Abstract
Microarray expression analysis is providing unprecedented data on gene expression in humans and mammalian model systems. Although such studies provide a tremendous resource for understanding human disease states, one of the significant challenges is cross-referencing the data derived from different species, across diverse expression analysis platforms, in order to properly derive inferences regarding gene expression and disease state. To address this problem, we have developed RESOURCERER, a microarray-resource annotation and cross-reference database built using the analysis of expressed sequence tags (ESTs) and gene sequences provided by the TIGR Gene Index (TGI) and TIGR Orthologous Gene Alignment (TOGA) databases [now called Eukaryotic Gene Orthologs (EGO)].
Rationale
Microarray expression analysis [1] has become one of the most widely used techniques for the assessment of gene expression on a genomic scale, allowing tens of thousands of genes to be assayed in a single experiment. Although the results that have emerged from microarray profiling have been impressive, as the technique has become more widespread the proliferation of platforms and reagents has made comparisons of results from disparate experimental groups a significant challenge. A further, and possibly more important, need is the ability to make comparisons of gene expression patterns between species. Analysis of gene expression in model organisms, particularly mouse and rat, has become a fundamental tool for the study of human development and disease. The challenge is linking the genes surveyed in these animal models to the corresponding human genes.
To address these issues, we have developed RESOURCERER [2], a database designed to provide annotation for widely used microarray platforms and to allow the genes represented to be compared within and across species. RESOURCERER is built as an extension of the TIGR Gene Indices (TGI) [3,4] and TOGA, the TIGR Orthologous Gene Alignment database ([5] and Y.L., R.S., G.P., J.C., S.K., J.T., B.P., F.C., V.A. and J. White, unpublished), and provides information for the most widely used microarray mammalian gene resources, including the Research Genetics sequence-verified human cDNA clone set (information about which can be found through [6]), the Brain Molecular Anatomy Project (BMAP; generated through NIMH/NINDS contract N01 MH80014 awarded to the University of Iowa; M.B. Soares, PI) [7] and NIA [8,9] mouse clone sets, the TIGR Rat Gene Index cDNA collection, human and mouse 70-mer oligonucleotide sets from Operon (information available through [10]), and the Affymetrix human, mouse, and rat GeneChip™ sets [11]. Additional resource sets from these species can quickly be added; a number have been, based on user requests, including the Affymetrix Mouse v2 GeneChip™. In addition, users can submit lists of GenBank accession numbers from a single species and find corresponding elements and their orthologs in any of the catalogued array resources.
Links to the TIGR Gene Index and TOGA databases
The relationships captured in RESOURCERER are based on the analysis of EST and gene sequences stored in the TGI and TOGA databases. The TGI databases [4] provide an analysis of publicly available EST and gene sequence data to identify transcripts, to place them into a genomic context, and to identify orthologs and paralogs where possible.
TGI treats ESTs and coding sequences as elements of a transcriptome shotgun sequencing project and uses them to assemble 'tentative consensus' (TC) sequences. ESTs are downloaded daily from dbEST [12], and are cleaned to remove untrimmed vector, linker, ribosomal, mitochondrial, low quality, and poly(T) sequences. Coding sequence (CDS) and CDS-join (coding sequences annotated as spanning multiple GenBank records) features are separately parsed from GenBank [13] records and stored locally. Gene and cleaned-EST sequences are compared pair-wise to identify overlaps using BLAST [14,15]; sequences with a minimum of 95% identity over a 45 base-pair (bp) or longer region are grouped into a cluster. The sequences within each cluster are assembled at high stringency using CAP 3 [16] to produce TC sequences, which are loaded into the appropriate species-specific database. TCs are annotated to provide a provisional functional assignment, and the resulting Gene Index database is released through the TGI website [4]. Gene Indices can be searched by TC number, the GenBank accession number of any EST contained within the dataset, or any gene used to build the Index. Users can perform a tissue-based search in which the library information in EST records is used to generate an 'electronic Northern Blot', identifying the tissue-specificity of expression on the basis of relative EST abundance. DNA and protein sequences can also be used to search the Gene Indices using WU-BLAST [17,18], a gapped BLAST program developed by Warren Gish [15,18]. The TIGR Gene Indices and the component TC assemblies are maintained within Sybase relational databases that allow versioning and heritability to be maintained. Each time a new version of the database is created, novel assemblies, caused by either the joining or the splitting of previous TCs, are assigned a new, unique TC identifier. Previously used identifiers are never reused and information regarding previous assemblies is never lost. Database queries using a TC identifier from a previous build return the most current version of that assembly, allowing assemblies to evolve while maintaining functional assignments across multiple releases.
We developed the TOGA database [5] to provide a cross-reference between the eukaryotic species highly sampled by EST and genomic sequencing projects. Starting with the assembled EST and gene sequences that comprise the 28 TGI databases, we use high-stringency pair-wise sequence searches and a reflexive, transitive closure process to associate sequence-specific best hits, generating 32,652 Tentative Orthologue Groups (TOGs). This allows us to identify putative orthologs and paralogs for known genes, as well as those that exist only as uncharacterized ESTs, and to provide links to additional information including genome sequence and mapping data. TOGA provides an important resource for the analysis of gene function in eukaryotes.
