Abstract
Prediction of the biological effect of missense substitutions has become important because they are often observed in known or candidate disease susceptibility genes. In this paper, we carried out a 3-step analysis of 1514 missense substitutions in the DNA-binding domain (DBD) of TP53, the most frequently mutated gene in human cancers. First, we calculated two types of conservation scores based on a TP53 multiple sequence alignment (MSA) for each substitution: (i) Grantham Variation (GV), which measures the degree of biochemical variation among amino acids found at a given position in the MSA; (ii) Grantham Deviation (GD), which reflects the ‘biochemical distance’ of the mutant amino acid from the observed amino acid at a particular position (given by GV). Second, we used a method that combines GV and GD scores, Align-GVGD, to predict the transactivation activity of each missense substitution. We compared our predictions against experimentally measured transactivation activity (yeast assays) to evaluate their accuracy. Finally, the prediction results were compared with those obtained by the program Sorting Intolerant from Tolerant (SIFT) and Dayhoff's classification. Our predictions yielded high prediction accuracy for mutants showing a loss of transactivation (∼88% specificity) with lower prediction accuracy for mutants with transactivation similar to that of the wild-type (67.9 to 71.2% sensitivity). Align-GVGD results were comparable to SIFT (88.3 to 90.6% and 67.4 to 70.3% specificity and sensitivity, respectively) and outperformed Dayhoff's classification (80 and 40.9% specificity and sensitivity, respectively). These results further demonstrate the utility of the Align-GVGD method, which was previously applied to BRCA1. Align-GVGD is available online at http://agvgd.iarc.fr.
INTRODUCTION
Analysis of mutations has become increasingly important due to their association with various diseases (1,2). Of the disease-associated variations present in the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/hgmd0.html), the majority are single base substitutions that result in an amino acid change (missense mutation). Specific examples where high frequencies of missense mutations are associated with disease include the tumor suppressor TP53 and sporadic cancers (3), CFTR and cystic fibrosis (4) and AVPR2 and neurogenic diabetes insipidus (http://www.medicine.mcgill.ca/nephros/). Large collections of mutation data are available via HGMD, the Human Genome Variation Society (http://www.genomic.unimelb.edu.au/mdi/) and OMIM (5). More specific to cancer, the Catalogue of Somatic Mutations in Cancer (Cosmic) holds over 23 000 mutations in 538 cancer-related genes (http://www.sanger.ac.uk/genetics/CGP/cosmic/).
Annotations of mutation effects are rarely found in these databases, mainly because mutagenesis experiments and functional assays are labor-intensive and data accrual does not follow the pace of the accumulation of descriptive, mutation data. In many instances, the nature and scope of such functional assays are still a matter of debate. To circumvent these limitations, more and more computational methods are being developed to predict the function of missense mutants and to identify residues that have a significant effect on maintaining wild-type function. Different approaches have been explored, including sequence-based methods (6,7), structure-based algorithms (8–12) and a combination of both (13–15).
The use of multiple sequence alignments (MSAs) to align either closely related sequences, distantly related sequences or both, have highlighted two major trends that are unique to disease-associated mutations. First, differences in biochemical properties between mutant and wild-type amino acids are larger for disease-associated mutations than for neutral mutations (16). This trend exists because large biochemical changes between mutant and wild-type are more likely to alter the structure and may hence change the function of the protein, explaining why such changes are generally not tolerated during natural selection. Second, mutations associated with disease tend to be located at residue positions that are highly conserved across species (17). In this sense, amino acids that are conserved across species are more likely to have an important structural or functional role. One popular method for measuring biochemical distances between pairs of amino acids is the Grantham Difference (18), which takes into account the composition, polarity and volume of mutant and wild-type amino acids.
In contrast to other tumor suppressor genes such as APC and BRCA1, the majority of TP53 mutations are missense mutations rather than truncating mutations. These missense mutations are concentrated in the DNA-binding domain (DBD) of the protein, comprising 194 amino acids. All mutations cited in the literature are compiled in the IARC TP53 database (3), which is the largest database of cancer associated mutations available for a single gene. The p53 protein is a transcription factor activated by various stress conditions, including DNA damage, oncogene activation or hypoxia. P53 regulates the transcription of several genes involved in DNA repair, cell cycle checkpoints or apoptosis (19). Kato et al. (20) used a yeast-based expression assay to measure the transactivation activity of all possible p53 missense mutations, that can arise due to a single nucleotide substitution, on eight different p53 response-elements (p53-RE) present in different p53 target-genes: BAX, CDKN1 (WAF1), GADD45A, MDM2, P53AIP1, PMAIP1 (NOXA), RRM2B (P53R2) and SFN (h1433S).
