Abstract
Gene expression is finely regulated at the post-transcriptional level. Features of the untranslated regions of mRNAs that control their translation, degradation and localization include stem-loop structures, upstream initiation codons and open reading frames, internal ribosome entry sites and various cis-acting elements that are bound by RNA-binding proteins.
The recent analysis of the human genome [1,2] and the data available about other higher eukaryotic genomes have revealed that only a small fraction of the genetic material - about 1.5% - codes for protein. Indeed, most genomic DNA is involved in the regulation of gene expression, which can be exerted at either the transcriptional level, controlling whether a gene is transcribed or not and to what extent, or the post-transcriptional level, controlling the fate of the transcribed RNA molecules, including their stability, the efficiency of their translation and their subcellular localization. This article will review the structure, functions and mechanisms of mRNA untranslated regions.
Transcriptional control is mediated by transcription factors, RNA polymerase and a series of cis-acting elements located in the DNA, such as promoters, enhancers, silencers and locus-control elements, organized in a modular structure and regulates the production of pre-mRNA molecules, which undergo several steps of processing before they become functional mRNAs. Introns are removed, a 7-methyl-guanylate (m7G) cap structure is added at the 5' end of the first exon, and a stretch of 100-250 adenine residues (the poly(A) tail) is added at the 3' end of the last exon, which is itself generated by endonucleolytic cleavage of the primary transcript. Sometimes the sequence of the mRNA is also altered in a process called mRNA editing, and the resulting coding sequence of the mature RNA differs from the corresponding sequence in the genome. The resultant mature mRNA, in eukaryotes, has a tripartite structure consisting of a 5' untranslated region (5' UTR), a coding region made up of triplet codons that each encode an amino acid and a 3' untranslated region (3' UTR). Figure 1 shows these and other features of mRNAs.
Figure 1.
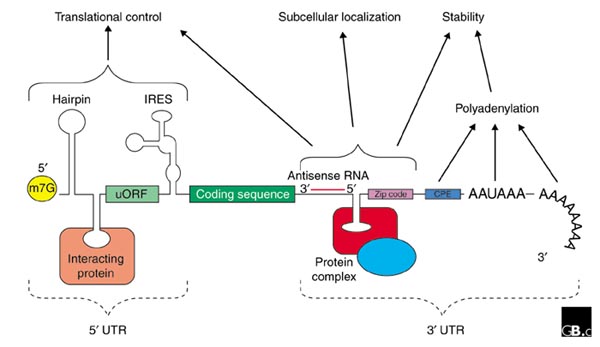
The generic structure of a eukaryotic mRNA, illustrating some post-transcriptional regulatory elements that affect gene expression. Abbreviations (from 5' to 3'): UTR, untranslated region; m7G, 7-methyl-guanosine cap; hairpin, hairpin-like secondary structures; uORF, upstream open reading frame; IRES, internal ribosome entry site; CPE, cytoplasmic polyadenylation element; AAUAAA, polyadenylation signal.
UTRs are known to play crucial roles in the post-transcriptional regulation of gene expression, including modulation of the transport of mRNAs out of the nucleus and of translation efficiency [3], subcellular localization [4] and stability [5]. This article focuses mainly on these three functions, but UTRs may also play other roles, such as the specific incorporation of the modified amino acid selenocysteine at UGA codons of mRNAs encoding selenoproteins in a process mediated by a conserved stem-loop structure in the 3' UTR [6]. The importance of UTRs in regulating gene expression is underlined by the finding that mutations that alter the UTR can lead to serious pathology [7].
Regulation by UTRs is mediated in several ways. Nucleotide patterns or motifs located in 5' UTRs and 3' UTRs can interact with specific RNA-binding proteins. Unlike DNA-mediated regulatory signals, however, whose activity is essentially mediated by their primary structure, the biological activity of regulatory motifs at the RNA level relies on a combination of primary and secondary structure. Interactions between sequence elements located in the UTRs and specific complementary non-coding RNAs have also been shown to play key regulatory roles [8]. Finally, there are examples of repetitive elements that are important for regulation at the RNA level. For example, CUG-binding proteins may bind to CUG repeats in the 5' UTR of specific mRNAs (such as that encoding the transcription factor C/EBPβ), affecting their translation efficiency [9].
Many RNA-binding proteins involved in the cytoplasmic post-transcriptional regulation of gene expression also participate in a wide variety of regulatory processes - such as alternative pre-mRNA splicing or 3'-end processing - within the nucleus, where they act as components of heterogeneous nuclear ribonucleoproteins (hnRNPs) [10]. This functional interconnection between post-transcriptional events in the nucleus and in the cytoplasm may explain experimental observations that the nuclear history of an mRNA can affect its cytoplasmic fate [11].
Structural features of untranslated regions
Comparison of the various completed and partial genome sequences reveals some conserved aspects of the structure of UTRs (see Table 1). The average length of 5' UTRs is roughly constant over diverse taxonomic classes and ranges between 100 and 200 nucleotides, whereas the average length of 3' UTRs is much more variable, ranging from about 200 nucleotides in plants and fungi to 800 nucleotides in humans and other vertebrates. It is striking that the length of both 5' and 3' UTRs varies a lot within a species, ranging from a dozen nucleotides to a few thousand [12]. In fact, it has been shown using a mammalian in vitro system that even a single nucleotide is a sufficient 5' UTR for the initiation of translation [13].