The RESOURCERER database
For each human, mouse, or rat microarray resource (full details of which are included in Table 1), including widely distributed clone sets [6,7,8,9,10] and the commercially available Affymetrix GeneChips™ [11], we loaded the clone or feature identifiers, as appropriate, into the RESOURCERER database, along with the associated GenBank accession numbers and other available annotation data, such as UniGene [19] cluster IDs, if provided by the creators of the primary resources. The GenBank accession numbers are then used as keys to link these resources to the TC sequences in the appropriate TGI database, and through those, to the orthologs captured in TOGA; as the TGI and TOGA databases are updated, annotations based upon them are updated as well, providing the most current annotation possible.
Table 1.
Array resources currently represented in RESOURCERER, with the total number of elements in each
| Species | Dataset | Total number of elements |
| Human | Research Genetics | 46,656 |
| Operon Human | 13,972 | |
| Affymetrix Human All | 63,175 | |
| Affymetrix_HG-U95A | 12,626 | |
| Affymetrix_HG-U95B | 12,620 | |
| Affymetrix_HG-U95C | 12,646 | |
| Affymetrix_HG-U95D | 12,644 | |
| Affymetrix_HG-U95E | 12,639 | |
| Rat | TIGR 13K Rat Set | 12,362 |
| Affymetrix Rat All | 26,379 | |
| Affymetrix_RG-U34A | 8,799 | |
| Affymetrix_RG-U34B | 8,791 | |
| Affymetrix_RG-U34C | 8,789 | |
| Mouse | NIA | 15,247 |
| BMAP | 11,136 | |
| NIA + BMAP | 26,383 | |
| Operon Mouse | 6,868 | |
| Affymetrix Mouse All | 38,018 | |
| Affymetrix_MG-U74A | 12,654 | |
| Affymetrix_MG-U74B | 12,636 | |
| Affymetrix_MG-U74C | 12,728 |
The Research Genetics human clone set is their sequence-verified collection [6]. The Operon oligonucleotide sets are available for human, mouse, and rat [10]. GeneChips™ from Affymetrix [11] are listed separately for each species, as well as collectively where, for example, 'Affymetrix Mouse All' is MU-74A + MU-74B + MU-74C. In mouse, the BMAP clone set was derived through the Brain Molecular Anatomy Project [7], and the NIA clone set was developed by M. Ko through a survey of mouse developmental stages [8,9]. The TIGR Rat 13K set was produced through an NHLBI funded project to generate a collection of unique, annotated rat cDNA clones.
As all data are stored in a relational database, we are then able to make a variety of complex queries. For each represented microarray resource, RESOURCERER allows creation of a table of TGI-based annotation for each element of the set, including TC numbers, putative functional assignments, and links to orthologous TCs in the other species (Figure 1). Further, one can use RESOURCERER to make comparisons between two resource sets derived from the same species, including identifying both the intersection and difference between those sets, with comparisons based on the appropriate species-specific TGI database, or UniGene identifiers, if available (see Table 2 for details of the relationships between the different array resources accessed through RESOURCERER). Finally, by using TOGA, orthologous genes represented in these microarray resources can be identified, facilitating comparisons of gene expression patterns between species (Figure 2).
Figure 1.
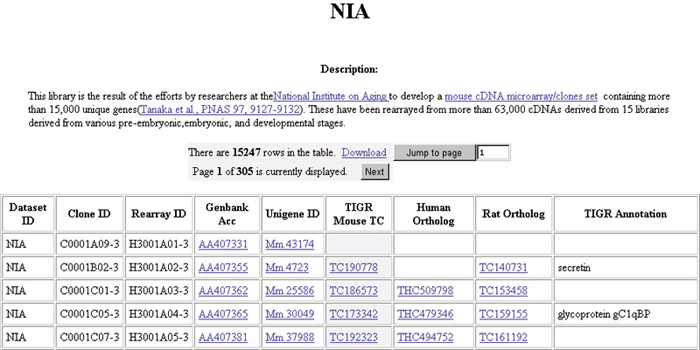
An example of the data provided for the NIA mouse cDNA collection [8,9]. Annotation for individual microarray resource sets is provided by the TGI databases, including functional assignments (where available), links to the TCs for the species in question, links to orthologous TCs in other mammalian species, and UniGene [19] cluster IDs where available. Text in blue is hot-linked to various databases through the RESOURCERER website.
Table 2.
