Abstract
Objective: To study the roles of different truncated hepatitis C virus (HCV) core proteins (CORE) in the pathogenesis of HCV persistent infection and hepatocellular carcinoma (HCC) and to assess intracellular localization in transiently transfected cells. Methods: Seven truncated CORE-GFP (green fluorescent protein) fusion protein expression plasmids were constructed, which contained HCV CORE sequences derived from tumor tissues (BT) and non-tumor tissues (BNT) from one patient infected with HCV. Amino acid (aa) lengths were BT: 1–172 aa, 1–126 aa, 1–58 aa, 59–126 aa, 127–172 aa; BNT: 1–172 aa and C191: 1–172 aa respectively. Subcellular localization of CORE-GFP was analyzed by con-focal laser scanning microscope. Apoptosis and necrosis were quantified by flow cytometry. Results: Different truncated CORE-GFP localized mainly in the cytoplasm, but nuclear staining was also observed. HCV CORE could induce apoptosis and necrosis, and different truncated COREs could induce cell apoptosis and necrosis at different levels. Among the same length 1–172 aa of BT, BNT and C191, the cell apoptosis and necrosis percentage of BT is highest, and C191 is the lowest (BT>BNT>C191). To the different fragment COREs of BT, N-terminal of CORE induced apoptosis and necrosis higher, compared with that of C-terminal (1–172 aa>1–126 aa>1–58 aa>127–172 aa>59–126 aa). Conclusion: These results suggest HCV CORE could induce apoptosis and necrosis of cells, which might play an important role in the pathogenesis of HCV persistent infection and HCC and the different CORE domains of different HCV quasi-species might have some difference in their pathogenesis.
Keywords: Hepatitis C virus, Core protein, Apoptosis, Necrosis
INTRODUCTION
The hepatitis C virus (HCV) is a major cause of chronic liver disease, liver cirrhosis and hepatocellular carcinoma (HCC). The pathogenesis of liver cell damage in hepatitis C (HC) and the role of viral proteins are partially understood (Suzuki et al., 1999). The HCV core protein (CORE) has been implicated in the regulation of cellular events, including apoptosis, but results have been conflicting and both proapoptotic and antiapoptotic effects were attributed to CORE in different experimental conditions. Different truncated forms of CORE have been described. Furthermore, different domains of CORE have different functions (Jin et al., 2000; Sabile et al., 1999). N-terminal 1–50 amino acid (aa) contains an RNA and DNA binding domain, nuclear localization signals (NSL). C-terminal 91–191 aa inactivates and binds to Leucine zipper (Jin et al., 2000) and 160–194 aa to apolipoprotein II (Sabile et al., 1999). The C-terminal domains contain hydrophobicity residues.
In most studies CORE was expressed in single clones of permanently transfected cell lines, while, more recently, transiently transfected cells were also used (Otsuka et al., 2002). These studies found that CORE affects normal cellular functions, such as cell proliferation and cell death, being involved, either directly or indirectly, in HCV hepatocarcinogenesis. Full-length CORE localizes in the cytoplasm and associates with endoplasmic reticulum (ER) and lipid droplets (Hope and McLauchlan, 2000). Truncated CORE forms, deleted of one or both C terminus hydrophobic domains, are localized in the nuclei (Yamanaka et al., 2002; Schwer et al., 2004; Xu et al., 2003). To further clarify the role of CORE in vivo, we have expressed several different truncated forms of CORE of different HCV strains derived from patient B-HCC tumour tissues (BT) caused by HCV infection, patient B-HCC non-tumour tissue (BNT) caused by HCV infection and C191 (HCV-J6), all belong to HCV genotype 1b, in transiently transfected HepG2 cells. The CORE proteins were fused with the green fluorescent protein (GFP) to allow analysis of subcellular localization in HepG2 by confocal laser scanning microscopy. Apoptosis and necrosis in transfected cells was quantified by flow cytometry.
MATERIALS AND METHODS
CORE gene amplification and plasmid construction
Seven different truncated CORE-GFP fusion plasmids were constructed (Figs.1 and 2). Different truncated CORE nucleotide sequences were amplified from pGEX 4T-1/HCV-CORE, containing CORE sequences from BT, BNT and C191 (HCV-J6) respectively. Sequence analysis revealed that BT, BNT and C191 were all HCV genotype 1b. The primers were designed according to the CORE gene sequence of BT, BNT and C191 (Table 1).