Table 1.
Features of complete UTR sequences derived from genomic entries annotated in UTRdb [47,48,50].
| 5' UTR | 3' UTR | |||||||
| Number of sequences | Average length | Maximum length | Minimum length | Number of sequences | Average length | Maximum length | Minimum length | |
| Humans | 1,203 | 210.2 | 2,803 | 18 | 1,247 | 1,027.7 | 8,555 | 21 |
| Other mammals | 142 | 141.3 | 936 | 20 | 148 | 441.1 | 3,324 | 37 |
| Rodents | 638 | 186.3 | 1,786 | 16 | 457 | 607.3 | 3,354 | 19 |
| Aves | 59 | 126.4 | 620 | 17 | 56 | 651.9 | 3,990 | 21 |
| Other vertebrates | 105 | 164.0 | 1,154 | 15 | 111 | 446.5 | 2,858 | 31 |
| Invertebrates | 5,464 | 221.9 | 4,498 | 14 | 3,736 | 444.5 | 9,142 | 15 |
| Liliopsidae | 144 | 129.8 | 715 | 17 | 127 | 273.3 | 1,605 | 22 |
| Other Viridiplantae | 1,471 | 103.0 | 1,355 | 12 | 1,699 | 207.7 | 1,911 | 13 |
| Fungi | 388 | 134.0 | 1,088 | 16 | 326 | 237.1 | 1,142 | 25 |
The genomic region corresponding to the UTRs of an mRNA may contain introns, more frequently in the 5' than in the 3' UTR. About 30% of genes in metazoa have fully untranslated 5' exons, whereas although 3' UTRs are much longer, they have a much lower intron frequency, in the range 1-11% depending on the taxon (Figure 2a). Alternative UTRs can be formed from the use of different transcription-start sites, polyadenylation sites or splice donor and/or acceptor sites. These have been shown to vary in abundance with the tissue, developmental stage or disease state and can affect the pattern of gene expression considerably [14].
Figure 2.
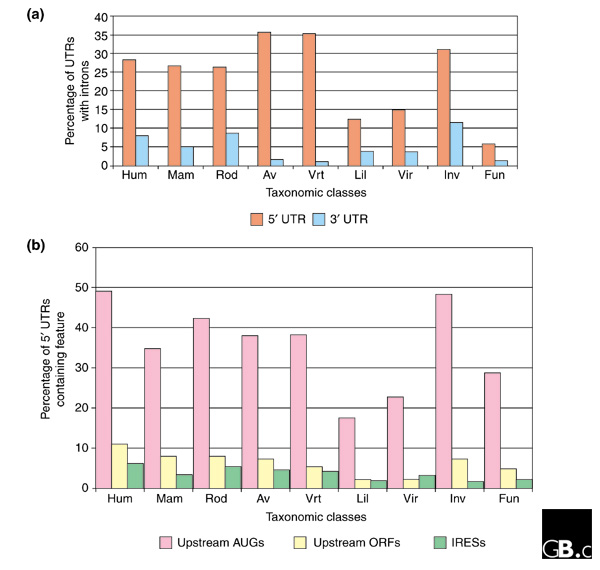
The percentage of complete UTR sequences in the different taxonomic classes that contain (a) introns or (b) upstream AUGs, upstream ORFs or IRES elements. Hum, human; mam, other mammals; rod, rodents; av, Aves; vrt, other vertebrates; lil, Liliopsidae; vir, other plants (Viridiplantae); inv, invertebrates; fun, fungi. Data are taken from UTRdb [47].
The base composition of 5' and 3' UTR sequences also differs; the G+C content of 5' UTR sequences is greater than that of 3' UTR sequences. This difference is more marked in mRNAs from warm-blooded vertebrates, whose G+C content is about 60% for 5' UTRs and 45% for 3' UTRs [15]. There is also an interesting correlation between the G+C content of 5' or 3' UTRs and that of the third codon positions of the corresponding coding sequences, and a significant inverse correlation has been observed between the G+C content of 5' and 3' UTRs and their lengths [16]. In particular, it has emerged that genes localized in large GC-rich regions of a chromosome (heavy isochores) have shorter 5' UTRs and 3' UTRs than genes located in GC-poor isochores. A similar correlation has been also shown for the coding sequence and introns [17].
Finally, eukaryotic mRNAs are also known to contain several types of repeat in the untranslated regions, including short interpersed elements (SINEs) such as Alu elements, long interspersed elements (LINEs), minisatellites and microsatellites. In human mRNAs, repeats are found in about 12% of 5' UTRs and 36% of 3' UTRs. A lower repeat abundance is observed in other taxa, including other mammals.
Control of translation efficiency
Translation of mRNAs can vary in efficiency, so that the amount of protein produced is modulated. This is an important level of gene regulation; indeed, a correlation between mRNA and protein abundance is seen only for secreted proteins, whereas for intracellular proteins the differing rates of translation of different mRNAs removes this correlation [18]. Features all along the mRNA can affect translation efficiency.
