones randomly selected from the libraries, 30 contained unique sequences whose analysis showed that the majority of them belonged to bacteria (95.9%), with Proteobacteria (63.5%) being the dominant division. One archaeal sequence and one eukaryotic sequence were found as well. Bacterial sequences belonging to the following phylogenic groups were identified: Bacteroidetes (20.3%), β, γ, δ and ε-subdivisions of Proteobacteria (47.3%, 9.5%, 5.4% and 1.3%, respectively), Firmicutes (1.4%), Actinobacteria (2.7%), Cyanobacteria (2.7%). The percentages of Proteobacteria and Bacteroides in seawater were greater than those in the groundwater from a non-seashore landfill, indicating a possible influence of seawater. Quite a few sequences had close relatives in marine or hypersaline environments. Many sequences showed affiliations with microbes involved in anaerobic fermentation. The remarkable abundance of sequences related to (per)chlorate-reducing bacteria (ClRB) in the groundwater was significant and worthy of further study.
Keywords: Saline groundwater, Landfill, 16S rRNA, Clone library, Phylogenetic analysis
INTRODUCTION
In some coastal cities of China, municipal solid waste (MSW) is ultimately landfilled at seashore zones with cheaper land-value and less disturbance to nearby residents. These seashore landfills were usually constructed without appropriate liners to prevent percolation of leachate into underlying aquifers. Dramatic hydrochemical changes in these aquifers have been detected as a result of the high organic load of leachate. Furthermore, these aquifers were inevitably infiltrated by seawater in the inflowing tide responsible for the high salinity (up to 18.5 g/L) of the groundwater.
It is often difficult and expensive to remediate a leachate-polluted environment; although microbes that naturally live in the subsurface may degrade, detoxify, or immobilize contaminants in a process called intrinsic bioremediation (Lovley, 2001). Research showed that the spreading of pollution in landfill aquifers is often less than expected due to natural attenuation (Christensen et al., 1994). It was shown that the composition of the microbial community at petroleum polluted aquifers was indicative of the occurrence of intrinsic bioremediation of benzene (Rooney-Varga et al., 1999). Likewise, composition of microbial communities could also be indicative of (potential for) intrinsic bioremediation at leachate-polluted aquifers. The ability to predict the potential for natural attenuation and the ability to monitor on-going degradation processes should help limit the number of landfills and aquifers that have to be actively remediated.
For this purpose, knowledge of the relationship between microbial community structure and hydrochemistry in leachate-polluted aquifers is required. Phylogenetic studies of microbial communities in several groundwater samples from a landfill were described by Röling et al.(2001). Members of the family Geobacteraceae were found to be strong contributors to the microbial communities in the iron-reducing environment. The author is not aware of investigations on seashore landfill aquifers which are environments of special physicochemical conditions (high organic load and high salinity). Microbes living there may have special adaptations, some of which may involve enzymes or other physiological properties with potential commercial significance.
This paper describes a phylogenetic study of the microbial community in a typical groundwater sample obtained from a seashore landfill by extraction of DNA and comparison of 16S rRNA gene sequences.
MATERIALS AND METHODS
Sample collection and microbes concentration
The Laogang landfill (120.97° E, 31° N), located along the shore of the East China Sea, is a primary example of the seashore landfills. This landfill is one of the two on-going landfills in Shanghai and began operations in 1989. In January 2003, sterile screw-cap bottles were used to obtain groundwater samples from a 17 m depth monitor well at the downstream of the landfill. After 3 volumes of standing water was removed with a special device, samples were immediately placed on ice for transport to laboratory and stored for less than 24 h at 4 °C. The microbial biomass of the groundwater was harvested by centrifugation at 4 °C for 10 min at 17000×g (Heraeus Varifuge 20RS, Germany) and frozen at −80 °C until DNA extraction.
Chemical analysis
Oxygen content, temperature and pH were measured in the field. Hydrochemical parameters were determined by using “China’s national measurement standards for oceanic environment GB17378-1998” and laboratory procedures.
