Abstract
Alternative splicing enables expression of functionally diverse protein isoforms. The structural and functional complexity of zinc-finger transcription factor ZBP-89 suggests that it may be among the class of alternatively spliced genes. We identified a human ZBP-89 splice isoform (ZBP-89ΔN), which lacks amino terminal residues 1–127 of the full-length protein (ZBP-89FL). ZBP-89ΔN mRNA was co-expressed with its ZBP-89FL cognate in gastrointestinal cell lines and tissues. Similarly, ZBP-89ΔN protein was expressed. To define its function in vivo, we generated ZBP-89ΔN knock-in mice by targeting exon 4 that encodes the amino terminus. Homozygous ZBP-89ΔN mice, expressing only ZBP-89ΔN protein, experienced growth delay, reduced viability and increased susceptibility to dextran sodium sulfate colitis. We conclude that ZBP-89ΔN antagonizes ZBP-89FL function and that over-expression of the truncated isoform disrupts gastrointestinal homeostasis.
INTRODUCTION
Alternative pre-mRNA splicing and multiple promoter usage are common mechanisms for increasing genetic complexity in humans (1) and mice (2). Genome-wide analyses indicate that the majority of human genes express alternative splice isoforms (3) and some variants contribute to neoplasia or other disease processes (4,5). For example, truncated isoforms of the p53 gene family, including p63 (6) and p73 (7–9), oppose the tumor suppressor activity of their full-length cognates and are over-expressed in tumors.
ZBP-89 (ZNF148; Zfp148; BFCOL1), a Krüppel-type zinc-finger protein (10), is both structurally (11) and functionally complex (12–14). It regulates diverse biological functions through direct promoter binding (10,14) and through multiple protein–protein interactions. It interacts directly with the tumor suppressor p53 through its zinc-finger domain (13) and indirectly with the histone acetyltransferase and transcriptional co-activator p300 through its amino terminus (12). ZBP-89 induces cell growth arrest through a p53-dependent mechanism (13) and apoptosis through a p53-independent mechanism (12). Mice haploinsufficient for ZBP-89 (Zfp148) are sterile owing to aberrant spermatogenesis (15). In addition, embryonic stem cells harboring a single ZBP-89 allele fail to exhibit p53 phosphorylation at Ser 15 (15).
ZBP-89 forms a complex with p300 during butyrate induction of p21waf1 in human colorectal cell lines (12). The amino terminus of ZBP-89 contains an acidic domain that is required for p300-mediated induction, since an expression construct lacking this domain (Δ amino acids 1–113) loses its ability to enhance butyrate induction of p21waf1 (12). The acidic domain of ZBP-89 is contained entirely within exon 4 of the human and mouse genes (11). Given the heterogeneous ZBP-89 mRNA expression pattern (10), the functional importance of the p300 interaction domain, and the fact that the ZBP-89 null mouse is likely an embryonic lethal (15), we generated a mouse homozygous for the ZBP-89ΔN isoform. Homozygous ΔNter mice experienced growth delay, reduced viability and increased susceptibility to Dextran sodium sulfate (DSS) induced colitis, suggesting that over-expression of the ΔNter isoform disrupts normal gastrointestinal homeostasis.
MATERIALS AND METHODS
Database deposition
National Center for Biotechnology Information (NCBI) reference sequences for ZBP-89 genomic loci are Homo sapiens chromosome 3 locus NC_000003 and BAC RP11-775J23 (AC108688); Mus musculus chromosome 16 contig NT_039624 and Pan troglodytes chromosome 3 genomic locus NW_104931. The human ZBP-89ΔN mRNA and exon 4B genomic sequences reported here are found in GenBank as entries DQ090088 and DQ090089, respectively. The P.troglodytes genomic sequence homologous to human exon 4B is deposited in Genbank DQ144540.
Organization of the human (ZNF148), chimp and mouse (Zfp148) ZBP-89 loci
A human genomic clone contig encompassing the ZBP-89 locus was previously described (14). Bacterial artificial chromosome (BAC) clones were used as templates to sequence intron/exon boundaries spanning the ZNF148 locus. Exon 4B was identified by gene prediction sequence analysis (16,17). Similarly, a BAC and bacteriophage lambda clone contig spanning the mouse Zfp148 locus was assembled and sequenced to determine the genomic organization of the Zfp148 locus. Primers Ptr-4B-F: 5′-TTCACCTCCCTGTCCTGTTC-3′ and Ptr-4B-R: 5′-TATCTGTCCCGTTTGCCTG-3′ were used to amplify and sequence chimp (P.troglodytes) genomic DNA containing exon 4B, after BLAST analysis (18) had revealed a sequence gap in this region. Chimp genomic DNA was obtained from the Integrated Primate Biomaterials and Information Resource through the Coriell Institute for Medical Research (Camden, NJ).