Structural features of the 5' UTR have a major role in the control of mRNA translation. Messenger RNAs encoding proteins involved in developmental processes, such as growth factors, transcription factors or proto-oncogenes, all of which need to be strongly and finely regulated, often have 5' UTRs that are longer than average [19], with upstream initiation codons or open reading frames (ORFs) and stable secondary structures that hamper translation efficiency (Table 2). Other specific motifs and secondary structures in the 5' UTR can also modulate translation efficiency.
Table 2.
Examples of genes with 5' UTRs longer than average and with upstream ORFs and/or repeat elements
| 5' UTR features | |||||
| UTR* | EMBL† | Gene description | Length | Repeats | uORFs |
| 5HSA002333 | M13994 | B-cell leukemia/lymphoma 2 (Bcl-2) proto-oncogene | 1,458 | 2 | 7 |
| 5HSA017553 | X63547 | Tre oncogene | 2,858 | 1 | 10 |
| 5SSC000518 | AJ000928 | c-Myc proto-oncogene (Sus scrofa) | 1,330 | 3 | 13 |
| 5HSA016490 | AF074913 | Transcription factor Pax-5 | 1,125 | 0 | 2 |
| 5HSA024311 | AJ297406 | Transcription factor II B-related factor | 1,437 | 2 | 7 |
| 5HSA001903 | AF006822 | Myelin transcription factor 2 (MYT2) | 1,155 | 0 | 8 |
| 5HSA004086 | M62302 | Growth/differentiation factor 1 (GDF-1) | 1,346 | 2 | 1 |
| 5HSA004101 | M22373 | Insulin-like growth factor (IGF-II) | 1,169 | 3 | 0 |
Accession numbers in the *UTRdb [47] and †EMBL databases [52]. Abbreviations: uORFs, upstream ORFs.
Under normal conditions, following the transport of an mRNA from the nucleus to the cytoplasm, the eIF4F protein complex assembles at the cap. This complex consists of three subunits: eIF4E, the cap-binding protein; eIF4A, which has RNA helicase activity; and eIF4G, which interacts with various other proteins, including polyadenylate-binding protein. The ATP-dependent helicase activity of eIF4A, stimulated by the RNA-binding protein eIF4B, unwinds any secondary structure in the mRNA, thus creating a 'landing platform' for the small (40S) ribosomal subunit [20]. When concentration of ribosomes or translation factor are limiting, the poly(A) tail can cooperate with 5' cap to enhance translation initiation through the intervention of a polyadenylate-binding protein that can physically interact with eIF4F complex [21].
In most eukaryotic mRNAs, it is thought that translation initiates at the first AUG codon encountered by the 40S ribosomal subunit as it moves, or scans, 3' along the mRNA from the 5' m7G cap. Sequences flanking the AUG initiation codon are not random but fit a consensus sequence; in mammals, this sequence is GCCRCCaugG, and the most conserved nucleotides are the purine (R), usually A, in position -3 with respect to the AUG start codon and the guanine in position +4. The strong preference for A at position -3 and G at position +4 is also conserved in other animals and in plants and fungi. The sequence context of the first AUG codon, in particular the part located in the untranslated region, may modulate the efficiency with which it is recognized as a translation initiation codon.
It is noteworthy that a large fraction of 5' UTRs contain upstream AUGs, from 15% to nearly 50% depending on the organism (Figure 2b), suggesting that the 'first AUG rule' predicted by the scanning model of ribosome start-site selection is disobeyed in a large number of cases. This implies that the 40S ribosomal subunit can sometimes bypass the most upstream AUG codon, possibly because its sequence context makes it a poor initiation codon, to initiate translation at a more distal AUG. With this mechanism, called 'leaky scanning', multiple different proteins can be obtained from the same mRNA [22]. Moreover, it has been calculated that the presence of an upstream AUG correlates with a long 5' UTR and with a 'weak' start codon context of the AUG that is usually used, whereas transcripts with an optimal start-codon context have short 5' UTRs without upstream AUGs [23], suggesting that upstream AUGs may have a role in keeping the basal translational level of a gene low.
If an in-frame stop codon is found following the upstream AUG and before the main start codon, it creates an upstream ORF. After translation of the upstream ORF and the detachment of the large (60S) ribosomal subunit, the small ribosomal subunit has multiple alternative fates, which affect translation efficiency and mRNA stability. The 40S subunit may hold onto the mRNA, resume scanning, and reinitiate translation at a downstream AUG codon, or it may leave the mRNA, thus impairing translation of the main ORF. The ability of a ribosome to reinitiate is limited in eukaryotes by the stop codon context [24] and by the length of the upstream ORF; if the upstream ORF is longer than around 30 codons [25], the ribosome cannot reinitiate. This process is known to down-regulate translation of the mRNAs for the yeast transcription factors GCN4 and YAP1, which contain upstream ORFs [26].