Fig. 1.
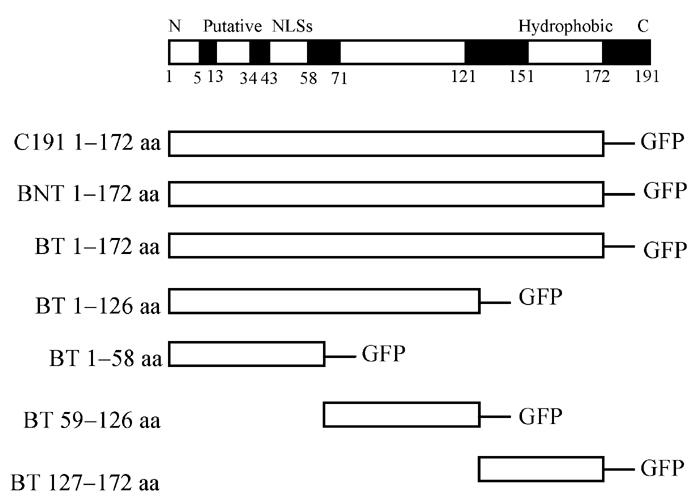
Schematic representation of the open reading frame encoding various truncated CORE. Functional domains are shown in shadowed boxes along the diagrams. All of them fused to GFP N-terminal
Fig. 2.
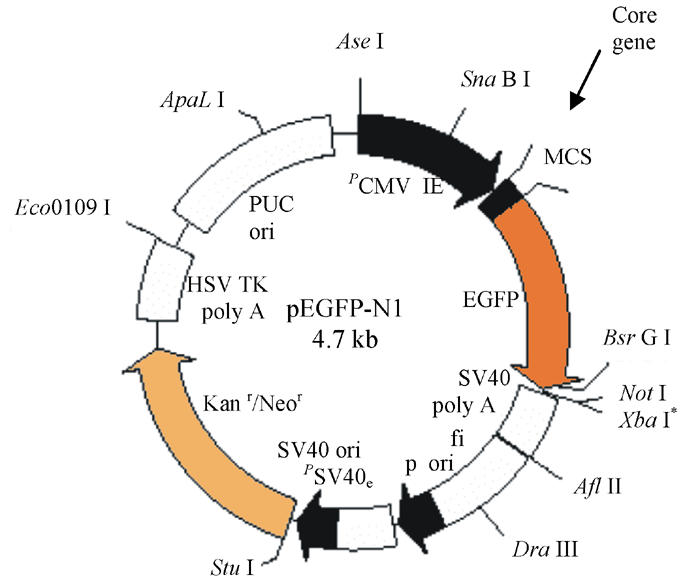
Schematic representation of different plasmid construction: Different truncated CORE were inserted into MCS: Nhe I and EcoR I to yield CORE-GFP fusion protein expression plasmids
Table 1.
Primers of different truncated CORE gene amplification
| Name | Primer up (location) | Primer down (location) |
| BT: 1–172 aa | CGCGCTAGCATGAGGCACGAATCC (1–14) | CCGGAATTCGCAACCGGGCAG (505–516) |
| BT: 1–126 aa | CGCGCTAGCATGAGCACGAATCC (1–14) | CCGGAATTCGAGGGTATCG (369–378) |
| BT: 1–58 aa | CGCGCTAGCATGAGCACGAATCC (1–14) | CCGGAATTCAGGTTGTGACCGC (162–174) |
| BT: 59–126 aa | CGCGCTAGCATGCCGCGTGGATCC (175–185) | CCGGAATTCGAGGGTATCG (369–378) |
| BT:127–172 aa | CGCGCTAGCATGCCGCGTGGATCC (379–391) | CCGGAATTCGCAACCGGGCAG (505–516) |
| BNT: 1–172 aa | CGCGCTAGCATGAGCACGAATCC (1–14) | CCGGAATTCGGCAACCGGGCAGATTC (505–516) |
| C191: 1–172 aa | CGCGCTAGCATGAGCACAAATCC (1–14) | CCGGAATTCGGCAACCGGGCAAATTC (505–516) |
GCTAGC: Nhe I; GAATTC: EcoR I
Polymerase chain reaction (PCR) reaction system (50 μl): H2O 40.5 μl, Buffer (10x) 5 μl, Template (45 ng) 1 μl, Primer up (20 pmol/L) 1 μl, Primer down (20 pmol/L) 1 μl, dNTP (20 mmol/L) 1 μl, Expand high enzyme 0.25 μl, 94 °C 2 min, 94 °C 30 s, 50 °C 30 s, 72 °C 30 s, 10 cycles, 94 °C 30 s, 55 °C 30 s, 72 °C 30 s, 20 cycles, 72 °C 7 min. PCR products were purified and cleaved with restriction enzymes Nhe I and EcoR I, then re-purified, and cloned into Nhe I and EcoR I-digested and purified pEGFP-N1 to yield CORE-GFP fusion protein plasmids (Fig.2). Positive clone were selected by PCR and sequence analysis.