RT–PCR analysis
Total cellular RNA was isolated from cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA). Archived whole cell RNA samples from paired colon cancer and normal mucosal samples were also analyzed. Two-step RT–PCR was performed using SuperScript III RT (Invitrogen), however reverse transcription was performed at 55°C in order to obtain adequate yields of human exon 4B-containing cDNA. RT reactions were primed with random nonamers. Human primer sequences (5′ to 3′) were 4A-F: ATGAACATTGACGACAAACTGGAAG; 5-R: TTCCATATGATATTTTTGTATGAAT; 4B-F: TAGGGATGGTCAGCACTG; 6-R: GTTCTTTTGTGCCTTTCC. Mouse primer sequences (5′ to 3′) included Ex2-F: GCGGATAGAAGAGAAGAATCAGTGG; Ex4-F: CATTGACGACAAACTGGAAGG; Ex4-R: ACTTCGATCTTGAAGTACTGACTC; Ex5-R: CAGGAGAGCGTTGTTTCCG; Ex3/5fusion-F: CAGCCTCAGATAAGTGTA and Ex9-R: TTGTGGCATCTGGTGAAG. RT–PCR products were purified and subjected to DNA sequence analysis (University of Michigan DNA Sequencing Core).
5′-RACE of human exon 4B
The 5′ end of the exon 4B variant was identified using the GeneRacer™ system (Invitrogen, Carlsbad, CA), gene specific primer (5′ to 3′) 4B211-Rev: CAGTGCTGACCATCCCTATC CTACTTG and 2 µg of Jurkat whole cell RNA. Nested PCR products, generated with adaptor primers included with the GeneRacer™ system, were cloned using the TOPO-TA system (Invitrogen, Carlsbad, CA), and sequenced.
Targeting vector
We constructed vector pΔEx-4 to replace ZBP-89 exon 4 with a PGK-Neo cassette by homologous recombination. High fidelity long-range PCR was used to amplify targeting arms from mouse BAC clone pBmZBP-89, using primers (5′ to 3′): (Kpn)Int3-F, GATAGGTACCGCATTGGATGGCACAAGTGACTGAGAGG with (Xho)Int3-R, CTCGAGCCCGGGCTTAAGTATAACTGCCTAGAAAG for the left arm and (Apa)Int4-F, CTCGAGGGGCCCGTAAGTACTAAACTAGAAATG with (Apa)Int4-R, CTCGAGGGGCCCAAGAGCCTTGCTGACTCATAG for the right arm. The left targeting arm, encompassing exon 3 and intron 3, was 8 kb in length. The right targeting arm consisted of the proximal 2 kb of intron 4. The neomycin cassette, including a transcriptional stop signal, was isolated from pPNT (19), generously provided by Dr Richard Mulligan.
Generation of targeted ES cells and ΔNter mice
Electroporation of embryonic stem (ES) cells with the pΔEx-4 targeting vector and microinjection of blastocysts with targeted ES cells were performed by the University of Michigan Transgenic Animal Model Core (www.med.umich.edu/tamc). Genomic PCR was utilized to genotype targeted 129 Sv/J ES clones, chimeric founders, and progeny resulting after germline transmission. Primer sequences (5′ to 3′) were Int3-F1, GGAGTATTCTCTGTCCGTT ATG; Int4-326R, GCAAGAACTACACAGAGAAACCAC and R506Neo, TGAGGAA GAGGAGAACAGCG.
Two-dimensional western blot analysis
Whole cell protein extracts were prepared from mouse spleen using T-PER tissue protein extraction reagent (Pierce, Rockford, IL), supplemented with Complete Mini protease inhibitors (Roche, Indianapolis, IN) according the manufacturers' recommendations. Whole cell protein extracts from human Jurkat cells were similarly prepared using M-PER mammalian protein extraction reagent (Pierce, Rockford, IL). Isoelectric focusing (IEF) was performed with 7 cm ZOOM® Strip ph3-10L (linear) immobilized pH gradient gels (Invitrogen, Carlsbad, CA), as recommended by the manufacturer. The focused proteins were then separated on NuPAGE® 4-12% acrylamide gels (Invitrogen). Electroblot transfer to PVDF membrane and immunoblot procedures with ZBP-89 antiserum are as previously described (10,12).
Dextran sodium sulfate colitis
DSS colitis was induced in 6–9 month old ZBP-89ΔN and littermate control mice by the addition of 4% DSS to drinking water for a period of 5 days (20). Treated mice were returned to normal drinking water for 2 days prior to necropsy for histopathological scoring. Hematoxylin and eosin (H&E) stained colon sections were prepared by the Swiss roll method (21) and were scored for colitis index by a modification of a previously described method (22). Briefly, crypt damage was scored from 0 to 3 as none, basal only, moderate damage and complete erosion, respectively. Inflammation was scored from 0 to 3 as none, minor, moderate and severe leukocyte infiltration. Submucosal edema was scored from 0 to 3 as none, minor, moderate and severe, respectively. Similarly, hemorrhage was scored from 0 to 3 as none, minor, moderate and severe. Each parameter was multiplied by an extent factor (1–3), <10%, up to 25%, 25 to 50% and >50%, respectively. Samples with transmural involvement received an additional four points. Therefore, the maximum possible colitis index score was 40. Animals that died during treatment because of colitis injury were also given the maximum score. Individuals scoring the samples were blinded to the genotype.