Secondary structures in 5' UTRs are also important in the regulation of translation. Experimental data suggest that moderately stable secondary structures (a change in free energy (ΔG) above -30 kcal/mol) directly involving the AUG start codon do not stall the migration of 40S ribosomal subunit; a significant decrease in the efficiency of translation is observed only when very stable structures (ΔG below -50 kcal/mol) are formed. UTR sequences with such very stable secondary structures are reported in Table 3. The inhibitory effects of these structures can be overcome by an increase in the level of eIF4A, the subunit of the eIF4F complex that promotes the unwinding of RNA secondary structures in cooperation with eIF4B and eIF4H [27].
Table 3.
Examples of 5' UTR sequences with highly stable stem-loop structures
| UTR | EMBL | Gene description | UTR length | Stem length | ΔG |
| 5hsa007030 | X12949 | Ret | 963 | 129 | -125.4 |
| 5hsa034512 | AF274954 | PNAS-29 | 323 | 95 | -71.7 |
| 5hsa019215 | AF139980 | LW-1 (LW-1) | 716 | 72 | -66.1 |
| 5hsa019416 | AF152961 | Chromatin-specific transcription elongation factor | 291 | 97 | -61.9 |
| 5hsa022262 | S95936 | Transferrin | 79 | 65 | -60.1 |
| 5hsa000763 | U19144 | GAGE-3 protein | 99 | 72 | -54.6 |
| 5hsa022576 | AF116649 | PRO0566 | 2,011 | 72 | -51.6 |
Highly stable structures are defined as those with ΔG ≤ -50 kcal/mol. Free energy was calculated with 'foldrna' program (GCG [53]) on stem-loop elements found with the PatSearch program on human UTRdb [47]. Stem length represents the total number of nucleotides involved in the structure.
An alternative mechanism for translation initiation, which occurs independently of the 5' cap, was discovered for the first time in picornaviruses [28]: a sequence element in the 5' UTR acts as an internal ribosome entry site (IRES). IRES elements have been found in many cellular mRNAs encoding regulatory proteins, such as proto-oncogene products like c-Myc, homeodomain proteins, growth factors (like the fibroblast growth factor FGF-2) and their receptors. The concept of IRESs has been very critically reviewed by Kozak [29], who originally defined the importance of initiation codon context. Comparative analysis of known cellular IRESs leads to the identification of a common structural motif shared by many mRNAs, including those encoding the immunoglobulin heavy chain binding protein BiP and FGF2: a Y-shaped stem-loop just upstream of the AUG initiation codon [30] (see Table 4 and Figure 2b). It has recently been discovered that short sequence motifs complementary to the small ribosomal RNA may also act as IRESs [31].
Table 4.
5' UTR sequences with experimentally proved IRES elements
| UTRdb* | EMBL† | Gene description | UTRsite | Reference |
| 5HSA004100 | J04513 | Human FGF-2 | Y | [54] |
| 5HSA007092 | X87949 | Human BiP | Y | [55] |
| 5DVI000022 | M95825 | Drosophila Antennapedia | [56] | |
| 5HSA007484 | M12783 | Human PDGF2/c-sis | Y | [57] |
| 5HSA011699 | AF025841 | Acute myeloid leukemia 1 protein (AML1) | Y | [58] |
| 5HSA000138 | AF013263 | Apoptotic protease activating factor 1 (Apaf-1) | [59] | |
| 5HSA001903 | AF006822 | Myelin transcription factor 2 (MYT2) | [60] | |
| 5CGR000096 | M17169 | Chinese hamster glucose-regulated protein GRP78 | Y | [30] |
| 5BTA000471 | M13440 | Bovine basic fibroblast growth factor (FGF) | Y | [30] |
| 5RNO001555 | M22427 | Rat basic fibroblast growth factor (FGF) | Y | [30] |
| 5HSA015336 | D14838 | Human FGF-9 | [30] | |
| 5HSA005291 | M17446 | Human Kaposi's sarcoma oncogene (fibroblast growth factor) | [30] |
The elements recognized by the IRES detection algorithm in UTRsite [47,48,50] are marked 'Y'. Accession numbers in the *UTRdb [47] and †EMBL databases [52].
Sequence elements that are the target of trans-acting RNA binding proteins can also regulate translation. For example, the iron-responsive element (IRE) located in the 5' UTR of mRNAs encoding proteins involved in iron metabolism (ferritin, 5-aminolevulinate synthase and aconitase) may inhibit translation through the iron-dependent binding of iron regulatory proteins, which impede the normal scanning process of the small ribosomal subunit in translation initiation. In addition, most vertebrate mRNAs that encode ribosomal proteins and translation elongation factors analyzed to date contain a 5' terminal oligopyrimidine tract (TOP) consisting of 5-15 pyrimidines immediately adjacent to the m7G cap. This tract is required for coordinated translational repression during growth arrest, differentiation, development and certain drug treatments [32].