DNA extraction, clone library construction and RFLP grouping
Total DNA was extracted from the biomass pellet by using a lysozyme/proteinase K/SDS treatment followed by standard phenol/chloroform extractions (Bond et al., 1995). The DNA were used for PCR amplification with the GeneAmp PCR System 2400 (Applied Biosystems, USA). All treatments were in triplicates. Two clone libraries were constructed from the DNA sample using two sets of different universal primers, S926f/L189r (Yu and Mohn, 2001) and 27f/1492r (Martin-Laurent et al., 2001). The primers S926f and L189r anneal to positions 910–926 of the 16S rRNA gene and positions 189–207 of the 23S rRNA gene (E. coli numbering), respectively. The PCR-amplified fragments containing approx. 600 bp of 16S rRNA gene and the corresponding library were referred to as JA-1. The primers 27f and 1492r anneal to positions 8–27 and positions 1492–1513 of the 16S rRNA gene (E. coli numbering). The resultant PCR products contained nearly complete 16S rRNA genes and the corresponding library was referred to as JA-2. PCR reaction mixtures and thermal cycling were performed under the conditions described by Yu and Mohn (2001) and Martin-Laurent et al.(2001), respectively.
The JA-1 and JA-2 libraries were constructed using the T-Vector PCR product cloning kit (Sangon, China). Clones were screened with PCR for the presence of the inserts using the corresponding primer pair in separate PCR reactions. The PCR products were purified using the 3S PCR product purification kit (Biocolor, China), and two aliquots were digested separately with MspI and RsaI for 2 h at 37 °C. The above digests were resolved on 2% agarose gels (Bio-Rad) and the resultant RFLP patterns were compared and grouped. One clone from each RFLP group within each clone library was chosen for sequencing.
Phylogenetic analysis of rRNA gene sequences
Sequencing was performed on an ABI PRISM 377 sequencer (Perkin-Elmer). The 16S rDNA region of those recombinant plasmids was sequenced using the T7 primer flanking the 16S rDNA region. The determined partial 16S rRNA gene sequences were compared with those available in GenBank by use of the BLAST network service to determine their approximate phylogenetic affiliations. Nucleotide sequences were aligned and phylogenetic trees were constructed, with the software ClustalX v.1.81 by using evolutionary distances (i.e., Jukes-Cantor distances) and the neighbor-joining method. Bootstrap analysis was applied to each generated tree to assign confidence levels to the nodes in the tree.
Nucleotide sequence accession numbers
Nucleotide sequences have been deposited in the GenBank under accession numbers AY381282 to AY381297 and AY391488 to AY391501.
RESULTS
Hydrochemical data of the groundwater sample are presented in Table 1.
Table 1.
Hydrochemical data on the groundwater sample
| Hydrochemical parameter | Value |
| Dissolved oxygen (mg/L) | <0.1 |
| pH | 7.6 |
| Total dissoluble solid (mg/L) | 15540 |
| CODCr (mg/L) | 32.2 |
| Total N (mg/L) | 15.6 |
| NO3−-N (mg/L) | 1.15 |
| Fe3+ (mg/L) | 21 |
| Hardness (mg/L) | 2362 |
| F− (mg/L) | 0.25 |
| LAS (mg/L) | <0.05 |
| Phenol (mg/L) | <0.002 |