Cell culture and transfection
Human hepatoma HepG2 was grown in Dulbeco’s modified eagle medium (DMEM, Life Technologies) supplemented with 10% fetal calf serum (FCS: Life Technologies), Cells were incubated for 10 h and then transfected with Effectene Reagent (Lipofectamine 2000) following the manufacturer’s instructions. Two μg of different truncated plasmids were used to achieve transfection. After 6 h, the medium was changed and cells were incubated for a further 48 h.
Confocal microscopy
Forty-eight hours after transfection, images of living cells were obtained with direct confocal microscope and argon or HeNe laser excitation. Subcellular localization of CORE-GFP was analyzed.
Flow cytometric analysis of apoptosis and necrosis
The apoptosis and necrosis percentage of cells transfected with different truncated CORE was compared 48 h after transfection. Cells were detached with 0.25% trypsin and washed twice with phosphorate buffered saline . To cells in 100 μl of PBS was added anti-annex-V at room temperature for about 10 min, then propidine iodide for 10 min. Finally 400 μl of the binding buffer was added. Cells were analyzed by flow cytometry.
RESULTS
Subcellular localization of different truncated HCV CORE
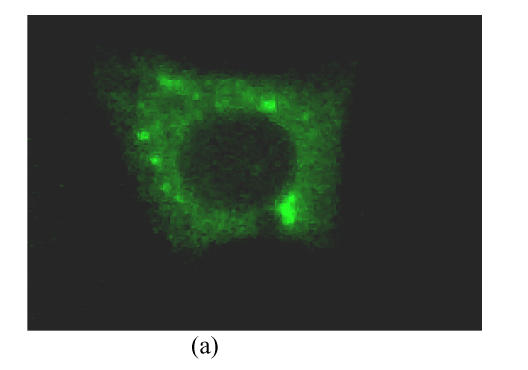
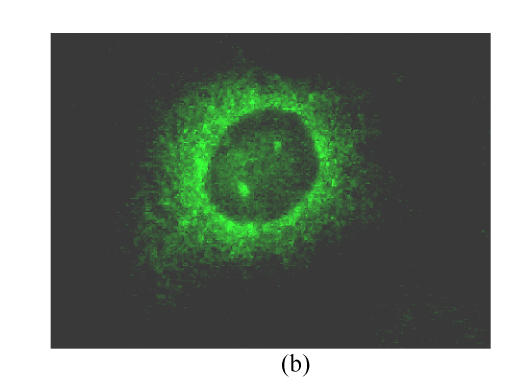
To investigate the intracellular distribution of different truncated CORE-GFP chimeric proteins in living cells, images of single transfected cells were obtained with confocal microscopy as shown in Fig.3. Cells transfected with different truncated CORE fused with GFP mainly showed granular cytoplasmatic pattern associating with ER (Fig.3a), also with a subpopulation of cells also showing nuclear staining (Fig.3b).
Fig. 3.