Regulation of mRNA stability
The turnover of mRNAs is another crucial step in post-transcriptional regulation of gene expression, as changes in mRNA abundance may alter the expression of specific genes by affecting the abundance of the corresponding protein. Several mechanisms have been proposed to describe how mRNA degradation takes place: decay can be preceded by shortening or removal of the poly(A) tail at the 3' end and/or by removal of the m7G cap at the 5' end [33]. The turnover of an mRNA is mostly regulated by cis-acting elements located in the 3' UTR, such as the AU-rich elements (AREs), which promote mRNA decay in response to a variety of specific intra- and extra-cellular signals. AREs have been experimentally grouped into three classes: class I and II AREs are characterized by the presence of multiple copies of the pentanucleotide AUUUA, which is absent from class III AREs [34]. Class I AREs control the cytoplasmic deadenylation of mRNAs by the degradation of all parts of the poly(A) tail at the same rate, generating intermediates with poly(A) tails of 30-60 nucleotides, which are then completely degraded. These elements are found mainly in mRNAs encoding nuclear transcription factors such as c-Fos and c-Myc (the products of 'fast response' genes) and also in mRNAs for some cytokines, such as interleukins 4 and 6. The presence of one or more copies of the pentanucleotide AUUUA next to a U-rich region is the structural characteristic of class I AREs. Class II AREs mediate asynchronous cytoplasmic deadenylylation, in other words the poly(A) tail is degraded at different rates in different transcripts, generating mRNAs without poly(A) tails. Among mRNAs containing this signal are those encoding the cytokines GM-CSF, interleukin 2, tumor necrosis factor α (TNF-α) and interferon-α. Class II AREs are characterized by tandem reiterations of the AUUUA pentamer, and an AU-rich region is usually found upstream of these repeats. The mRNAs containing class III AREs, such as those encoding c-Jun, do not contain the pentanucleotide AUUUA but have only a U-rich segment; they show degradation kinetics similar to those of mRNAs containing class I AREs.
Degradation of mRNAs can also take place following endonuclease activity, in a mechanism independent of both deadenylation and decapping. Such a mechanism has been observed for the mRNA encoding the transferrin receptor, a protein that mediates iron transfer in the cell. The degradation pathway of this mRNA involves an endonucleolytic cleavage in the 3' UTR region that is mediated by the recognition of IRE structures and is regulated by the level of intracellular iron [35].
Upstream initiation codons and ORFs may also play a role in mRNA decay through the nonsense-mediated mRNA decay (NMD) pathway. The signal that triggers NMD is a nonsense codon followed by a splicing junction (the junction between two removed exons) [36]; the presence of the splicing junction may be how normal stop codons are distinguished from premature termination codons. Indeed, normal stop codons and the 3' UTR are usually located in the last exon of the sequence and thus are not followed by a splicing junction. Exon junctions are recognized because a marker protein binds to the intron-containing transcript in the nucleus, remains bound to the exon junction after the splicing event has finished and is translocated to the cytoplasm with the processed mRNA [11]. The translation machinery usually displaces the marker protein, preventing the degradation of wild-type mRNAs. But if the ribosome encounters a stop codon that is either premature or due to the presence an upstream ORF, it disassembles and the marker proteins at the exon junction direct the aberrant mRNA towards NMD [37]. In Saccharomyces cerevisiae (which uses a downstream exonic element, DSE, as the second signal that triggers NMD), mRNAs containing functionally active upstream ORFs, like those encoding GCN4 or YAP1, are not degraded through the NMD pathway because they contain an mRNA-specific stabilizer sequence elements between the upstream ORF and the coding sequence that prevents the activation of the NMD pathway by interacting with the RNA-binding ubiquitin ligase Pub1 [38].
Upstream ORFs can also regulate mRNA stability through an NMD-independent mechanism. The 5' UTR of the S. cerevisiae gene YAP2 contains two upstream ORFs that inhibit ribosomal scanning and promote mRNA decay [26]. The destabilizing effect relies on the termination codon context, which modulates translation efficiency and mRNA stability. Table 5 reports some genes in which upstream ORFs have been demonstrated to affect gene expression.
Table 5.
Genes with experimentally characterized upstream ORFs in their 5' UTR
| Gene | Organism | Number of upstream ORFs | Effects | Reference |
| AdoMetDC | Mammalian | 1 | Spermidine-dependent translation control | [61] |
| GCN4 | Yeast | 4 | Starvation-dependent translation/stability regulation | [62] |
| CD36 | Human | 3 | Glucose-mediated translation control | [63] |
| YAP2 | Yeast | 2 | NMD-independent mRNA destabilization | [26] |
| YAP1 | Yeast | 1 | Weak translation inhibition | [26] |
| V(1b) Vasopressin receptor | Rat | 5 | Translation inhibition (no destabilization) | [64] |
| Connexin-41 | Xenopus | 3 | Translation inhibition | [65] |
| Mdm2 long transcript | Human | 2 | Translation inhibition | [66] |
Several studies have provided evidence that many hnRNPs not only function in the nucleus but also are involved in the control of mRNA fate in the cytoplasm [10] and can regulate translation, mRNA stability and cytoplasmic localization [37]. One example is the regulation of the amyloid precursor protein (APP); increasing the level of APP is an important contributing factor to the development of Alzheimer's disease. Stability of APP mRNA is dependent on a highly conserved 29-nucleotide element located in the 3' UTR that interacts with several cytoplasmic RNA-binding proteins [39]. Very interestingly, although some of these proteins are fragments of nucleolin (which is known to shuttle between the nucleus and cytoplasm), two proteins of 39 kDa and 38 kDa are subunits of hnRNP C, seen in this study for the first time in the cytoplasm [40].