Cell culture
The following cell lines were obtained from ATCC (Manassas, VA) and maintained on the recommended growth media: Human gastric epithelial cells Kato III and MKN45; human colon cancer cells Colo 320DM, CaCo-2 and HCT116; and human Jurkat leukemia cells. Cells were cultured in a humidified atmosphere of 5% CO2 and 95% air at 37°C. All culture media were supplemented with penicillin G (100 U/ml) and streptomycin (100 µg/ml).
RESULTS
Identification of human ZBP-89ΔN isoform
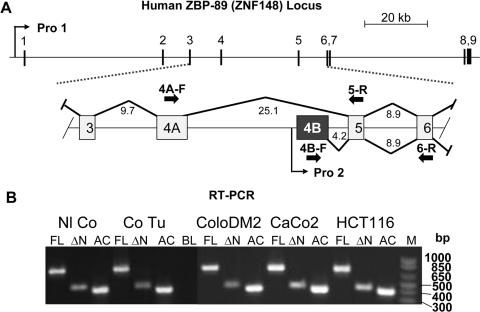
To determine if isoforms of ZBP-89 exist, we screened the human ZNF148 genomic locus in silico (16,17). After localizing candidate alternative exon 4B within 4 kb upstream of exon 5 (Figure 1A), we used RT–PCR analysis to show that exon 4B is expressed (Figure 1B). Exon 4B mRNA (ΔN) was co-expressed with exon 4A-containing message (FL) in colon cancer cells (ColoDM2, CaCo2, HCT116), and in primary tissue from normal colon and colon adenocarcinoma. Both forms also were abundantly expressed in Jurkat cells (data not shown). This suggested that at least two forms of ZBP-89 exist.
Figure 1.
Identification of a novel exon within the human ZBP-89 (ZNF148) locus. (A) The human ZBP-89 (ZNF148) locus spans 142 kb (upper panel) and encompasses three untranslated (1–3) and six coding (4–9) exons. A potential alternative exon 4B (shaded box) was identified at a position 4.2 kb upstream of exon 5. Arrows indicate the locations of forward (F) and reverse (R) RT–PCR primers used for expression analysis of FL ZBP-89 and the exon 4B variant. Selected intron sizes (kb) are indicated. (B) RT–PCR analysis of full-length (FL) and variant (ΔN) ZBP-89 mRNA expression, using actin (Ac) as control. The cDNAs were generated with cultured cell lines using primers 4A-F/5-R (726 bp product); primers 4B-F/6-R (432 bp product) and (Ac), Actin control primers (400 bp product). Whole cell RNA was isolated from ColoDM2, CaCo 2, and HCT116 human colon cell lines as well as paired normal colon and colon adenocarcinoma tissue. Full-length message is expressed from promoter 1 (Pro 1) and variant (ΔN) ZBP-89 mRNA is expressed from promoter 2 (Pro 2), as described in Figure 2.
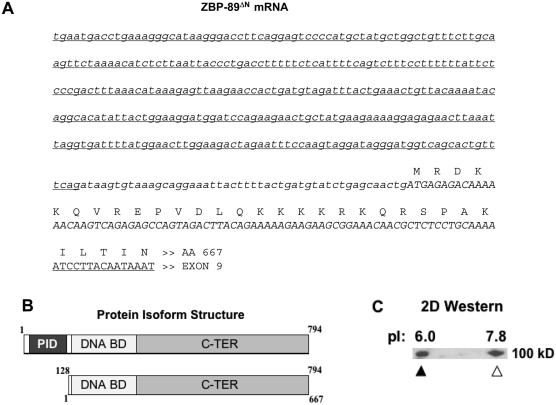
To better understand the function of the isoform, exon 4B-containing cDNA was sequenced. In addition to exon 4B, the variant mRNA included exons 5 and 6 (Figure 2A), as well as exons 7–9 (not shown). In contrast, RT–PCR with forward primers from exons 1-4A failed to generate products with exon 4B antisense primers, suggesting that an independent promoter regulates exon 4B expression. This was confirmed by 5′-rapid amplification of cDNA ends (RACE), which showed that transcription was initiated immediately upstream of exon 4B. The cDNA sequence showed that exon 4B was spliced to exon 5 resulting in an alternative reading frame relative to the cDNA encoded by exon 4A (Figure 2A). Exon 4B was 329 nt in length and composed of untranslated sequences when fused to exon 5. These data predicted that alternative promoter usage upstream of exon 4B resulted in the expression of an amino-terminally truncated ZBP-89 isoform, ZBP-89ΔN, with an alternative initiation codon corresponding to M128 of full-length mRNA (Figure 2B). This isoform lacks the acidic domain and p300-interaction region (12) found in full-length (ZBP-89FL) protein (10). Identical results were obtained with cDNA derived from esophagus, stomach, colon and Jurkat T-cells, suggesting that the exon 4B alternative promoter mechanism is common.