| As (mg/L) | 0.019 |
| Hg (mg/L) | <0.00005 |
| Temperature (°C) | 13.5 |
| Cl− (mg/L) | 7468 |
| Dissolved organic carbon (mg/L) | 12.5 |
| CODMn (mg/L) | 10.3 |
| NH4+-N (mg/L) | 13.2 |
| NO2−-N (mg/L) | 0.004 |
| Fe2+ (mg/L) | 11.2 |
| SO42−-N (mg/L) | <8.0 |
| Mn (mg/L) | 0.485 |
| S2− (mg/L) | <0.01 |
| Pb (mg/L) | 0.136 |
| Ni (mg/L) | 0.0922 |
| Cr6+ (mg/L) | <0.004 |
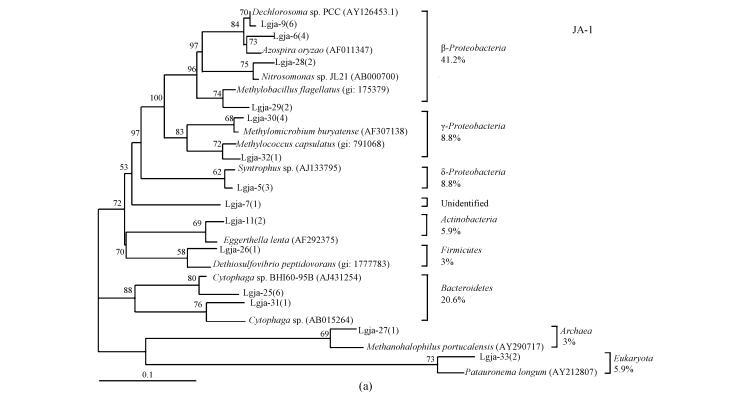
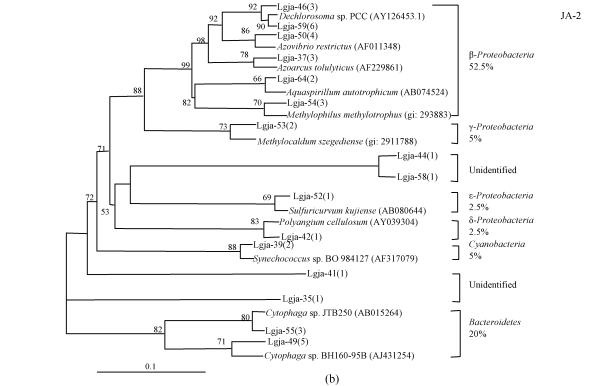
To reduce problems arising from primer efficiency and specificity, two sets of universal primers (S926f/L189r and 27f/1492r) were chosen to construct two libraries, JA-1 and JA-2, from the same sample. From 34 and 40 clones randomly selected in the JA-1 and JA-2 libraries, 14 and 16 gave unique RFLP patterns, respectively. Inserts of plasmids from each of the RFLP patterns were successfully sequenced producing in average 600-bp-long good quality sequences. Sequencing analysis showed that these 74 clones contained 30 unique sequences altogether. Closest known relatives of these sequences were identified by a search in GenBank databases, and two phylogenetic trees were produced for the JA-1 and JA-2 libraries (Fig.1).
Fig. 1.
Phylogenetic tree showing the affiliations of the partial 16S rRNA sequences, using nucleotides 926~1476 (JA-1) (a) and 27~567 (JA-2) (b) corresponding to E. coli 16S rRNA numbering, determined from 14 and 16 unknown groundwater clones found in the JA-1 and JA-2 libraries. The closest known microbe in GenBank is included as a reference for each sequence. Numbers in parentheses following clone identifications indicate the number of clones of each RFLP pattern. The relative abundances of different phylogenetic groups are also shown. Bootstrap values of more than 50 are reported as percentage. The scale bar corresponds to an estimated 0.1 mutation per nucleotide position
The majority of retrieved sequences belonged to bacteria (95.9%), with Proteobacteria (63.5%) being the dominant division. However, one archaeal sequence and one eukaryotic sequence were retrieved as well. Bacterial sequences belonging to the following phylogenic groups were identified: Bacteroidetes (20.3%), β, γ, δ and ε-subdivisions of Proteobacteria (47.3%, 9.5%, 5.4% and 1.3%, respectively), Firmicutes (1.4%), Actinobacteria (2.7%), Cyanobacteria (2.7%). Members of the β-subdivision were relatively most abundant among Proteobacteria representing 41.2% of the JA-1 library and 52.5% of the JA-2 library.
Most sequences in the Laogang landfill groundwater libraries were not closely affiliated with presently determined sequences of cultured bacteria. These results confirm suspicions raised by previous studies (Wise et al., 1997), i.e., that the vast majority of microbial diversity is uncharacterized.
DISCUSSION
In this study, hydrochemical data on dissolved organic carbon, CODCr, CODMn, NH4 +-N and Pb indicate that the groundwater sample was polluted by the leachate since a landfill represent a large source of organic carbon, nitrogen, and heavy metals. Additionally, the contents of Cl−, total dissoluble solid and hardness may be used as a relative measure of the influence of seawater infiltration.