Control of mRNA subcellular localization
UTRs have a fundamental role in the spatial control of gene expression at the post-transcriptional level, which is particularly important during development. The asymmetric localization of some mRNAs leads to an asymmetry of cellular distribution of the encoded proteins; such a situation is clearly more efficient than other possible mechanisms of protein localization, because the same mRNA molecule can serve as a template for multiple rounds of translation. In many cases, mRNAs are localized as ribonucleoprotein complexes along with proteins of the translational apparatus, thus ensuring efficient localized translation.
There are three main mechanisms for the asymmetric distribution of mRNAs: active directed transport, requiring a functional cytoskeleton and specific motor proteins interacting with the targeted mRNAs; local stabilization of transcripts; and diffusion of the mRNA followed by its local entrapment. Myelin basic protein (MBP) mRNA is localized to the myelin produced by oligodendrocytes of the central nervous system through an active transport mechanism. A 21-nucleotide sequence, termed the RNA-transport signal, and an additional element, the RNA-localization region, both in the 3' UTR of MBP mRNA, are required for its transport and localization in mouse [41]. Many examples of local stabilization come from Drosophila early development: transcripts encoding the RNA-binding protein Nanos or the heat-shock protein Hsp83 are degraded everywhere in the embryo except in the posterior polar plasm. Distinct cis-acting elements located in the 3' UTR of these mRNAs mediate both degradation in the embryo as a whole and the stabilization at the pole [5]. The diffusion and entrapment mechanism is well represented by localization of Bicoid mRNA in Drosophila. The elements that regulate the anchoring of the transcript, the key step of the process, are not all characterized, but one protein involved is Staufen, a double-stranded RNA-binding protein that is essential for the immobilization of Bicoid mRNA in the anterior pole of the egg [42].
In all these cases, subcellular localization of mRNA is mediated by cis-acting elements located in the 3' UTR, but there are also examples of elements in the 5' UTR or even in the coding sequence; these are known as mRNA zip codes and interact with zip-code-binding proteins (such as Staufen). Zip codes lack any apparent similarity in their primary or secondary structure; they can have a complex secondary or tertiary structure, as in the Bicoid localization element, in which primary sequence is less important than the overall structure [43], or they can be short, defined nucleotide sequences [44], sometimes in repeated elements (such as in the case of the Xenopus localized transcript Vg1 [45]).
In conclusion, untranslated regions of mRNAs have crucial roles in many aspects of gene regulation. Further information on the structures and functions of UTRs, including the cis-acting elements found in them (Table 6) [46], can be found at our UTR home page [47] and from the UTRdb and UTRsite databases, which can be downloaded from our ftp site [48] or accessed with SRS [49] from our website [50] or the European Bioinformatics Institute [51].
Table 6.
Functional elements in UTRsite collection annotated in UTRdb entries
| Functional elements | Localization (UTR) | Number of annotated entries* |
| 15-lipoxygenase differentiation control element (15-LOX-DICE) | 3' | 94 |
| Adh mRNA down-regulation element | 3' | 61 |
| Amyloid precursor protein 3' UTR destabilizing element | 3' | 15 |
| Class 2 AU-rich elements (ARE2) | 3' | 70 |
| Bruno-responsive element (BRE) | 3' | 199 |
| Barley yellow dwarf virus (bydv) | 5' and 3' | 6 |
| Cytoplasmic polyadenylation element (CPE) | 3' | 5,186 |
| GLUT1 mRNA-stability control element | 5' and 3' | 66 |
| Histone mRNA 3' UTR stem loop | 3' | 38 |
| Iron responsive element (IRE) | 5' and 3' | 121 |
| Internal ribosome entry site (IRES) | 5' | 7,356 |
| Msl-2 3' UTR control element | 3' | 19 |
| Msl-2 5' UTR control element | 5' | 5 |
| Nanos translation control element | 3' | 1 |
| Ribosomal S12 mRNA translational control element | 5' | 2 |
| Selenocysteine insertion sequence type 1 (SECIS-1) | 3' | 1,773 |
| Selenocysteine insertion sequence type 2 (SECIS-2) | 3' | 355 |
| Tra-2 and GLI element (TGE) | 3' | 81 |
| TNF-α mRNA stability control element | 3' | 8 |
| Terminal oligopyrimidine tract (TOP) | 5' | 272 |
| Upstream ORF | 5' | 71,438 |
| Vimentin mRNA 3' UTR control element | 3' | 6 |
*The number of genes in UTRdb [47] in which the structure is found as of June 2001.
Acknowledgments
Acknowledgements
This work was supported by Telethon, Ministero dell'Istruzione e Ricerca, Italy (projects: Programma "Biotecnologie" (legge 95/95 - 5%), Programma "Studio di geni di interesse biomedico e agroalimentare", CEGBA).