CORE-GFP localization in HepG2 cells. The confocal microscope setting for GFP detection was the same for all captured images: 488 nm excitation emission, laser power 10%, Iris1.8, Gain 75, background −10, emission filter 514/30. (a) CORE is only expressed in the cytoplasm associating with ER; (b) CORE is expressed not only in the cytoplasmic associating with ER but also in nuclear
Apoptosis and necrosis quantification in CORE expressing cells
The percentage of transfected cells showing apoptosis and necrosis was at least fifteen times as high among cells compared to control group. Therefore, HCV (genotype 1b) CORE could induce apoptosis and necrosis in transiently transfected HepG2. With the same length (N terminal 1–172 aa) among the different HCV strains BT, BNT and C191, CORE from the BT and BNT induced apoptosis higher than that of C191, and BT induced apoptosis and necrosis higher than those of BNT (BT>BNT>C191). In the same HCV strain, different truncated CORE induced apoptosis and necrosis at different levels, the longer the fragment, the higher the apoptosis and necrosis, and with N-terminal being higher than that of the C-terminal, the middle domain of CORE (59–126 aa) induced apoptosis and necrosis lower than those of the other truncated COREs derived from the same BT strain (1–172 aa>1–126 aa>1–58 aa>127–172 aa>59–126 aa). The results are summarized in Table 2.
Table 2.
Apoptosis and necrosis induced by the different truncated HCV-CORE
| Apoptosis (annex V+/PI−) | Necrosis (annex V+/PI+) | |
| Control | 2.13 | 0.2 |
| C191: 1–172 aa | 36.6 | 0.57 |
| BNT: 1–172 aa | 53.9 | 1.68 |
| BT: 1–172 aa | 70.0 | 3.19 |
| BT: 1–126 aa | 60.4 | 1.61 |
| BT: 1–58 aa | 62.1 | 1.25 |
| BT: 59–126 aa | 40.2 | 0.41 |
| BT: 127–172 aa | 59.5 | 0.76 |
DISCUSSION
The mechanisms leading to liver cell injury, inflammation, and fibrosis in chronic HC, are not fully understood. However, there is evidence to suggest that apoptosis of liver cells may play a significant role in the pathogenesis of HC (Sabile et al., 1999). In this study, we used a novel approach of transient expression of different truncated GFP-CORE to assess the function of these viral products. By using flow cytometry, we found that HCV-CORE exerted proapoptotic and pronecrosis effects.
Since HCV genome exhibits a considerable degree of variation, it is currently classified into 6 genotypes and more than 60 subtypes. Even in the same subtype, there exist quasi-species. However, CORE encoding genome among different genotypes is highly conserved. The nucleotide (nt) (Delhem et al., 2000) and aa sequences of BT, BNT, C191, have high homology at aa and nucleotide (nt) levels (data not shown). BT did not contain specific motif, but CORE containing the same length (1–172 aa) from BT, BNT and C191 induced apoptosis and necrosis at different levels. CORE from BT and BNT induced apoptosis higher than those of C191, and BT induced apoptosis and necrosis higher than those of BNT. Factors leading to CORE function differences of different HCV strains might be due to not only the CORE nt and aa sequence differences, but also to the conformation differences and host immune environment differences. But the exact mechanisms of BT and BNT induced apoptosis and necrosis being higher than those of C191, and those of BT being higher than those of BNT require further study.
Different domains of CORE have different functions (Jin et al., 2000; Sabile et al., 1999). Versatile functions of HCV CORE have been mapped to its N-terminal half. N-terminal is hydrophilic and contains clusters of basic aa residues which serve as NLS. The C-terminal domains contain hydrophobicity residues. CORE affects normal cellular functions, such as cell proliferation and cell death, being involved, either directly or indirectly in HCV hepatocarcinogenesis. We found that in different truncated CORE from the same strains derived from BT, the longer the truncated fragment, the higher the apoptosis and necrosis. N-terminal of CORE induced apoptosis and necrosis higher than those of C-terminal. The middle domain has the lowest induced apoptosis and necrosis percentages, which further showed that different domains of CORE indeed have some difference in their functions, especially those affecting cell apoptosis and necrosis, and that the function of CORE might depend on its conformation.