Under subsurface environments, microbes use electron acceptors such as oxygen, nitrate, Fe3+, or sulfate to oxidize contaminants. Once these electron acceptors are depleted, anaerobic metabolism proceeds by converting organic matter to methane and carbon dioxide (Lovley, 2001). In regard to hydrochemical data on the groundwater sample, the contents of dissolved oxygen (<0.1 mg/L), nitrate (1.15 mg/L) and sulfate (<8.0 mg/L) were remarkably low. Fe3+ (21 mg/L) was likely to be the dominant electron acceptor and iron-reducing could be a major redox process.
The compositions of the JA-1 and JA-2 libraries were not significantly different from each other due to the common dominance of β-Proteobacteria and Bacteroidetes. However, four JA-1 phylogenic groups were not found in the JA-2 library and two JA-2 phylogenic groups were not found in the JA-1 library. The difference was not unexpected, although it is more reasonable to conduct an integrated analysis of the two libraries to get results close to the actual situation.
Phylogenetic analysis revealed the presence of Archaea and Eukaryota, and the overwhelming dominance of Bacteria. Among 74 clones, 63.5% clustered with Proteobacteria and 20.3% clustered with Bacteroides. The percentages of Proteobacteria and Bacteroides were greater than the results of Röling et al.(2001), who reported 45.7% and 5.8% abundance of the two groups in the groundwater clone library from the corresponding site of the Banisveld landfill. Many studies (Wise et al., 1997; Bowman et al., 1997) showed members of Proteobacteria dominate seawater (68.2%), and the widespread occurrence of members of the Bacteroides was also described in phylogenetic studies of sea sediments (Yanagibayashi et al., 1999; Li et al., 1999). It appeared that seawater influenced the microbial communities in the Laogang aquifers. Of course, due to PCR biases (Suzuki et al., 1998), the proportion of a 16S rDNA sequence in a library does not necessarily correspond to the same proportion in which a microbe with this 16S rRNA is present in a natural community. However, the qualitative complexity of a microbial community can be assessed.
A phylogenetic description of the microbial community importantly helps understanding of microbes participation in intrinsic bioremediation in aquifers. However, phylogenetic affiliation alone is not sufficient to firmly connect certain microbial taxa with a biochemical reaction (Todorov et al., 2000).
Keeping in mind these limitations, we can only hypothesize about metabolic potentials and hence, a possible biochemical role of bacteria in the groundwater. Hydrochemical data (Table 1) can be used to support the validity of our speculations. A significant number of sequences retrieved from the libraries have close relatives in marine or hypersaline environments. This finding is consistent with the high salinity content (15.5 g/L) of the groundwater contaminated by seawater. It may be surmised that only halophilic or halo-adapted bacteria could remain active in such an environment.
It appears that many Laogang groundwater sequences are related to anaerobic fermentative microbes. Lgja-27 is affiliated with halophilic methanogenic archaeon (Methanohalophilus portucalensis); Lgja-30 and Lgja-53 are associated phylogenetically with the methanotrophic bacteria (Methylomicrobium buryatense and Methylocaldum szegediense); Lgja-29 is closely related to Methylobacillus flagellatus, which is an obligate methylotroph; and Lgja-37 is related to potential denitrifier (Azoarcus tolulyticus), which is capable of anaerobic halobenzoate-degrading (Song et al., 2000).
Although iron reduction is deduced to be a major redox process, none of the sequences yielded by the groundwater sample were related to bacteria known to be iron-reducing. One group of sequences stands out as being particularly interesting and deserves further discussion. These sequences showed affiliations with (per)chlorate-reducing bacteria (ClRB), which couple growth to the reduction of chlorate or perchlorate under anaerobic conditions, and were found in relatively high abundance in the JA-1 libraries (Lgja-9, 17.6%) and the JA-2 library (Lgja-46 and Lgja-59, 22.5%). In general, oxyanions of chlorine are not formed naturally, but are introduced into the environment in large quantities in the form of disinfectants, bleaching agents and herbicides (Coates et al., 1999). In regard to the environment of this study, the most likely source of oxyanions was supposedly pesticides. Workers routinely sprayed pesticides in significant doses to kill flies, as there were significant amounts of flies present throughout the site due to the lack of soil cover. The remarkable abundance of ClRB-related sequences in the groundwater is significant and worthy of further study.