In this study, we have constructed a p53 protein MSA and combined a conservation score (GV) with a measure of biochemical difference between wild-type and mutant residues with respect to the alignment (GD). This extension of the Grantham Difference, called Align-GVGD, has previously been applied to BRCA1 and contributed to the clinical categorization of eight previously unclassified missense mutations (21). To demonstrate the significance of the classifications obtained with Align-GVGD, we have assessed them against functional categories derived from experimental measurements of transactivation activities of a subset of 1514 missense p53 mutants (20). Next, we have compared the resulting prediction accuracies with those yielded by two well known prediction methods. The first is SIFT which calculates normalized probabilities that specific substitutions would be tolerated at a given position, and assigns the mutation effect from a specific probabilities cutoff value (13,14). The second is based on Dayhoff's relatedness odds matrix (22), which gives the probability that a substitution will occur by chance in a second sequence using the PAM matrix.
MATERIALS AND METHODS
MSA
The p53 protein MSA was constructed with 3D-Coffee (23), a web-based tool for aligning multiple sequences, which takes into account relevant protein structure(s) to improve the alignment. The following nine sequences were used for the MSA: Homo sapiens (sp|P04637), Macaca mulatta (monkey, sp|P56424), Bos taurus (bovine, sp|P67939), Canis familiaris (dog, sp|Q29537), Mus musculus (mouse, sp|P02340), Rattus norvegicus (rat, sp|P10361), Gallus gallus (chicken, sp|P10360), Xenopus laevis (frog, tr|P53_XENOPUS), Brachydanio rerio (zebrafish, sp|P79734). The X-ray solved structure of the DBD of human p53 [PDB (24) id 1tsr, chain B] was also input. All default parameters when running 3D-Coffee were used with the exception of excluding the Msap_pair option, which performs structural alignments (only one PDB structure was used), and including the ‘Mclustalw_aln’, an option for MSA.
Align-GVGD, SIFT and Dayhoff's classification
GV and GD calculations in the Align-GVGD program (21) are an extension of the Grantham Difference, which takes into account the composition (C), polarity (P) and volume (V) of amino acids (18). Conceptually, each amino acid can be plotted on a 3D graph, having C, P, V as the three axes, with different weights (18) applied to each axis. All amino acids at a given position in the MSA then form a cloud of points when plotted. This cloud of points can be enclosed within a box, the coordinates of which are defined by the minimum and maximum values of C, P, V, for the observed amino acids. GV is computed as the Euclidian length of the main diagonal of the box. GV is thus a measure of the amount of observed biochemical variation in a particular position in the alignment. Next, the GD is calculated by plotting a given mutation on the composition-polarity-volume graph, and measuring the Euclidian distance between that mutation and the closest point on the GV box. If the substitution lies within the box, then GD = 0. Otherwise, GD is greater than 0. GD is thus a measure of the biochemical difference between the mutant and the observed variation at that position according to the MSA.
The freely available web-based tool SIFT (http://blocks.fhcrc.org/sift/SIFT.html) (13) was used with default settings and the MSA obtained by 3D-Coffee as input. SIFT calculates the probabilities of having an amino acid at a specific position relative to the most frequent amino acid at that position. A cutoff for these probabilities is used to classify the mutations as tolerated and non-tolerated.
Mutants were classified according to Dayhoff's substitution matrix. The Dayhoff matrix highlights groups of amino acids with common chemical and physical properties. The following groups derived from the log odds ratio of the 250 PAM matrix were used (22): (V,L,I,M), (R,K,H), (D,E,N,Q), (F,Y,W), (C), (A,S,T,G,P). When the mutant and wild-type amino acid fall within a group, the mutation is considered conservative, if they fall into different groups, the mutation is classified as non-conservative.
P53 functional dataset
Kato et al. (20) measured the transactivation activity of all possible missense mutations (2314) in p53 (codons 2 to 393), resulting from a single nucleotide substitution, on eight different p53-RE derived from the following p53 target-genes BAX, CDKN1, GADD45A, MDM2, P53AIP1, PMAIP1, RRM2B and SFN. The transactivation activity of each mutant on each p53-RE was expressed as a percentage of the transactivation activity of the wild-type protein on the corresponding p53-RE. Mutants that showed variations in transactivation activity depending on the p53-RE were disregarded in our analysis because of their ambiguous overall activity. A subset of 1514 mutants, showing a similar activity across all eight promoters, was considered where mutants with percent activity below 45% on all promoters were categorized as non-functional (446 mutants), and those with percent activity above 45% and below 200% on all promoters were classified as functional (1068 mutants). We refer to these mutants as ‘consistent mutants’ (listed in Supplementary Table S1). In addition, mutations at codon 72 were omitted from further analysis due to the uncertainty of the wild-type amino acid at that position.