Figure 2.
Variant ZBP-89 transcript encodes a truncated protein, ZBP-89ΔN. (A) The 5′ end of exon 4B-variant cDNA, determined by 5′-RACE, is shown. Underlined italics, exon 4B-encoded; lower case italics, exon 5 untranslated (frameshift relative to ZBP-89FL cDNA); upper case italics, exon 5 coding sequence commencing at M128 relative to FL protein; upper case underlined, beginning of exon 6-encoded. The remainder of the cDNA and amino acid sequences are identical to the corresponding segments of FL cDNA and protein. (B) Comparison of FL (upper) and ΔN (lower) protein isoforms. The amino terminus of ZBP-89FL includes an acidic and p300-interaction domain (PID), which is absent in ZBP-89ΔN. The ZBP-89ΔN isoform retains the DNA binding and C-terminal domains. (C) 2D western blot analysis of Jurkat whole cell protein extract. ZBP-89FL (solid triangle) and ZBP-89ΔN (open triangle) peptides share an electrohoretic mobility of ∼100 kDa and are separated on the basis of their distinct isoelectric points (pI). The acidic end of the focusing gel is at the left.
Detection of human ZBP-89ΔN protein
We found that the electrophoretic mobility of ZBP-89ΔN protein, despite its shorter length, overlapped with the mobility of its ZBP-89FL cognate (Figure 2C). Both forms, isolated from Jurkat cells, migrated at 100 kDa on a 4–20% gradient gel. Similar SDS–PAGE anomalies have been reported with other proteins, including CTCF (23) and XPA (24). An alternative approach to separate the protein isoforms was suggested by comparing their predicted (25) isoelectric points (pI). Loss of the acidic domain in ZBP-89ΔN protein predicted a pI of 7.8, compared to 6.0 for ZBP-89FL. This difference could be revealed by two-dimensional (2D) gel electrophoresis, followed by western blot analysis (Figure 2C). ZBP-89 antiserum detected two protein species, with apparent electrophoretic mobilities of 100 kDa, but with a pI difference of ∼1.5, confirming that the more basic form is ZBP-89ΔN.
ZBP-89ΔN alternative promoter mechanism is restricted to hominids
Although the human (ZNF148) and mouse (Zfp148) ZBP-89 loci share many features of genomic organization (11), exon 4B is absent in mice and most other mammals (data not shown). In contrast, we identified a 133 bp segment of chimpanzee (P.troglodytes) chromosome 3 sequence with striking DNA sequence homology to human exon 4B, but there was a gap in the existing online genomic sequence. Sequencing across the gap after genomic DNA amplification, we found that the chimp shares 99% DNA sequence identity to the 329 bp human exon 4B. This suggested that the ZBP-89ΔN alternative promoter mechanism is restricted to hominids.
Generating a mouse model of ZBP-89ΔN expression
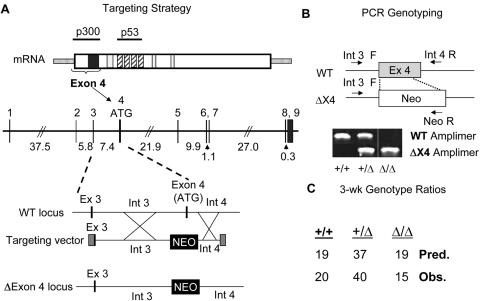
Since mice lack exon 4B, and therefore do not express a ZBP-89ΔN isoform, we targeted the mouse exon 4 domain by homologous recombination (Figure 3A). The left targeting arm contained exon 3 and intron 3, and the right targeting arm contained 1.8 kb of the 5′ margin of intron 4. Homologous recombination occurred in 4 of 1200 ES clones identified by PCR (Figure 3B). Of the four homologous recombinants, three demonstrated stable karyotypes and were therefore injected into blastocysts. Germline transmission of the exon 4-targeted locus resulted in two founder lines, 4E10 and 8C6, while a third founder, 9C6, failed to transmit the recombinant locus. Germline transmission was obtained first in the 4E10 lineage. Near-Mendelian ratios were observed at the age of weaning (3 weeks) in offspring of heterozygous intercrosses (Figure 3C). The data suggested that ∼20% of Δexon4 homozygous embryos die in utero or perinatally, however the difference between predicted and observed ΔExon4/ΔExon4 yields was not statistically significant (χ2-square analysis; P > 0.05). A similar pattern was observed in the 8C6 lineage. These results also suggest that the mutant locus was at least partially functional, since heterozygosity for a null ZBP-89 (Zfp148) allele results in failed germ cell development and therefore is not transmitted (15).
Figure 3.