CORE could contribute to the development of HCC by promoting and perpetuating chronic inflammation and continuous regeneration of hepatic cells resulting in tumorigenesis. e.g., CORE may promote the apoptosis of immune cells during HCV infection via the Fas signaling pathway. CORE causes anti-apoptosis via activation of NF-kappaB (Kountouras et al., 2003) and may be implicated in a mechanism by which HCV may evade the host’s immune surveillance leading to viral persistence and possibly to hepatocarcinogenesis; CORE inhibits p53-mediated apoptosis by blocking the interaction between p53 and ASPP2, without modulating the transcriptional activity of p53, which plays a role in oncogenesis of HCC; CORE may also inhibit apoptosis through augmentation of Bcl-x expression, resulting in inhibition of caspase 3 activation (Kountouras et al., 2003). In this study, we found that the truncated COREs have proapoptotic and pronecrosis effects on transiently transfected HepG2 cells. CORE induced apoptosis and necrosis of infected hepatocytes, might reflect another important role of CORE in liver damage during chronic infection. It was proposed that the proapoptotic effect of CORE could contribute to the development of HCC by promoting continuous regeneration of hepatic cells (Delhem et al., 2000). On the other hand, apoptosis induced by different truncated COREs may also represent a mechanism for viral shedding rather than a mechanism for viral elimination. Recently, a different ability of nuclear and cytoplasmic forms of CORE in regulating p21 expression was described (Yamanaka et al., 2002), suggesting that the amount of nuclear protein could be important not only in inducing apoptosis but also in the regulation of the cell cycle. Even if we have not been able to demonstrate a significant effect of CORE on cell growth, it cannot be excluded that cells resistant to CORE mediated apoptosis could have enhanced growth and a replicative cycle. This is in agreement with the demonstration of an accelerated cell cycle coexisting with apoptosis induction in cell clones expressing CORE.
The apoptotic process appears to be a host defence mechanism against viral infections. However, many viral genomes encode proteins which repress apoptosis so as to escape from immune attack by the host. Therefore, virus-host interactions may determine viral persistence, extent and severity of liver inflammation. HCV E2 and NS5A proteins of HCV inhibit the activity of protein kinase R (PKR) (Ray and Ray, 2001; Taylor et al., 1999). Recent studies found that CORE increases PKR activity (Delhem et al., 2000; Francois et al., 2000; Realdon et al., 2004) and expression of 2′–5′ oligoadenylate synethase in the liver (Naganuma et al., 2000). Eventually CORE can decrease the suppression effect of E2 and NA5A on PKR, which keep PKR activity to a suitable level. Firstly, it is advantageous to keep viral replication at a lower level and to prevent persistent infection. Otherwise, if all proteins of HCV inhibit high PKR activity, HCV could replicate rapidly, leading to much hepatocyte apoptosis and necrosis and much HCV that is disadvantageous for escaping host immune surveillance. It is also advantageous for HCV to select the hepatocarcinogenesis virus strains that can resist the anti-viral and pro-apoptotic action of PKR and resist the host immunity aggression (Delhem et al., 2000).
Regarding the HCV-induced apoptotic mechanism, the core protein, although a structural component of the virus, may have a regulatory function in modulating apoptosis by either enhancing or inhibiting it. In particular, the core protein exhibits both proapoptotic or antiapoptotic actions, and different experient found different expression sites in the subcellular level, depending on experimental conditions such as CORE from different HCV strains, different truncated forms of CORE and types of cells used (Sabile et al., 1999).
By using confocal analysis, we found that CORE of genotype 1b mainly expressed in the cytoplasm associating with ER. To CORE of the same strains from HCC tissue, different truncated COREs localized mainly in the cytoplasm, although nuclear staining was also observed. Without comparing the mean fluorescence intensity of cytoplasm and nuclear staining of different truncated COREs, we did not find the expression site difference of different truncated COREs. Since some studies showed that nuclear localization of CORE has been previously shown to occur only with C-terminus deleted forms (Yamanaka et al., 2002; Delhem et al., 2000; Naganuma et al., 2000; Schwer et al., 2004; Xu et al., 2003), this would suggest that apoptosis could be mediated also in such conditions by truncated forms of COREs generated by in vivo cleavage.
In conclusion, we have shown that COREs are mainly localized in the cytoplasm and induce apoptosis and necrosis in HepG2.