The number of orthologous and/or corresponding genes shared between array resources across and/or within species, based on the TOGA and TGI databases
| Research Genetics | Operon human | Affymetrix human all | TIGR 13K rat | Affymetrix rat all | NIA | BMAP | Operon mouse | Affymetrix mouse all | |
| Research Genetics | 11,854 | 39,573 | 10,923 | 5,840 | 7,331 | 7,056 | 5,375 | 10,060 | |
| Operon human | 18,969 | 3,949 | 7,372 | 4,825 | 4,479 | 7,037 | 5,404 | ||
| Affymetrix human all | 7,849 | 15,219 | 9,372 | 9,266 | 8,639 | 12,802 | |||
| TIGR 13K rat | 8,434 | 3,877 | 3,816 | 2,395 | 4,521 | ||||
| Affymetrix rat all | 6,739 | 6,588 | 4,499 | 8,199 | |||||
| NIA | 5,543 | 3,016 | 8,744 | ||||||
| BMAP | 2,997 | 8,715 | |||||||
| Operon mouse | 2,039 |
The array resources are those listed in Table 1.
Figure 2.
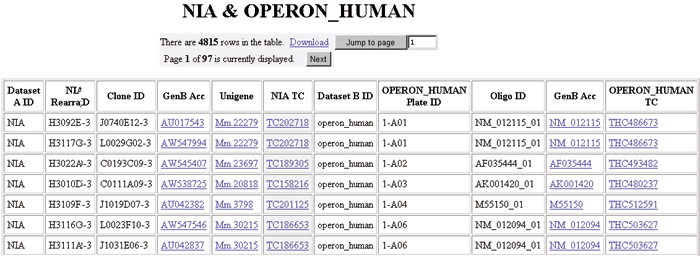
Using RESOURCERER, one can identify corresponding orthologous elements in various microarray resources, providing the information necessary to facilitate cross-species comparisons. Note that in this comparison between the NIA mouse cDNA collection [8,9] and the Operon human oligos [10], redundancy in the clone set is captured by multiple rows in the table.
It is this final feature that provides the greatest utility in RESOURCERER. Mouse and rat are ideal organisms for comparative analysis of mammalian coding sequences. Mouse is the premier organism for the study of mammalian genetics and development, while rat has been extensively used for physiological and pharmacological studies. Mouse and rat genome projects, involving genetic and physical mapping, EST sequencing, and genomic sequencing, are underway and progressing rapidly. Consequently, there is a tremendous opportunity to understand disease processes in humans by comparing and contrasting gene expression profiles in both mice and rats, and linking these to patterns observed in human patients. RESOURCERER provides a crucial tool for making such comparisons. Already, RESOURCERER has been used to facilitate comparisons between patterns of expression observed in rodent models of tumor metastasis and those seen in patients (N.H. Lee, personal communication). As expression analysis programs continue to expand, comparison between experiments and experimental systems will be increasingly important. RESOURCERER plays the critical role of facilitating these comparisons.
Accessing RESOURCERER
RESOURCERER is freely available from the TIGR website [2], which includes a 'readme' help file. Users can select a single, existing microarray resource and retrieve an annotation based on the TGI and TOGA, including functional assignments and links to putative orthologs. Selecting two resources derived from the same species allows users to identify either common elements shared by the set or those elements that are unique to either. If resources from two different species are selected, the user is provided with a set of the elements in each that are orthologous to each other as identified by TOGA. Finally, users submitting a list of GenBank accession numbers representing ESTs from a single species are provided with annotation as well as the corresponding elements and their orthologs in any of the catalogued array resources.
Acknowledgments
Acknowledgements
This work was supported by grants from the US Department of Energy, the National Science Foundation, and the National Heart, Lung, and Blood Institute. The authors thank N.H. Lee for valuable comments.
Disclaimer: Whilst every effort is made by the authors, editors and publishers to ensure that the software performs as described, they accept no liability whatsoever for the consequences of downloading or using it.
References
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- RESOURCERER http://pga.tigr.org/tigr-scripts/nhgi_scripts/resourcerer.pl
- Quackenbush J, Cho J, Lee Y, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana J, White J. The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acid Res. 2001;29:159–164. doi: 10.1093/nar/29.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TIGR Gene Indices http://www.tigr.org/tdb/tgi.shtml
- TIGR Orthologous Gene Alignment database http://www.tigr.org/tdb/tgi/ego/index.shtml [All references in this article to TOGA and TIGR Orthologous Gene Alignments have been changed to EGO and Eukaryotic Gene Orthologs, respectively].
- ResGen, an Invitrogen Corporation http://www.resgen.com
- Trans-NIH Brain Molecular Anatomy Project http://trans.nih.gov/resources/resources.htm
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, et al. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci USA. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIA Mouse cDNA Project http://lgsun.grc.nia.nih.gov/cDNA/cDNA.html
- Operon, a QIAGEN company http://www.operon.com
- Affymetrix http://www.affymetrix.com
- dbEST Expressed Sequence Tags database http://www.ncbi.nlm.nih.gov/dbEST/index.html
- GenBank http://www.ncbi.nlm.nih.gov/Genbank/
- BLAST http://www.ncbi.nlm.nih.gov/BLAST/
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU-BLAST of The TIGR Gene Indices http://www.tigr.org/cgi-bin/BlastSearch/blast_tgi.cgi
- WU-BLAST http://blast.wustl.edu
- UniGene http://www.ncbi.nlm.nih.gov/UniGene/