Our present work can be used as a starting point for in-depth studies. For example, their sequences can be used to develop new specific probes for quantitative analyses such as fluorescence in situ hybridization (FISH) or rRNA slot blot hybridization among more groundwater samples. Enough knowledge about the relationship between microbial community structure and hydrochemistry in leachate-polluted aquifers would be of help in predicting the potential for intrinsic bioremediation.
Acknowledgments
We are grateful to Dr. Wellington for his intellectual contributions and constructive criticism on the manuscript.
Footnotes
Project (No. 20377030) supported by the National Natural Science Foundation of China
References
- 1.Bond PL, Hugenholtz P, Keller J, Blackall L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Applied and Environmental Microbiology. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowman JP, McCammon SA, Brown MV, Nichols DS, McMeekin TA. Diversity and association of psichrophilic bacteria in antarctic sea ice. Applied and Environmental Microbiology. 1997;63:3068–3078. doi: 10.1128/aem.63.8.3068-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen TH, Kjeldsen P, Albrechtsen HJ, Heron G, Nielsen PH, Bjerg PL, Holm PE. Attenuation of landfill leachate pollutants in aquifers. Critical Review on Environmental Science and Technology. 1994;24:119–202. [Google Scholar]
- 4.Coates JD, Michaelidou U, Bruce RA, O’connor SM, Crespi JN, Achenbach LA. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Applied and Environmental Microbiology. 1999;65:5234–5241. doi: 10.1128/aem.65.12.5234-5241.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L, Kato C, Horikoshi K. Bacterial diversity in the deep-sea sediments from different depths. Biodiversity Conservation. 1999;8:659–677. [Google Scholar]
- 6.Lovley DR. Anaerobes to the rescue. Science. 2001;293:1444–1446. doi: 10.1126/science.1063294. [DOI] [PubMed] [Google Scholar]
- 7.Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon JC, Soulas G, Catroux G. DNA extraction from soils: old bias for new microbial diversity analysis methods. Applied and Environmental Microbiology. 2001;67:2354–2359. doi: 10.1128/AEM.67.5.2354-2359.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Röling WFM, Breukelen BM, Braster M, Lin B, Verseveld HW. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Applied and Environmental Microbiology. 2001;67:4619–4629. doi: 10.1128/AEM.67.10.4619-4629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rooney-Varga JN, Anderson RT, Fraga JL, Ringelberg D, Lovley DR. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Applied and Environmental Microbiology. 1999;65:3056–3063. doi: 10.1128/aem.65.7.3056-3063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song B, Palleroni NJ, Haggblom MM. Isolation and characterization of diverse halobenzoate-degrading denitrifying bacteria from soils and sediments. Applied and Environmental Microbiology. 2000;66:3446–3453. doi: 10.1128/aem.66.8.3446-3453.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki M, Rappe MS, Giovannoni SJ. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogenity. Applied and Environmental Microbiology. 1998;64:4522–4529. doi: 10.1128/aem.64.11.4522-4529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Todorov JR, Chistoserdov AY, Aller JY. Molecular analysis of microbial communities in mobile deltaic muds of Southeastern Papua New Guinea. FEMS Microbiology Ecology. 2000;33:147–155. doi: 10.1111/j.1574-6941.2000.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 13.Wise MG, Mcarthur JV, Shimkets LJ. Bacterial diversity of a carolina bay as determined by 16S rRNA gene analysis: confirmation of novel taxa. Applied and Environmental Microbiology. 1997;63:1505–1514. doi: 10.1128/aem.63.4.1505-1514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanagibayashi M, Nogi Y, Li L, Kato C. Changes in the microbial community in Japan Trench sediment from a depth of 6292 m during cultivation without decompression. FEMS Microbiology Letter. 1999;170:271–279. doi: 10.1111/j.1574-6968.1999.tb13384.x. [DOI] [PubMed] [Google Scholar]
- 15.Yu Z, Mohn WW. Bacterial diversity and community structure in an aerated lagoon revealed by ribosomal intergenic spacer analysis and 16S ribosomal DNA sequencing. Applied and Environmental Microbiology. 2001;67:1565–1574. doi: 10.1128/AEM.67.4.1565-1574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]