Targeting ZBP-89 exon 4 in mice. (A) Replacement of exon 4 with a Neomycin resistance cassette (NEO) by homologous recombination in ES cells. Top panel: bracket shows location of features encoded by exon 4, including initiation codon, acidic domain (black rectangle) and p300 interaction domain. Open boxes, basic domains; hatched boxes, zinc-fingers; stippled region, untranslated. Middle panel; Zfp148 genomic locus. Intron sizes (kb) are shown. Lower panel; targeting strategy. (B) Tail biopsy DNA genotyping. WT (Int 3 F/Int 4 R) and Δexon4 (Int 3 F/Neo R) amplimers. (C) Near-Mendelian inheritance of ΔExon-4 alleles. The differences between predicted and observed values were not significant.
Alternative splicing of exon 3 to exon 5 in recombinant mice
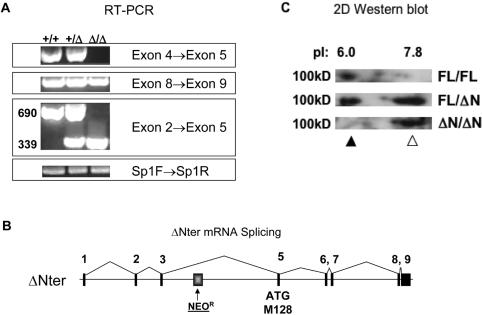
To determine the effect of exon 4 deletion on ZBP-89 expression, we analyzed cDNA derived from normal, heterozygous and exon 4-targeted animals (Figure 4A). Whereas exon 4 sequences were absent from recombinant mRNA, both upstream and downstream exons were expressed. RT–PCR with primers spanning exon 4 generated a 690 bp cDNA from wild type and a 339 bp cDNA from mutant alleles. The size difference between WT and targeted cDNA was 351 bp, the size of exon 4. This suggested that exon 3 was spliced directly to exon 5, excluding the intervening Neo cassette, and this was confirmed by DNA sequencing (Figure 4B). Deletion of exon 4 removed the initiation codon found in FL message and also resulted in an alternative reading frame. Gene feature sequence analysis (16,17) predicted that the alternative initiation codon corresponded to M128 of the FL protein (Figure 4B), analogous to the human ZBP-89ΔN variant (Figure 2B).
Figure 4.
ΔExon4 locus encodes mouse ZBP-89ΔN. (A) RT–PCR analysis was used to determine the variant mRNA structure in recombinant mice. Spleen whole-cell RNA was reverse-transcribed and amplified with primers within the indicated exons. RT–PCR of another ubiquitously expressed zinc-finger transcription factor, Sp1, was used for comparison and showed that the RNA samples were of uniform quality. Control experiments showed that PCR products were RT-dependent (data not shown), indicating that they were indeed derived from mRNA rather than possibly resulting from amplification of ZBP-89 related processed pseudogene sequences, such as ΨBERF1 (32). (B) DNA sequencing of recombinant cDNA showed that deletion of exon 4 resulted in direct splicing of exon 3 to exon 5, excluding the Neo cassette. The resulting reading frameshift predicts an alternative initiation codon corresponding to M128 of FL protein, similar to the naturally occurring human ZBP-89ΔN variant. (C) 2D immunoblot analysis of whole-cell spleen protein extracts from FL/FL, FL/ΔN and ΔN/ΔN mice, using conditions identical to those used for analysis of human samples (Figure 2C).
Mouse ZBP-89ΔN protein expression
2D western blot analysis (Figure 4C) showed that ZBP-89FL and ZBP-89ΔN proteins are expressed in an allele-dependent manner. The two forms have similar electrophoretic mobilities of 100 kDa, but are readily separated by their pI differences, similar to the human isoforms (Figure 2C). As mentioned above, similar electrophoretic anomalies have been observed with other protein isoforms (23,24). The immunoblot data also suggest that the ΔN protein isoform may be present at higher levels than the FL form in spleen tissue, however mRNA levels appear comparable, suggesting a possible difference in protein stability. Collectively, the mRNA and protein expression data confirmed that we knocked the ZBP-89ΔN variant into the mouse genome. Thus, the mouse model offers the advantage of assessing the biological effect of ZBP-89ΔN expression exclusively.