References
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- van der Velden AW, Thomas AA. The role of the 5' untranslated region of an mRNA in translation regulation during development. Int J Biochem Cell Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- Jansen RP. mRNA localization: message on the move. Nat Rev Mol Cell Biol. 2001;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Cooperstock RL, Lipshitz HD. Spatial and temporal control of RNA stability. Proc Natl Acad Sci USA. 2001;98:7025–7028. doi: 10.1073/pnas.111145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak R, Westhof E, Carbon P, Krol A. A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA. 1996;2:367–379. [PMC free article] [PubMed] [Google Scholar]
- Conne B, Stutz A, Vassalli JD. The 3' untranslated region of messenger RNA: a molecular 'hotspot' for pathology? Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- Sweeney R, Fan Q, Yao MC. Antisense ribosomes: rRNA as a vehicle for antisense RNAs. Proc Natl Acad Sci USA. 1996;93:8518–8523. doi: 10.1073/pnas.93.16.8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko LT. Myotonic dystrophy: the role of RNA CUG triplet repeats. Am J Hum Genet. 1999;64:360–364. doi: 10.1086/302268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Chen CY, Shyu AB. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol Cell Biol. 2001;21:6960–6971. doi: 10.1128/MCB.21.20.6960-6971.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- Hughes MJ, Andrews DW. A single nucleotide is a sufficient 5' untranslated region for translation in an eukaryotic in vitro system. FEBS Lett. 1997;414:19–22. doi: 10.1016/s0014-5793(97)00965-4. [DOI] [PubMed] [Google Scholar]
- Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Prog Neurobiol. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- Pesole G, Liuni S, Grillo G, Saccone C. Structural and compositional features of untranslated regions of eukaryotic mRNAs. Gene. 1997;205:95–102. doi: 10.1016/s0378-1119(97)00407-1. [DOI] [PubMed] [Google Scholar]
- Pesole G, Bernardi G, Saccone C. Isochore specificity of AUG initiator context of human genes. FEBS Lett. 1999;464:60–62. doi: 10.1016/s0014-5793(99)01675-0. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D, Gautier C. Statistical analysis of vertebrate sequences reveals that long genes are scarce in GC-rich isochores. J Mol Evol. 1995;40:308–317. doi: 10.1007/BF00163235. [DOI] [PubMed] [Google Scholar]
- Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18:533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra U, Stringer EA, Chaudhuri A. Initiation factors in protein biosynthesis. Annu Rev Biochem. 1982;51:869–900. doi: 10.1146/annurev.bi.51.070182.004253. [DOI] [PubMed] [Google Scholar]
- Michel YM, Poncet D, Piron M, Kean KM, Borman AM. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J Biol Chem. 2000;275:32268–32276. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- Xiong W, Hsieh CC, Kurtz AJ, Rabek JP, Papaconstantinou J. Regulation of CCAAT/enhancer-binding protein-beta isoform synthesis by alternative translational initiation at multiple AUG start sites. Nucleic Acids Res. 2001;29:3087–3098. doi: 10.1093/nar/29.14.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Kochetov AV, Kondrashov FA, Koonin EV, Milanesi L. Presence of ATG triplets in 5' untranslated regions of eukaryotic cDNAs correlates with a 'weak' context of the start codon. Bioinformatics. 2001;17:890–900. doi: 10.1093/bioinformatics/17.10.890. [DOI] [PubMed] [Google Scholar]
- Cassan M, Rousset JP. UAG readthrough in mammalian cells: effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol Biol. 2001;2:3. doi: 10.1186/1471-2199-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen BG, Tan W, Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela C, Ramirez CV, Linz B, Rodrigues-Pousada C, McCarthy JE. Post-termination ribosome interactions with the 5' UTR modulate yeast mRNA stability. EMBO J. 1999;18:3139–3152. doi: 10.1093/emboj/18.11.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5' secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Kaplan G, Racaniello VR, Sonenberg N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5' noncoding region. Mol Cell Biol. 1988;8:1103–1112. doi: 10.1128/mcb.8.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. New ways of initiating translation in eukaryotes? Mol Cell Biol. 2001;21:1899–1907. doi: 10.1128/MCB.21.6.1899-1907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SY, Maizel JV., Jr A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res. 1997;25:362–369. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shama S, Meyuhas O. The translational cis-regulatory element of mammalian ribosomal protein mRNAs is recognized by the plant translational apparatus. Eur J Biochem. 1996;236:383–388. doi: 10.1111/j.1432-1033.1996.00383.x. [DOI] [PubMed] [Google Scholar]
- Brown CE, Sachs AB. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Shyu AB. Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: evidence for a novel class of AU-rich elements. Mol Cell Biol. 1996;16:1490–1499. doi: 10.1128/mcb.16.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Kuhn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc Natl Acad Sci USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF. The double lives of shuttling mRNA binding proteins. Cell. 2000;102:135–138. doi: 10.1016/s0092-8674(00)00018-0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, Peltz SW. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 2000;101:741–751. doi: 10.1016/s0092-8674(00)80886-7. [DOI] [PubMed] [Google Scholar]