RESULTS
GV values were calculated for amino acid positions 2 to 393 (except codon 72) in p53, and GD values were computed for the 1514 consistent missense mutations. GV values provide a quantitative measure of the range of biochemical properties for a given amino acid position, based on all residues found at that position in a given MSA. GD values measure the distance between the mutant amino acid (described by its polarity, volume and composition) and the allowed variation as calculated by GV (see Materials and Methods for more details). GV and GD values were calculated for four different MSA: (i) Placental level: includes six placental mammal sequences, (ii) Chicken level: includes seven sequences from placental mammals to chicken, (iii) Frog level: includes eight sequences from placental mammals to frog, (iv) Fish level: includes nine sequences from placental mammals to fish. As more distantly related sequences are added to the alignment, GV is expected to increase for some residues. In addition, GD is expected to decrease for many of the substitutions observed at residues where GV increases.
Correlations between GV, mutation frequencies and structural features
In this analysis, GV were calculated separately for all four MSAs. The MSA of p53 using all sequences (human, monkey, bovine, dog, mouse, rat, chicken, frog and zebrafish) with the corresponding GV values is shown in Figure 1. Positions with a GV = 0 (highlighted in green) have the same amino acid across all species and are thus invariant. A total of 126 ‘invariant’ residues out of 391 (32%) were found. Seven fully conserved regions (with at least four positions in a row with GV = 0) were identified and include the following residue positions: 117–122, 175–181, 196–200, 213–216, 218–221, 237–254 and 270–282. These regions lie within the DBD of p53 and are consistent with previously identified conservation domain (25).
Figure 1.
GV values and MSA constructed using 3D-coffee. GV values calculated with the complete MSA (fish level) are shown for each position in the alignment (the numbering follows that of the human sequence). Areas of total conservation are shown in green.
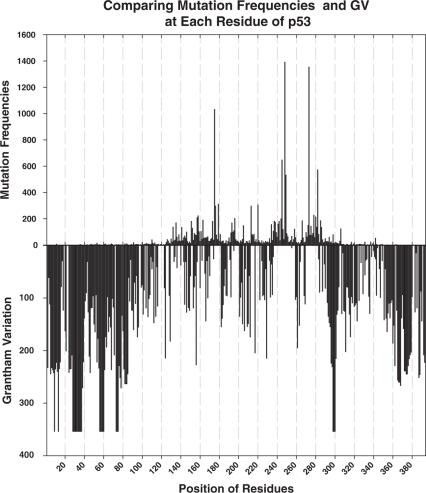
GV values were directly compared with frequencies of mutations found in cancers (data extracted from the IARC TP53 database, http://www-p53.iarc.fr/). These frequencies measure the association of a mutation with cancer, such that positions with high frequencies are often observed in cancer, making them likely to contribute to cancer development. Figure 2 depicts the distribution of these frequencies at each position in the p53 DBD with the corresponding GV values. The graphs show an overall correlation between positions that are mutated at high frequency and those that have a low GV, indicating that substitutions at positions that are highly conserved are strongly selected for during tumorigenesis. This is particularly true in areas of p53 that contain the so-called ‘mutation hotspots’ (residues 171–181, 237–258 and 270–282). However, not all conserved residues were frequently mutated, as is shown by the low mutation frequencies at residues 117–122 and 125–127. In addition, some areas of relatively low conservation appear to contain minor mutation hotspots, such as residues 157 and 158.
Figure 2.
Comparison between GV and mutation frequency, extracted from the IARC TP53 database, for a given position. Generally, areas comprising residues with low GV values (<61.3, high conservation) correspond to areas with frequently mutated residues (171–181, 237–258 and 270–282). Exceptions of this trend include residues 117–122 and 125–127, which describe areas with low GV values (high conservation) but very low mutation frequency.
