Abstract
The fission yeast Schizosaccharomyces pombe has long been a model organism for studies of eukaryotic cells, winning renown especially for studies of the cell cycle. Now that its genome has been sequenced, S. pombe is ready to assume its rightful place in the pantheon of small eukaryotic giants.
For those molecular cell biologists who count the fission yeast Schizosaccharomyces pombe as their favorite model organism, 2001 has justly been celebrated as a most memorable year. Paul Nurse, the widely acknowledged leader of the S. pombe field, was one of the recipients of the Nobel Prize in Physiology or Medicine for his studies of cell-cycle control - along with Lee Hartwell, who founded cell-cycle genetics using the budding yeast Saccharomyces cerevisiae, and Tim Hunt who discovered cyclin. Also in 2001, the whole genome sequence of S. pombe was completed, by a team headed by Nurse at the Imperial Cancer Research Fund (now Cancer Research UK) in London and Bart Barrell at the Wellcome Trust Sanger Institute (Hinxton, UK). The paper describing key features and interpretation of the entire genome sequence of S. pombe has now been published in Nature [1] (an event that will be celebrated at the forthcoming 2nd International Fission Yeast Meeting, March 25-30 in Kyoto, Japan).
S. pombe is the sixth model eukaryotic organism whose genome has been fully sequenced, following the budding yeast S. cerevisiae, the nematode worm Caenorhabditis elegans, the fruit fly Drosophila melanogaster, mustard weed Arabidopsis thaliana and (in draft) the human, Homo sapiens. In this article, I consider some of the key features of the genome as identified by the complete sequence, as well as the place S. pombe occupies in past and future studies of eukaryotic cells.
How many genes does it take to make a eukaryote?
The sequenced genome of S. pombe holds no hint of a large-scale genomic duplication of the type that occurred in budding yeast and many other organisms; there is a small (approximately 50 kilobase) duplication near the telomere, however. This 'lower' eukaryotic unicellular organism has 4,824 predicted genes, the smallest number recorded for a eukaryote and significantly fewer than the 5,570-5,651 genes predicted for budding yeast. As the bacterium Streptomyces coelicolor has 7,846 genes, two conclusions that can be drawn are that a free-living eukaryote can be constructed with fewer than 5,000 genes, and that it may even need fewer genes than some prokaryotes. It is a matter of judgment whether this number seems small or large for the task of creating a eukaryotic cell from molecular components; in any case, S. pombe will for some time to come hold an honorable position as the eukaryote with the fewest genes.
Around two-thirds of the predicted fission yeast proteins have homologs in both budding yeast and nematode, and only 14% are found in neither budding yeast nor nematode. 'All against all' sequence comparisons indicate that 4,515 genes among the total 4,876 have no homologs within the S. pombe genome itself, while the remaining 361 genes' products are distributed among groups of proteins that have two or more members. Using the same type of comparison in budding yeast, 5,061 genes are unique while 716 fall into groups that have two or more members. The fission yeast genome can thus be said to be 'less redundant' than that of budding yeast; this has been empirically known, but now we can put a number on redundant genes.
A 'typical' eukaryote?
One characteristic of the fission yeast genome is that 43% of its genes have introns (often rather short; a total of 4,730 introns), many more than are found in budding yeast. This difference was also previously well established on the basis of gene-by-gene experiments, and is borne out by the whole-genome sequence. From gene-sequence comparisons, the two yeasts are estimated to have diverged from each other around 330-420 million years ago, and from metazoa and plants around 1,000-1,200 million years ago. Because the rates of divergence seem to be fast within fungi, the sequences of the two yeasts are nevertheless as divergent from each other as either yeast is from higher eukaryotes. The morphology of the two yeasts is also very different: unlike the rounded mother and budding daughter cells of S. cerevisiae, S. pombe has rod-shaped cells that divide along the long axis (Figure 1). The gene density for the complete genome of S. pombe is one gene every 2,528 base pairs, as compared to one gene every 2,088 base pairs for budding yeast. Regions upstream of genes are also significantly longer than in budding yeast, possibly reflecting more extended control regions.
Figure 1.