- Zaidi SH, Malter JS. Amyloid precursor protein mRNA stability is controlled by a 29-base element in the 3'-untranslated region. J Biol Chem. 1994;269:24007–24013. [PubMed] [Google Scholar]
- Zaidi SH, Malter JS. Nucleolin and heterogeneous nuclear ribonucleoprotein C proteins specifically interact with the 3'-untranslated region of amyloid protein precursor mRNA. J Biol Chem. 1995;270:17292–17298. doi: 10.1074/jbc.270.29.17292. [DOI] [PubMed] [Google Scholar]
- Ainger K, Avossa D, Diana AS, Barry C, Barbarese E, Carson JH. Transport and localization elements in myelin basic protein mRNA. J Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Kerr K, Smith JL, Leask A. RNA regulatory element BLE1 directs the early steps of bicoid mRNA localization. Development. 1993;118:1233–1243. doi: 10.1242/dev.118.4.1233. [DOI] [PubMed] [Google Scholar]
- Chan AP, Kloc M, Etkin LD. fatvg encodes a new localized RNA that uses a 25-nucleotide element (FVLE1) to localize to the vegetal cortex of Xenopus oocytes. Development. 1999;126:4943–4953. doi: 10.1242/dev.126.22.4943. [DOI] [PubMed] [Google Scholar]
- Mowry KL, Melton DA. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- Pesole G, Liuni S, D'Souza M. PatSearch: a pattern matcher software that finds functional elements in nucleotide and protein sequences and assesses their statistical significance. Bioinformatics. 2000;16:439–450. doi: 10.1093/bioinformatics/16.5.439. [DOI] [PubMed] [Google Scholar]
- UTR home page http://bighost.area.ba.cnr.it/BIG/UTRHome/
- UTRdb and UTRsite download page ftp://area.ba.cnr.it/pub/embnet/database/utr
- Etzold T, Argos P. SRS - an indexing and retrieval tool for flat file data libraries. Comput Appl Biosci. 1993;9:49–57. doi: 10.1093/bioinformatics/9.1.49. [DOI] [PubMed] [Google Scholar]
- UTRdb and UTRsite SRS page http://bighost.area.ba.cnr.it/srs
- SRS at European Bioinformatics Institute http://srs.ebi.ac.uk:80/
- EMBL nucleotide sequence database http://www.ebi.ac.uk/embl/
- GCG is now Accelrys http://www.accelrys.com/about/gcg.html
- Creancier L, Morello D, Mercier P, Prats AC. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J Cell Biol. 2000;150:275–281. doi: 10.1083/jcb.150.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Sarnow P. Location of the internal ribosome entry site in the 5' non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA-protein interactions. Nucleic Acids Res. 1997;25:2800–2807. doi: 10.1093/nar/25.14.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Fong P, Iizuka N, Choate D, Cavener DR. Ultrabithorax and Antennapedia 5' untranslated regions promote developmentally regulated internal translation initiation. Mol Cell Biol. 1997;17:1714–1721. doi: 10.1128/mcb.17.3.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J, Sella O, Le SY, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES). J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- Pozner A, Goldenberg D, Negreanu V, Le SY, Elroy-Stein O, Levanon D, Groner Y. Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol Cell Biol. 2000;20:2297–2307. doi: 10.1128/mcb.20.7.2297-2307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coldwell MJ, Mitchell SA, Stoneley M, MacFarlane M, Willis AE. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905. doi: 10.1038/sj.onc.1203407. [DOI] [PubMed] [Google Scholar]
- Kim JG, Armstrong RC, Berndt JA, Kim NW, Hudson LD. A secreted DNA-binding protein that is translated through an internal ribosome entry site (IRES) and distributed in a discrete pattern in the central nervous system. Mol Cell Neurosci. 1998;12:119–140. doi: 10.1006/mcne.1998.0701. [DOI] [PubMed] [Google Scholar]
- Law GL, Raney A, Heusner C, Morris DR. Polyamine regulation of ribosome pausing at the upstream open reading frame of S-adenosylmethionine decarboxylase. J Biol Chem. 2001;276:38036–38043. doi: 10.1074/jbc.M105944200. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- Griffin E, Re A, Hamel N, Fu C, Bush H, McCaffrey T, Asch AS. A link between diabetes and atherosclerosis: glucose regulates expression of CD36 at the level of translation. Nat Med. 2001;7:840–846. doi: 10.1038/89969. [DOI] [PubMed] [Google Scholar]
- Nomura A, Iwasaki Y, Saito M, Aoki Y, Yamamori E, Ozaki N, Tachikawa K, Mutsuga N, Morishita M, Yoshida M, et al. Involvement of upstream open reading frames in regulation of rat V(1b) vasopressin receptor expression. Am J Physiol Endocrinol Metab. 2001;280:E780–E787. doi: 10.1152/ajpendo.2001.280.5.E780. [DOI] [PubMed] [Google Scholar]
- Meijer HA, Dictus WJ, Keuning ED, Thomas AA. Translational control of the Xenopus laevis connexin-41 5'-untranslated region by three upstream open reading frames. J Biol Chem. 2000;275:30787–30793. doi: 10.1074/jbc.M005531200. [DOI] [PubMed] [Google Scholar]
- Brown CY, Mize GJ, Pineda M, George DL, Morris DR. Role of two upstream open reading frames in the translational control of oncogene mdm2. Oncogene. 1999;18:5631–5637. doi: 10.1038/sj.onc.1202949. [DOI] [PubMed] [Google Scholar]