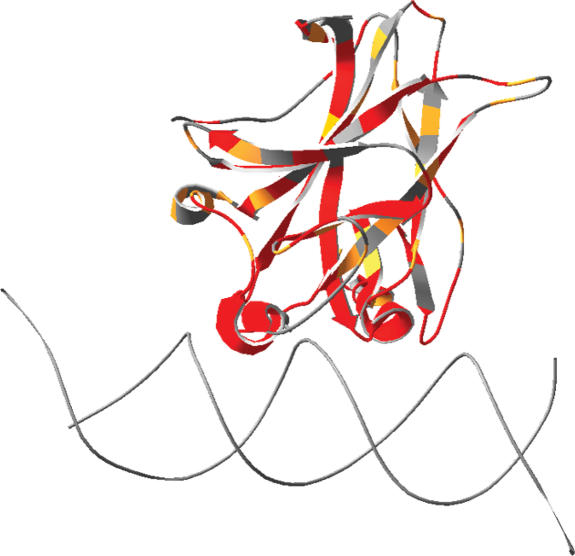
The GV values were mapped on to the 3D structure of the DBD of p53 (26), comprising residues 96 to 289 (Figure 3). Invariant positions have GV = 0 (red), conserved positions have GV < 61.3 (orange) and for variable positions GV ≥ 61.3 (grey). The value 61.3 corresponds to the variation across the polar amino acid set Asp, Asn, Glu and Gln. It was arbitrarily chosen as the greatest level of amino acid variation within a single position that one might consider ‘conservative substitution’. Importantly, the figure shows that low GV values (less than 61.3) tend to concentrate in the area of DNA-binding residues, which contains the majority of hotspot mutations (27). Some correlations between structural features of residues and their GV values are worth noting. First, the residues involved in the zinc-binding domain, Cys176, His179, Cys238 and Cys242 are all highly conserved positions (the zinc is shown as a grey sphere in the figure). Those residues are essential for zinc-binding and maintaining the structure. Second, all residues involved in DNA-binding are highly conserved: out of 14 residues, GV = 0 for 13 positions and GV = 26 for 1 position (residue 283). Those residues are functionally important as they either directly bind DNA or help maintain the proper conformation to allow DNA-binding. Third, most positions in the hydrophobic core, which are mainly responsible for maintaining the stability of the protein, have a high frequency of mutations and low GV values (less than or equal to 30.9, which is GV for the non-polar amino acid set Leu, Ile, Met and Val). Previously, eleven highly conserved positions have been identified in the hydrophobic core (156–158, 205, 215, 220, 232, 258, 259 and 266) (28). Only Arg156 showed a high GV value of 227.4. This high GV value is mainly due to the addition of the more distantly related sequences of chicken, frog and fish.
Figure 3.
Representation of the DBD of p53 (PDB ID 1tsr, chain B), color-coded by GV values with the following cutoffs: GV = 0, red; 0 < GV < 61.3, orange; GV > 61.3, grey. The figure shows that areas of high conservation (red) involve residues found in DBD and zinc-binding motif. Other areas of the protein do not show a particular conservation trend.
Predictions of transactivation activity using Align-GVGD
All missense mutants in the p53 DBD were predicted as neutral, deleterious, or unclassified using the following five Align-GVGD criteria:
if GD = 0: the composition, polarity and volume of the mutant amino acid fall within observed range of variation according to the alignment at that position so the mutant is predicted as neutral;
if (GV > 61.3) and (0 < GD ≤ 61.3): the position tolerates more than ‘conservative’ substitution and the composition, polarity and volume of the mutant amino acid fall close to the observed range of variation according to the alignment at that position so the mutant is predicted as neutral;
if (GV = 0) and (GD > 0): the position of interest is invariant (100% conservation) so any mutation at the position is predicted as deleterious;
if (0 < GV ≤ 61.3) and (GD > 0): there is a small variation in amino acids at a given position (the residues encountered are biochemically similar), yet the mutant amino acid does not fall within that range of variation. The mutant is predicted as deleterious;
if the mutant does not fall in the previous categories, it is unclassified.
Table 1 shows the distribution of the classifications derived from Align-GVGD values using the four different MSA described in the previous section. Our results show that adding more distantly related sequences increased the number of mutants predicted as neutral while the number of mutants predicted as deleterious decreased. Because adding more distantly related sequences to the MSA introduces more sequence variability, mutants previously classified as deleterious may now fall into the neutral category or into the unclassified category (when GV and GD both exceed 61.3).
Table 1.
Distribution of classifications made by Align-GVGD
| Eutheriana | Chickenb | Frogc | Fishd | |
|---|---|---|---|---|
| Neutral | 765 | 766 | 794 | 800 |
| Deleterious | 637 | 637 | 608 | 607 |
| Unclassified | 112 | 111 | 112 | 107 |
aMSA used includes all sequences but chicken, frog and fish.
bMSA used includes all sequences but chicken and frog.
cMSA used includes all sequences but frog.
dMSA used includes all sequences.
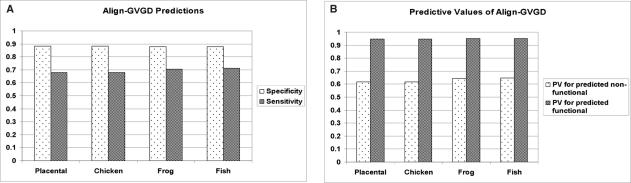
These predictions were directly compared with transactivation categories derived from results of experimental assays performed in yeast (20) on 1514 ‘consistent mutants’ (see Materials and Methods). Figure 4A shows the specificity (ratio of correctly predicted deleterious mutants versus the total number of observed non-functional mutants) and sensitivity (ratio of correctly predicted neutral mutants versus the total number of observed functional mutants) of the predictions obtained for each MSA on the ‘consistent’ mutants. The results show three major trends. First, the prediction accuracies for deleterious mutants are high (high specificity) while they are lower for neutral mutants (low sensitivity). Second, the addition of more distantly related species increases the sensitivity. Because adding more divergent sequences to the MSA increases sequence variability in the alignment, GV for many of the positions increases, and this will in turn decrease the GD for certain mutants (a mutated residue is more likely to fit in a larger allowable variation at a given position). Third, adding more sequences does not notably decrease the specificity. This important observation suggests that the addition of more distantly related sequences is essential for more accurate predictions of transactivation activity.