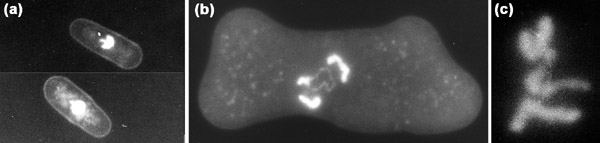
S. pombe cells. (a) Wild-type interphase cells; (b,c) β-tubulin mutant cells at the restrictive temperature. The condensed chromosomes are stained with DAPI; individual chromosomes can each be recognized by their distinctive sizes.
Unlike most eukaryotic sequencing papers, the S. pombe genome paper [1] reports sequencing of most of each of the three centromeres (35, 65 and 110 kilobases on chromosomes I, II and III, respectively), revealing a complex organization. It was my personal delight that the structures determined in my laboratory [2,3,4,5] and also by Louise Clarke and John Carbon [6,7] proved to be correct. For our part, the task took eight years of very hard work by the brightest students in Kyoto University. The nucleotide sequences of S. pombe centromeres were determined by Takahashi et al. [5] (unfortunately not cited in [1]), and now the complete, detailed sequence is available. The large centromeres of S. pombe are a prominent symbol within the S. pombe genome, much as is the Cdc2 protein kinase within fission-yeast cell-cycle control, or the Kiyomizu-temple and Sanjyu-sangendo in the city of Kyoto. A frequently asked question in the past when I gave talks about the centromeres of S. pombe was, "Why are the centromeres of S. pombe so large?". But now people tend to ask instead why the centromeres of budding yeast are so small, at only several hundred base-pairs.
The issue of centromere size is addressed in the paper by Wood et al. [1], as it is not clear why fission yeast centromeres are 300-1,000 times larger than their budding yeast equivalents. One possibility is that their kinetochore structures are very different, particularly with respect to the sizes of two specialized regions - the repetitious centromeric heterochromatin containing the HP1 and Swi6 proteins and the more central, CENP-A-containing region. A related but distinct possibility is that the difference arises because kinetochore microtubules in budding yeast may have the unusual feature of being maintained in association with the centromeres throughout the cell cycle (see, for example [8]). The two-domain structure - the central and the outer repetitive domains - of the centromeres in fission yeast [5,9,10] are now popularly seen as a model by researchers working on higher eukaryotic centromeres (although the similarity has not been proven). The S. pombe centromere was once considered to be exceptional because it has a two-domain structure, but the field now seems to think that it may represent the general form of centromeric DNA. We must now await a complete centromeric analysis of higher eukaryotes, which has hardly been attempted so far, for resolution of this point.
In the S. pombe genome milestone paper [1], the authors also focus on the problem of which genes are responsible for the unique characteristics of eukaryotic cells. Some of the most highly conserved genes among sequenced eukaryotes include those encoding proteins required for the cytoskeleton, vesicular compartmentation, cell-cycle control, proteolysis, protein phosphorylation and dephosphorylation, and RNA splicing. These genes seem therefore to have originated with the appearance of eukaryotic life. A total of 62 genes have been found to be highly conserved in all eukaryotes and are not present in prokaryotes; these can be divided into distinct functional groups, yielding a number of important insights. One important idea emphasized in the paper [1] is that the evolutionary transition from prokaryotes to eukaryotes took much longer than that from single to multi-cellular eukaryotes. The highly conserved genes specific to eukaryotes were necessary for eukaryotic organization to be established, but sequence analyses indicate that the transition from unicellular eukaryotes to multicellularity may not have required many new genes. The authors speculate [1] that the transition to multicellular organization may have utilized genes that were already present in unicellular eukaryotes, perhaps by shuffling functional domains to give rise to novel combinations.
A place in history
Looking at the large number of authors (131) on the fission yeast genome paper [1], it is difficult to appreciate that only a handful of laboratories around the world engaged in fission yeast research 20 years ago; needless to say, only a very few researchers were then seriously interested in the genome of fission yeast. In 1977, Jurg Kohli and Pierre Thuriaux at the University of Bern reported that S. pombe has three linkage groups (three chromosomes) after years of genetic analyses (mutant isolation and mapping) [11]. The genetics of S. pombe is extremely clean: Urs Leupold, also at Bern, initiated genetic studies in the middle of the last century (see, for example, [12,13]), and all the S. pombe workers since then have used derivatives of Leupold's original strains.