ZBP-89ΔN/ΔN mice experience growth delay and reduced viability
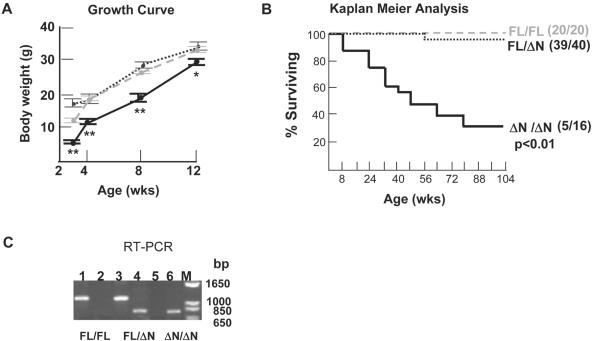
At weaning, by age 3 weeks, male ZBP-89ΔN/ΔN mice weighed an average of 5.9 ± 0.5 g (Figure 5A), 34% smaller than ZBP-89FL/FL (12.1 ± 0.7 g) and 49% smaller than ZBP-89FL/ΔN heterozygous littermates (15.1 ± 0.8 g). From ages 4–8 weeks, the size differential between ZBP-89ΔN/ΔN and control mice was diminished. A similar effect was observed with female mice (data not shown). As shown above (Figure 3C), ZBP-89ΔN/ΔN mice experienced 20% perinatal mortality as determined by genotype ratios at weaning. Reduced viability persisted and became statistically significant (Figure 5B), with 50% mortality at 48 weeks and 69% mortality by 104 weeks. The 2 year survival ratios were 20:20 ZBP-89FL/FL, 39:40 ZBP-89FL/ΔN and 5:16 ZBP-89ΔN/ΔN mice. Histopathological organ and tissue surveys revealed no obvious abnormalities in ZBP-89ΔN/ΔN mice, with the possible exception of the colon, where we noted a trend toward slightly increased lymphocytic infiltrates (data not shown). Semi-quantitative RT–PCR analysis of colon mRNA levels (Figure 5C), suggested that FL and ΔN forms were expressed in an allele-dependent manner. Collectively, these data demonstrated that balanced expression of ZBP-89FL and ZBP-89ΔN isoforms, as observed in heterozygous mice, supports normal growth and viability. In contrast, exclusive expression of the ZBP-89ΔN isoform in transgenic mice resulted in growth delay and reduced viability.
Figure 5.
Growth delay and decreased survival in ΔNter mice. (A) Growth curve for male ZBP-89FL/FL (gray dashed line, n = 20), ZBP-89FL/ΔN (black dotted line, n = 36), and ZBP-89ΔN/ΔN (black solid line, n = 16) offspring. A similar pattern was seen with female mice (not shown). ** P < 0.01; * P < 0.05. A trend toward lower body weights in ZBP-89ΔN/ΔN persisted after 12 weeks, but was no longer statistically significant (data not shown). (B) Kaplan–Meier analysis of survival interval to the onset of morbidity or death. (C) Semi-quantitative RT–PCR analysis of ZBP-89FL and ZBP-89ΔN mRNA levels in the colon.
ZBP-89ΔN confers DSS colitis susceptibility
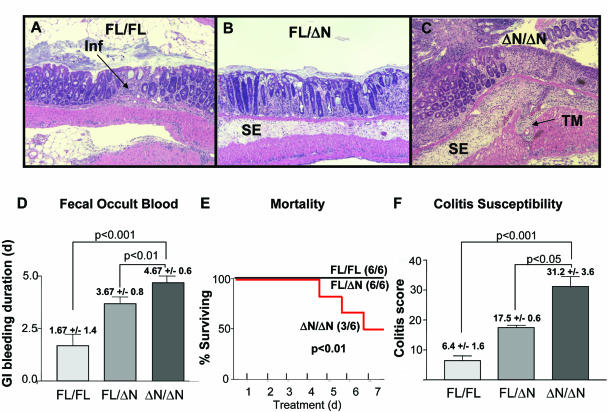
We previously showed that the amino terminal region of ZBP-89 (amino acids 1–111) is required to potentiate butyrate induction of p21waf1, suggesting that the amino terminal region mediates the butyrate-dependent anti-proliferative activities of ZBP-89 in the mammalian gastrointestinal tract (12). Butyrate suppresses colonic inflammation when induced by DSS (26). Since DSS accentuates a tendency to develop colitis, we challenged ZBP-89ΔN/ΔN mice acutely with 4% DSS for 5 days, as previously described (20). Histopathogical analysis (hematoxylin and eosin stained sections) demonstrated the presence of severe colitis that correlated with a ZBP-89ΔN gene dosage effect (Figure 6A–C). ZBP-89FL/FL mice (Figure 6A) had localized areas of lymphocytic infiltration and minimal submucosal edema. ZBP-89FL/ΔN mice (Figure 6B) exhibited more extensive infiltration, with displaced normal crypt architecture in addition to submucosal edema. ZBP-89ΔN/ΔN mice (Figure 6C) had areas with complete erosion of crypt architecture, grossly visible hemorrhage and extensive submucosal edema. The ZBP-89ΔN gene dosage effect also correlated with the duration of gastrointestinal bleeding (Figure 6D). Moreover, only ZBP-89ΔN/ΔN mice died during DSS treatment, with 50% mortality by the conclusion of the 7 day regimen (Figure 6E). Composite colitis scoring similarly paralleled the ZBP-89ΔN gene dosage (Figure 6F). These data suggest that increased expression of ZBP-89ΔN is associated with increased susceptibility to colitis.
Figure 6.
Increased susceptibility to DSS colitis in ZBP-89ΔN mice. (A–C) Representative H&E stains of normal (A), heterozygous (B) and ZBP-89ΔN/ΔN (C) mice. Inf = infiltrating lymphocytes; SE = submucosal edema; TM = transmural inflammation. (D) ΔNter expression accelerates onset of gastrointestinal bleeding, measured by fecal occult blood screening. (E) Mortality during DSS treatment: 50% of ZBP-89ΔN/ZBP-89ΔN mice die during 4% DSS treatment; all ZBP-89FL/ZBP-89FL and ZBP-89FL/ZBP-89ΔN mice survived (six in each group). (F) Colitis index scoring of DSS-treated mice, as described in Materials and Methods; six mice in each genotype cohort.