Figure 4.
Prediction of experimentally measured transactivation activity by the Align-GVGD scoring method. (A) The specificity (ratio of correctly predicted deleterious mutants versus the total number of observed non-functional mutants) and sensitivity (ratio of correctly predicted neutral mutants versus the total number of observed functional mutants) of the predictions are shown for four different MSA. As more divergent sequences are added to the MSA, the predictions improve and show a substantial increase in specificity with only a comparatively slight decrease in sensitivity. (B) PV for predicted deleterious and predicted neutral mutants at the four sequence levels are depicted. While the PV for predicted neutral mutants is relatively unchanged, the PV for predicted deleterious mutants increases when more divergent sequences are added to the MSA.
To evaluate the efficacy of our predictions, predictive values (PV) for deleterious (percent of mutations predicted as deleterious that are observed non-functional) and neutral predictions (percent of mutations predicted as neutral that are observed functional) were calculated (Figure 4B). The figure demonstrates that when Align-GVGD predicts mutants as neutral, about 95% actually are functional, according to the transactivation activities. On the other hand, when Align-GVGD predicts mutants as deleterious, 61.7 to 64.6% are actually non-functional. As more divergent sequences are added to the MSA, the predictive value for predicted deleterious mutants increases while the predictive value for the predictive neutral mutants remains nearly constant.
Predictions of transactivation activity using SIFT and Dayhoff's classification
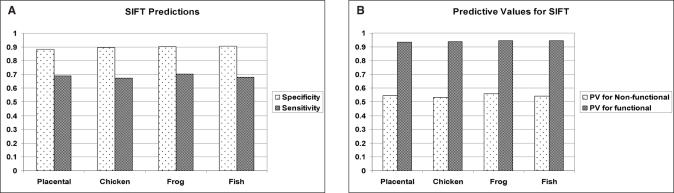
SIFT was used to classify the p53 missense mutants using our four different MSA as input. The categories obtained with SIFT (tolerant/intolerant) were compared to the transactivation categories as performed above with Align-GVGD. Mutants classified as non tolerant by SIFT were expected to be non-functional while those classified as tolerant were expected to be functional. The specificity/sensitivity and PV of the SIFT predictions are shown in Figure 5A and B, respectively. In comparison with Align-GVGD predictions, SIFT yielded similar specificity (88.3 to 90.6% versus ∼88% for Align-GVGD) and similar sensitivity values (67.4 to 70.3% versus 67.9 to 71.2% for Align-GVGD). In addition, although the PV for the predicted tolerant/neutral mutants are very similar (about 95%), those for intolerant/deleterious mutants are higher for Align-GVGD (61.7 to 64.6%) than for SIFT (53.4 to 55.9%).
Figure 5.
Prediction of experimentally measured transactivation activity using SIFT software (13) and four different MSA as input. (A) The specificity and sensitivity, and (B) PV of the predictions are shown. No noticeable correlation is observed between the addition of more divergent sequences in the MSA and the diagnostic values.
Finally, categories based on Dayhoff's conservation rules (see Materials and Methods) were compared with transactivation activity. Table 2 shows the resulting predictions when associating conservative mutations with non-functional mutants and non-conservative mutations with functional mutants. The sensitivity/specificity and PV are lower compared to those obtained with either the Align-GVGD or SIFT approaches.
Table 2.
Transactivation prediction results using Dayhoff conservation categories
| Specificity | 80 |
| Sensitivity | 40.9 |
| PV for predicted deleterious | 36.1 |
| PV for predicted neutral | 83.1 |
Align-GVGD as a freely available and web-based software
A web interface has been developed to access the Align-GVGD program, written in Perl (available at http://agvgd.iarc.fr). As input, users must provide their own MSA and a list of mutations either by uploading the appropriate files or by copying/pasting. Both input files must be simple text files (Word documents are not recommended) and the MSA must be in FASTA format. The program will then output a table containing GV and GD for all mutations given in the input file, along with functional predictions: deleterious 1 (GV = 0 and GD > 0), deleterious 2 (0 < GV ≤ 61.3 and GD > 0), neutral 1 (GD = 0), neutral 2 (GV > 61.3 and 0 < GD ≤ 61.3) and unclassified. Users may download the results in a tab delimited simple text file which may easily be imported into Excel for further analysis. The program uses all sequences in the alignment when performing the calculations. Error messages appear when the MSA is not in the correct format or when unknown amino acid letter codes (other then the 20 letters for the naturally occurring amino acids and the gap symbol ‘-’) are found in the MSA or list of mutations.