I myself started to employ fission yeast as a model eukaryote for studying chromosome dynamics in the cell cycle around 1977, after reading articles written by Murdoch Mitchison and Urs Leupold. I had previously worked on head morphogenesis in the bacteriophage T4 and considered T4 as a model chromosome. I quickly decided to change the subject of my study from T4 to S. pombe at the time when gene cloning by transformation of mutant cells became in principle possible. The fission yeast's cell shape looked like large Escherichia coli - the organism I was most familiar with - and the cells divide by fission, again just like E. coli. I liked the simplicity of the S. pombe cell cycle, and within a few years of starting to work on fission yeast ourselves I was fascinated with the beautiful nuclear chromatin and condensed chromosomes that could easily be seen by DAPI (diamidino phenylindole) staining. Some years after the identification of the three S. pombe linkage groups by genetics, three condensed chromosomes were visualized using DAPI staining of mutant cells arrested because of a defect in β tubulin [14]. I remember vividly the excitement when we could first see the three DAPI-stained chromosomes (Figure 1b,1c); the use of DAPI revolutionized the isolation of mutants with defects in the nucleus and chromosomes [15].
Technology for handling large pieces of DNA was also developing around the mid-1980s, and Charles Cantor and colleagues were interested in applying the newly developed technique of pulsed-field gel electrophoresis to the genome of fission yeast. In collaboration with my group, Smith et al. [16] reported successful separation of the individual chromosomes and determined the size of each: chromosome I, 5.7 megabases; chromosome II, 4.6 megabases; and chromosome III, 3.5 megabases. All of the NotI restriction enzyme sites in the genome were also determined. After that, any cloned fission yeast gene could be physically mapped onto individual chromosomes and NotI fragments. With these technical developments and the advent of chromosome-walking technology, physical mapping of the whole fission yeast genome was undertaken. Two papers from the laboratories of Hans Lehrach and colleagues [17] at the ICRF in London and David Beach at Cold Spring Harbor Laboratory (in collaboration with us) [18] were published in 1993, reporting the physical map of the fission yeast genome. Although these physical maps had numerous gaps and false assignments, they were nevertheless vital to the start of the whole-genome sequencing project that was completed in 2001 thanks to the tireless efforts of the 131 authors [1].
The value of fission yeast as a model organism will be upheld - or even strengthened - with the completion of whole-genome sequencing. The rise of S. pombe in the past 20 years was due to the ease of genetics and cell biology and the great cell-cycle discoveries that followed the identification of Cdc2. Then, developments in chromosome biology, mitosis, cytokinesis, meiosis and cell-shape control built solid research foundations to make S. pombe attractive to general cell biologists who read papers from S. pombe labs, and many newcomers actually began working on S. pombe. The publication of the whole genome sequence will now further lure biologists and others working in such diverse fields as bioinformatics, pharmacology, and evolution. Researchers who are seriously interested in the evolution and establishment of eukaryotic organisms must consider fission yeast as a premier organism for study: it did not experience large genome rearrangements in its evolution; it has a typical mitochondrial genome, like humans (mitochondria are essential in fission yeast while dispensable in budding yeast, which can grow under anaerobic conditions); and it has the smallest eukaryotic genome, with only three chromosomes.
For all these reasons, fission yeast seems bound to be highly popular - at least in the United Kingdom, Europe and Japan, for the next ten to fifteen years, judging by the enthusiasm and high caliber of studies of the many young group leaders in these countries. But S. pombe has found it difficult to proliferate in the USA, particularly in the Boston and San Francisco Bay areas, arguably the two key centers of power in the world for life sciences. I gave lectures in Boston last October, and strongly felt I was seen there as a heretic. The S. pombe community can already boast some prominent researchers in the USA, but we must make a special effort to establish more S. pombe labs on the two coasts. I am rather optimistic about this task, as I recall there were few respectable cell biologists in Harvard 20 years ago, but now it is full of excellent cell biologists - and the current task should be much easier than trying to convince Americans that soccer is more popular and widespread than American football. The success of S. pombe whole-genome sequencing is one more reason why the S. pombe field should enjoy a solid global position - and the field of S. pombe research will continue to welcome any life scientists of any nationality and cultural background who choose to work with S. pombe - the model eukaryotic organism.