DISCUSSION
Through structural and functional analyses, we identified a human-specific ZBP-89 splice isoform, ZBP-89ΔN, which was generated by alternative promoter usage upstream of an alternative exon 4B. As a consequence, ZBP-89ΔN protein lacks a transcriptional domain (12) found in its full-length ZBP-89 cognate. This is the first characterization of a ZBP-89 isoform and is consistent with previous observations that gene complexity is lower in mice than in humans. As many as 59% of human genes are alternatively spliced (27), while the highest estimate to date for the mouse is 33% (28). Comparative genomic analysis suggested that the ZBP-89ΔN splicing mechanism has been conserved between humans and chimps, indicating that this specific mechanism of regulating ZBP-89 function is restricted to hominids.
Alternative mRNA expression increases genetic diversity through regulatory mechanisms that also are implicated in cancer (1,3–5,29) and other disease processes (30,31). The human ZBP-89ΔN variant described here renders the colonic mucosa more susceptible to injury. Our previous studies showed that the N-terminal domain is required for ZBP-89 to mediate butyrate-dependent activation of p21waf1 by cooperating with p300 (12). This result demonstrated that the N terminal domain is functionally important in vitro. Generating a mouse model in which only the truncated form was expressed revealed that loss of the p300-interacting domain results in delayed growth and a shortened lifespan. Although the overall effect on the health of the organism was impressive, an initial survey of several organs did not reveal obvious abnormalities, with the exception of the colon, which appeared to exhibit slightly more inflammatory infiltrates than the wild type mice. The propensity of the ZBP-89ΔN expressing mice to develop colitis was subsequently uncovered when the animals were challenged with DSS. Therefore, we concluded from these studies that expression of ZBP-89FL tends to protect against colitis. The gene dosage dependence of colitis susceptibility further suggests that the ZBP-89ΔN isoform functions as a dominant negative antagonist of ZBP-89FL. Similar antagonistic interactions have been reported between FL and ΔN isoforms within the p53 gene family, including ΔNp63 (29) and ΔNp73 (7–9).
The ZBP-89ΔN knock-in model reported here, along with an earlier Zfp148+/− model (15), help to elaborate how individual protein domains mediate the in vivo functions of ZBP-89. Mice heterozygous for a null ZBP-89 allele demonstrate complete failure of male germ cell development and are defective in p53-dependent embryogenesis (15). This finding suggests that the ability of ZBP-89 to interact with p53 is exquisitely sensitive to gene dosage. In contrast, homozygous ZBP-89ΔN expression is compatible with embryonic and postnatal survival, albeit at reduced levels. We previously showed that the DNA-binding, zinc-finger region of ZBP-89 mediates its interaction with p53 (13), and this domain is retained in the ZBP-89ΔN isoform. Therefore, the Nter domain, which includes the p300 interaction domain, is dispensable for p53-dependent embryonic development and postnatal survival, but is essential for normal gastrointestinal function.
Acknowledgments
The authors gratefully acknowledge the expertise of the University of Michigan Transgenic Animal Model Core, especially Thom Saunders, Linda Samuelson and Elizabeth Hughes. The authors also thank members of the DNA Sequencing Core and the Unit for Laboratory Animal Medicine at the University of Michigan, and Kathy McClinchey for assistance with histology. Proteomics data were provided by the Michigan Proteome Consortium (www.proteomeconsortium.org) which is supported in part by funds from the Michigan Life Sciences Corridor. Specifically, valuable assistance with 2D gel electrophoresis was provided by Jennifer Callahan. This research is supported in part by the National Institutes of Health through the University of Michigan's Cancer Center Support Grant (5 P30 CA46592) and the University of Michigan Gastrointestinal Peptide Research Center (DK-34533). Art Tessier and Gail Kelsey provided administrative support. Stacey Ehrenberg provided technical assistance. This work was supported by grants from the National Institutes of Health to D.J.L. (R21 DK65004-01) and J.L.M. (RO1-DK55732). Funding to pay the Open Access publication charges for this article was provided by NIH grant RO1-DK55732 (J.L.M.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Xu Q., Lee C. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 2003;31:5635–5643. doi: 10.1093/nar/gkg786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zavolan M., Kondo S., Schonbach C., Adachi J., Hume D.A., Hayashizaki Y., Gaasterland T. RIKEN GER Group and GSL Members. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 2003;13:1290–1300. doi: 10.1101/gr.1017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalnina Z., Zayakin P., Silina K., Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–357. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- 4.Hui L., Zhang X., Wu X., Lin Z., Wang Q., Li Y., Hu G. Identification of alternatively spliced mRNA variants related to cancers by genome-wide ESTs alignment. Oncogene. 2004;23:3013–3023. doi: 10.1038/sj.onc.1207362. [DOI] [PubMed] [Google Scholar]