DISCUSSION
Previous studies have found a strong correlation between highly conserved residues and intolerance of mutations, which are likely to cause disease (16,17). The majority of mutations associated with disease show larger biochemical differences between mutant and wild-type amino acids than between amino acids observed in the MSA for a given position (17). These observations provide the basis for our Align-GVGD approach, which considers both the biochemical variability for a position in the MSA and the distance from a mutant amino acid to the observed variability. Application of the Align-GVGD method to p53, which has the largest mutation dataset available on a single gene, along with a comprehensive dataset of transactivation activities measured in experimental yeast assays, provides an opportunity to further assess the significance of the correlation between conservation and functional effect of mutations.
Validation of our MSA and GV values was performed by comparing GV values with conservation and structural features previously reported. Seven fully conserved regions (117–122, 175–181, 196–200, 213–216, 218–221, 237–254 and 270–282) with GV = 0 were found, consistent with previously identified conserved clusters (28,29). To validate our MSA and GV values, we compared our correlations between conservation and structural features of residues with those previously reported (30) and concordantly found that residues in the zinc- and DNA-binding motifs are highly conserved. We found that GV values at each position in p53 strongly correlated with the frequencies of mutants observed in human cancer as reported in the IARC TP53 database (3). In the DBD, low GV values (less than 42.8) were observed for the vast majority of residues located in the previously identified five ‘mutation hotspot’ regions, including residues 132–143, 151–159, 172–179, 237–249 and 272–286 (29).
A divergence between GV values and mutation frequency was noted for the region of the p53 DBD comprising residues 117 to 122. This cluster corresponds to a conserved portion of the L1 loop that directly binds to the major groove of the DNA. These residues have low GV values but are rarely found mutated in human cancer. It is thus likely that substitution at these residues does not affect p53 function in a way compatible with loss of tumor suppressive activities. Indeed, studies by Resnick and colleagues (31) have found that mutations in this area often resulted in mutant proteins with increased transactivation activities (called ‘supertrans mutants’). Such mutants would be counter-selected, and therefore very rare, in human cancer.
Functional predictions based on appropriate GV and GD cutoff values were then compared with the experimentally measured transactivation activities. We accurately predicted the activity of up to 88.1% deleterious mutants (unable to transactivate) and 71.2% neutral mutants (able to transactivate). The accurate prediction of neutral mutants increased as more distantly related sequences were added to the MSA, while only slightly decreasing the accurate prediction of deleterious mutants. The Align-GVGD values are thus highly dependant on the MSA. Previous studies suggest that using both closely and distantly related sequences in appropriate proportion is most suitable for accurate construction of MSA (16,32). This is because relying mainly on closely related sequences will result in an apparent lack of sequence variation due to little divergence between the individual sequences. On the other hand, using sequences that are too divergent increases the risk of including a related sequence that codes for a protein that has a different function. In this case, the number of functionally constrained sites will be under-estimated. Because the choice of sequences to use in an MSA is highly dependent on the protein studied, it is best to make a systematic comparison between alignments of increasing divergence in order to find an optimal combination of sensitivity and specificity.
We found that our predictions, using Align-GVGD, were similar to those obtained from SIFT, and better than those obtained using Dayhoff's conservation rules. Although SIFTattempts to classify all mutants as either tolerated or not tolerated, Align-GVGD allows a third ‘unclassified mutants’ category which probably leads to a higher level of certainty in the predicted effect of categorized mutants. Indeed, the biggest performance difference between Align-GVGD and SIFT was in the predictive value for deleterious substitutions [PV(D)]. For Align-GVGD, PV(D) ranged from 61.7 to 64.6% and the highest value resulted from the complete alignment, whereas, for SIFT, PV(D) ranged from 53.4 to 55.9% and depended little on the alignment. Thus both programs over-predict false deleterious substitutions, which is the type of false-positive error that we would most likely to avoid. False-positive prediction of deleterious substitutions should be mostly due to two sources of error. The first is assay-based: p53 is involved in numerous pathways and has biochemical activities other than DNA-binding and transcriptional activation, thus some of the substitutions are likely to affect functions not measured by the transcriptional reporter assay. But in silico prediction methods have intrinsic false-positive error rates; e.g. SIFT is reported to have a ‘weighted false-positive error rate’ of ∼20% (14). As assay-based false positives should affect both algorithms equally, the PV(D) difference between the two algorithms should reflect a small Align-GVGD advantage towards prediction of deleterious substitutions. Interestingly, ∼87% of mutants unclassified by Align-GVGD are experimentally functional. This implies that some mutants (93 out of 1514) may have high GV and/or GD (both higher than 61.3) while being experimentally functional. These mutants then show a substantial variation across species and/or the mutant residue is substantially different from the residues observed at the respective position. This counter-intuitive observation demonstrates the limitations of using solely sequence information to make predictions.