Footnotes
Project (No. 2001BA705B06) supported by a grant from the National Key Technologies R&D program (10th Five-Year Plan) of China
References
- 1.Delhem N, Sabile A, Gajardo R, Podevin P, Abadie A, Balton MA, Kremsdorf D, Bretta L, Brechot C. Activation of the interferon-inducible protein kinase PKR by hepatocelluar carcinoma derived-hepatitis C virus CORE protein. Oncogene. 2000;20:5836–5845. doi: 10.1038/sj.onc.1204744. [DOI] [PubMed] [Google Scholar]
- 2.Francois C, Duverlie G, Rebouillat D, Khorsi H, Castelain S, Blum HE, Gatignol A, Wychowski C, Moradpour D, Meurs EF. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J Virol. 2000;74:5587–5596. doi: 10.1128/jvi.74.12.5587-5596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hope RG, McLauchlan J. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus CORE protein. J Gen Virol. 2000;81:1913–1925. doi: 10.1099/0022-1317-81-8-1913. [DOI] [PubMed] [Google Scholar]
- 4.Jin DY, Wang HL, Zhou Y, Chun AC, Kibler KV, Hou YD, Kung H, Jeang KT. Hepatitis C virus CORE protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 2000;19(4):729–740. doi: 10.1093/emboj/19.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kountouras J, Zavos C, Chatzopoulos D. Apoptosis in hepatitis C. J Viral Hepat. 2003;10(5):335–342. doi: 10.1046/j.1365-2893.2003.00452.x. [DOI] [PubMed] [Google Scholar]
- 6.Naganuma A, Nozaki A, Tanaka T, Sugiyama K, Takagi H, Mori M, Shimotohno K, Kato N. Activation of the interferon-inducible 2-5-oligoadenylate synethase gene by hepatitis C virus CORE protein. J Virol. 2000;74:8744–8750. doi: 10.1128/jvi.74.18.8744-8750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otsuka M, Kato N, Taniguchi H, Yoshida H, Goto T, Shiratori Y, Omata M. Hepatitis C virus CORE protein inhibits apoptosis via enhanced Bcl-xL expression. Virology. 2002;296:84–93. doi: 10.1006/viro.2002.1371. [DOI] [PubMed] [Google Scholar]
- 8.Ray RB, Ray R. Hepatitis C virus CORE protein: intriguing properties and functional relevance. FEMS Microbiol Lett. 2001;202:149–156. doi: 10.1111/j.1574-6968.2001.tb10796.x. [DOI] [PubMed] [Google Scholar]
- 9.Realdon S, Gerotto M, Dal Pero F, Marin O, Granato A, Basso G, Muraca M, Alberti A. Proapoptotic effect of hepatitis C virus CORE protein in transiently transfected cells is enhanced by nuclear localization and is dependent on PKR activation. J Hepatol. 2004;40:77–85. doi: 10.1016/j.jhep.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 10.Sabile A, Perlemuter G, Bono F, Kohara K, Demaugre F, Kohara M, Matsuura Y, Miyamura T, Brechot C, Barba G. Hepatitis C virus CORE protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology. 1999;30:1064–1076. doi: 10.1002/hep.510300429. [DOI] [PubMed] [Google Scholar]
- 11.Schwer B, Ren S, Pietschmann T, Kartenbeck J, Kaehlcke K, Bartenschlager R, Yen TS, Ott M. Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif. J Virol. 2004;78(15):7958–7968. doi: 10.1128/JVI.78.15.7958-7968.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki R, Suzuki T, Ishii K, Matsuura Y, Miyamura T. Processing and functions of hepatitis C virus proteins (review) Intervirology. 1999;42:145–152. doi: 10.1159/000024973. [DOI] [PubMed] [Google Scholar]
- 13.Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285(5424):107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 14.Xu Z, Choi J, Lu W, Ou JH. Hepatitis C virus f protein is a short-lived protein associated with the endoplasmic reticulum. J Virol. 2003;77(2):1578–1583. doi: 10.1128/JVI.77.2.1578-1583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamanaka T, Kodama T, Doi T. Subcellular localization of HCV CORE protein regulates its ability for p53 activation and p21 suppression. Biochem Biophys Res Commun. 2002;294:528–534. doi: 10.1016/S0006-291X(02)00508-9. [DOI] [PubMed] [Google Scholar]