Editor's note
Readers interested in the comparative genomics and evolutionary history of Schizosaccharomyces pombe might like to read the following articles previously published in Genome Biology:
Minireview: Where does fission yeast sit on the tree of life? Matthias Sipiczki. Genome Biology 2000, 1(2):reviews1011.1-1011.4http://genomebiology.com/2000/1/2/reviews/1011
Minireview: Membrane traffic between genomesJohn Armstrong. Genome Biology 2000, 1(1):reviews104.1-104.4http://genomebiology.com/2000/1/1/reviews/104
Acknowledgments
Acknowledgements
The author's laboratory is supported by the COE Research Fund from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Wood V, Gwilliam R, Rajandream M-A, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 1986;5:1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Chikashige Y, Yanagida M. Characterization of S. pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J. 1989;8:3045–3052. doi: 10.1002/j.1460-2075.1989.tb08455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Niwa O, Funabiki H, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in the fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Baum MP. Functional analysis of a centromere from fission yeast: a role for centromere-specific repeated DNA sequences. Mol Cell Biol. 1990;10:1863–1872. doi: 10.1128/mcb.10.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnenberger KM, Carbon J, Clarke L. Identification of DNA regions required for mitotic and meiotic functions within the centromere of Schizosaccharomyces pombe chromosome I. Mol Cell Biol. 1991;11:2206–2215. doi: 10.1128/mcb.11.4.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Takahashi K, Yanagida M. Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell. 1997;90:131–143. doi: 10.1016/s0092-8674(00)80320-7. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Kohli J, Hottinger H, Munz P, Strauss A, Thuriaux P. Genetic mapping of Schizosaccharomyces pombe by mitotic and meiotic analysis and induced haploidization. Genetics. 1977;87:471–489. doi: 10.1093/genetics/87.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leupold U. Studies on recombination in Schizosaccharomyces pombe. Cold Spring Harbor Symp Quant Biol. 1958;23:161–170. doi: 10.1101/sqb.1958.023.01.020. [DOI] [PubMed] [Google Scholar]
- Munz P, Amstutz H, Kohli J, Leupold U. Recombination between dispersed serine tRNA genes in Schizosaccharomyces pombe. Nature. 1982;300:225–231. doi: 10.1038/300225a0. [DOI] [PubMed] [Google Scholar]
- Umesono K, Hiraoka Y, Toda T, Yanagida M. Visualization of chromosomes in mitotically arrested cells of the fission yeast Schizosaccharomyces pombe. Curr Genet. 1983;7:123–128. doi: 10.1007/BF00365637. [DOI] [PubMed] [Google Scholar]
- Toda T, Yamamoto M, Yanagida M. Sequential alterations in the nuclear chromatin region during mitosis of the fission yeast Schizosaccharomyces pombe : video fluorescence microscopy of synchronously growing wild-type and cold-sensitive cdc mutants by using a DNA-binding fluorescent probe. J Cell Sci. 1981;52:271–287. doi: 10.1242/jcs.52.1.271. [DOI] [PubMed] [Google Scholar]
- Smith C, Matsumoto T, Niwa O, Klco S, Fan JB, Yanagida M, Cantor C. An electrophoretic karyotype for Schizosaccharomyces pombe by pulsed field gel electrophoresis. Nucleic Acids Res. 1987;15:4481–4489. doi: 10.1093/nar/15.11.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoheisel JD, Maier E, Mott R, McCarthy L, Grigoriev AV, Schalkwyk LC, Nizetic D, Francis F, Lehrach H. High resolution cosmid and P1 maps spanning the 14 Mb genome of the fission yeast. Cell. 1993;73:109–120. doi: 10.1016/0092-8674(93)90164-l. [DOI] [PubMed] [Google Scholar]
- Mizukami T, Chang WI, Gargavitsev I, Kaplan N, Lombardi D, Matsumoto T, Niwa O, Kounousu A, Yanagida M, Marr TG, Beach D. A 13 kb resolution cosmid map of the 14 Mb fission yeast genome by nonrandom sequence-tagged site mapping. Cell. 1993;73:121–132. doi: 10.1016/0092-8674(93)90165-m. [DOI] [PubMed] [Google Scholar]