One advantage of SIFT over our Align-GVGD method is that SIFT allows automatic generation of an MSA, either from the input of one sequence (of the species under study), or from the input of all sequences to be included in the alignment. However, Align-GVGD shows a stronger dependency on the input MSA than does SIFT, suggesting that Align-GVGD predictions may be improved with a more informative MSA as input.
A major incentive for using Align-GVGD is that the program provides quantitative measures of the range of biochemical variation of the amino acids present at the position of a missense substitution (GV) and the distance between the missense substitution and that range of variation (GD), and these measures are on the same scale as the original Grantham Difference. One can thus easily trace back the features of amino acids and help explain the reasons for strong or weak correlations between GV, GD and function. The current set of Align-GVGD cutoff values are based on biophysical reasoning rather than optimization over a dataset. Furthermore, this approach does not require entire sequences or entire structures as input. Although the structure may be used to construct the MSA, as was done in this study, it is not mandatory.
Several modifications may be applied to further improve the predictions. First, the ‘consistent mutants’ could be used as a training set to improve the cutoff values for GV and GD. However, the a priori rules that are currently used in Align-GVGD present an advantage in that the cutoffs are not dependant on functional data, which are often neither comprehensive nor fully representative of the range of activities displayed by a protein. Second, adding another distantly related sequence may further improve the results. Provided the trend seen in Figure 4A continues, it is likely that the sensitivity will increase. Moreover, the specificity and sensitivity will likely be closer to converging, indicating that the algorithm may accurately classify as many neutral as deleterious mutants. However, one must ascertain that the most distantly related sequence still has the same function as human p53. Over the last few years, the release of tunicate and sea urchin genome sequences in addition to a series of vertebrate sequences should lead to many more gene model and cDNA sequences of human gene orthologs, thereby leading to more appropriately informative MSA. With these new sequences and the simplicity and web-based availability of softwares such as Align-GVGD and SIFT, the accuracies of functional predictions of missense substitutions will undoubtedly increase.
Overall, we show that the GV values for each residue in p53 correlate well with the frequencies of mutations in human cancers extracted from the IARC TP53 database. Using pre-defined Align-GVGD cutoff values, we accurately predicted the transactivation activity of up to 88.1% of deleterious and 71.2% of neutral mutants, which have a similar transactivation activity across all p53-REs tested. The addition of more distantly related sequences increases the accuracy of predictions for neutral mutants substantially while only slightly decreasing the accuracy of predictions for deleterious mutants. The simplicity and web-based availability of Align-GVGD will allow functional classification of missense mutants for any genes with sufficient sequences available.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The work of E.M. was carried out during the tenure of a Special Training Award from the International Agency for Research on Cancer. E.M.'s STA was supported by National Institute of Environmental Health Sciences funding. The authors would also like to thank Catherine Voegele for creating the Align-GVGD website. Funding to pay the Open Access publication charges for this article was provided by the International Agency for Research on Cancer.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cooper D.N., Ball E.V., Krawczak M. The human gene mutation database. Nucleic Acids Res. 1998;26:285–287. doi: 10.1093/nar/26.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 3.Olivier M.L., Eeles R., Hollstein M., Khan M.A., Harris C.C., Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 4.Bobadilla J.L., Macek M., Jr, Fine J.P., Farrell P.M. Cystic fibrosis: a worldwide analysis of CFTR mutations–correlation with incidence data and application to screening. Hum. Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 5.Online Mendelian Inheritance in Man, OMIM (TM) McKusick-Nathans Institute of Genetic Medicine, John Hopkins University (Baltimore, MD), and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD) 2000. World Wide Web URL: http://www.ncbi.nlm.nih.gov/omim/
- 6.Sunyaev S., Hanke J., Aydin A., Wirkner U., Zastrow I., Reich J., Bork P. Prediction of nonsynonymous single nucleotide polymorphisms in human disease-associated genes. J. Mol. Med. 1999;77:754–760. doi: 10.1007/s001099900059. [DOI] [PubMed] [Google Scholar]
- 7.Yang Z., Ro S., Rannala B. Likelihood models of somatic mutation and codon substitution in cancer genes. Genetics. 2003;165:695–705. doi: 10.1093/genetics/165.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dambosky J., Prokop M., Koca J. TRITON: graphic software for rational engineering of enzymes. Trends. Biochem. Sci. 2001;26:71–73. doi: 10.1016/s0968-0004(00)01708-4. [DOI] [PubMed] [Google Scholar]
- 9.Ferrer-Costa C., Orozco M., de la Cruz X. Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. J. Mol. Biol. 2002;315:771–786. doi: 10.1006/jmbi.2001.5255. [DOI] [PubMed] [Google Scholar]
- 10.Prokop M., Damborsky J., Koca J. TRITON: in silico construction of protein mutants and prediction of their activities. Bioinformatics. 2000;16:845–846. doi: 10.1093/bioinformatics/16.9.845. [DOI] [PubMed] [Google Scholar]
- 11.Stitziel N.O., Binkowski T.A., Tseng Y.Y., Kasif S., Liang J. topoSNP: a topographic database of non-synonymous single nucleotide polymorphisms with and without known disease association. Nucleic Acids Res. 2004;32:D520–D522. doi: 10.1093/nar/gkh104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sunyaev S., Ramensky V., Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 13.Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng P.C., Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller M.P., Kumar S. Understanding human disease mutations through the use of interspecific genetic variation. Hum. Mol. Genet. 2001;10:2319–2328. doi: 10.1093/hmg/10.21.2319. [DOI] [PubMed] [Google Scholar]
- 17.Vitkup D., Sander C., Church G.M. The amino-acid mutational spectrum of human genetic disease. Genome Biol. 2003;4:R72. doi: 10.1186/gb-2003-4-11-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 19.Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 20.Kato S., Han S.Y., Liu W., Otsuka K., Shibata H., Kanamaru R., Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl Acad. Sci. USA. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tavtigian S.V., Deffenbaugh A.M., Yin L., Judkins T., Scholl T., Samollow P.B., de Silva D., Zharkikh A., Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J. Med. Genet. 2005 doi: 10.1136/jmg.2005.033878. 2005 July 13; [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dayhoff M.O. Atlas of Protein Sequence and Structure. Washington,D.C: National Biomedical Research Foundation; 1978. [Google Scholar]
- 23.O'Sullivan O., Suhre K., Abergel C., Higgins D.G., Notredame C. 3DCoffee: combining protein sequences and structures within multiple sequence alignments. J. Mol. Biol. 2004;340:385–395. doi: 10.1016/j.jmb.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 24.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soussi T., Caron de Fromentel C., May P. Structural aspects of the p53 protein in relation to gene evolution. Oncogene. 1990;5:945–952. [PubMed] [Google Scholar]
- 26.Cho Y., Gorina S., Jeffrey P.D., Pavletich N.P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 27.Hollstein M., Hergenhahn M., Yang Q., Bartsch H., Wang Z.Q., Hainaut P. New approaches to understanding p53 gene tumor mutation spectra. Mutat. Res. 1999;431:199–209. doi: 10.1016/s0027-5107(99)00162-1. [DOI] [PubMed] [Google Scholar]
- 28.Walker D.R., Bond J.P., Tarone R.E., Harris C.C., Makalowski W., Boguski M.S., Greenblatt M.S. Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 protein structural and functional features. Oncogene. 1999;18:211–218. doi: 10.1038/sj.onc.1202298. [DOI] [PubMed] [Google Scholar]
- 29.Caron de Fromentel C., Soussi T. TP53 tumor suppressor gene: a model for investigating human mutagenesis. Genes Chromosomes Cancer. 1992;4:1–15. doi: 10.1002/gcc.2870040102. [DOI] [PubMed] [Google Scholar]
- 30.Martin A.C., Facchiano A.M., Cuff A.L., Hernandez-Boussard T., Olivier M., Hainaut P., Thornton J.M. Integrating mutation data and structural analysis of the TP53 tumor-suppressor protein. Hum. Mutat. 2002;19:149–164. doi: 10.1002/humu.10032. [DOI] [PubMed] [Google Scholar]
- 31.Inga A., Monti P., Fronza G., Darden T., Resnick M.A. p53 mutants exhibiting enhanced transcriptional activation and altered promoter selectivity are revealed using a sensitive, yeast-based functional assay. Oncogene. 2001;20:501–513. doi: 10.1038/sj.onc.1204116. [DOI] [PubMed] [Google Scholar]
- 32.Greenblatt M.S., Beaudet J.G., Gump J.R., Godin K.S., Trombley L., Koh J., Bond J.P. Detailed computational study of p53 and p16: using evolutionary sequence analysis and disease-associated mutations to predict the functional consequences of allelic variants. Oncogene. 2003;22:1150–1163. doi: 10.1038/sj.onc.1206101. [DOI] [PubMed] [Google Scholar]