- 5.Venables J.P. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 6.Wu G., Osada M., Guo Z., Fomenkov A., Begum S., Zhao M., Upadhyay S., Xing M., Wu F., Moon C., et al. DeltaNp63alpha up-regulates the HSP70 gene in human cancer. Cancer Res. 2005;65:758–766. [PubMed] [Google Scholar]
- 7.Hashimoto Y., Zhang C., Kawauchi J., Imoto I., Adachi M.T., Inazawa J., Amagasa T., Hai T., Kitajima S. An alternatively spliced isoform of transcriptional repressor ATF3 and its induction by stress stimuli. Nucleic Acids Res. 2002;30:2398–2406. doi: 10.1093/nar/30.11.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stiewe T., Zimmermann S., Frilling A., Esche H., Putzer B.M. Transactivation-deficient DeltaTA-p73 acts as an oncogene. Cancer Res. 2002;62:3598–3602. [PubMed] [Google Scholar]
- 9.Zaika A.I., Slade N., Erster S.H., Sansome C., Joseph T.W., Pearl M., Chalas E., Moll U.M. DeltaNp73, A dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J. Exp. Med. 2002;196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merchant J.L., Iyer G.R., Taylor B.R., Kitchen J.R., Mortensen E.R., Wang Z., Flintoft R.J., Michel J.B., Bassel-Duby R. ZBP-89, a Kruppel-like zinc finger protein, inhibits epidermal growth factor induction of the gastrin promoter. Mol. Cell. Biol. 1996;16:6644–6653. doi: 10.1128/mcb.16.12.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feo S., Antona V., Cammarata G., Cavaleri F., Passantino R., Rubino P., Giallongo A. Conserved structure and promoter sequence similarity in the mouse and human genes encoding the zinc finger factor BERF-1/BFCOL1/ZBP-89. Biochem. Biophys. Res. Commun. 2001;283:209–218. doi: 10.1006/bbrc.2001.4753. [DOI] [PubMed] [Google Scholar]
- 12.Bai L., Merchant J.L. Transcription factor ZBP-89 cooperates with histone acetyltransferase p300 during butyrate activation of p21waf1 transcription in human cells. Mol. Cell. Biol. 2000;21:4670–4683. doi: 10.1074/jbc.M004249200. [DOI] [PubMed] [Google Scholar]
- 13.Bai L., Merchant J.L. ZBP-89 promotes growth arrest through stabilization of p53. J. Biol. Chem. 2001;275:30725–30733. doi: 10.1128/MCB.21.14.4670-4683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Law D.J., Tarle S.A., Merchant J.L. The human ZBP-89 homolog, located at chromosome 3q21, represses gastrin gene expression. Mamm. Genome. 1998;9:165–167. doi: 10.1007/s003359900711. [DOI] [PubMed] [Google Scholar]
- 15.Takeuchi A., Mishina Y., Miyaishi O., Kojima E., Hasegawa T., Isobe K. Heterozygosity with respect to Zfp148 causes complete loss of fetal germ cells during mouse embryogenesis. Nature Genet. 2003;33:172–176. doi: 10.1038/ng1072. [DOI] [PubMed] [Google Scholar]
- 16.Kulp D., Haussler D., Reese M.G., Eeckman F.H. A generalized hidden Markov model for the recognition of human genes in DNA. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1996;4:134–142. [PubMed] [Google Scholar]
- 17.Reese M.G., Eeckman F.H., Kulp D., Haussler D. Improved splice site detection in Genie. J. Comput. Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 18.McGinnis S., Madden T.L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tybulewicz V.L., Crawford C.E., Jackson P.K., Bronson R.T., Mulligan R.C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 20.Myers K.J., Murthy S., Flanigan A., Witchell D.R., Butler M., Murray S., Siwkowski A., Goodfellow D., Madsen K., Baker B. Antisense oligonucleotide blockade of tumor necrosis factor-alpha in two murine models of colitis. J. Pharmacol. Exp. Ther. 2003;304:411–424. doi: 10.1124/jpet.102.040329. [DOI] [PubMed] [Google Scholar]
- 21.Mähler M., Bristol I.J., Leiter E.H., Workman A.E., Birkenmeier E.H., Elson C.O., Sundberg J.P. Differential susceptibility of inbred mouse strains to dextran sulfate sodium-induced colitis. Am. J. Physiol. 1998;274:G544–G551. doi: 10.1152/ajpgi.1998.274.3.G544. [DOI] [PubMed] [Google Scholar]
- 22.Rachmilewitz D., Karmeli F., Takabayashi K., Hayashi T., Leider-Trejo L., Lee J., Leoni L.M., Raz E. Immunostimulatory DNA ameliorates experimental and spontaneous murine colitis. Gastroenterology. 2002;122:1428–1441. doi: 10.1053/gast.2002.32994. [DOI] [PubMed] [Google Scholar]
- 23.Klenova E.M., Nicolas R.H., Sally U., Carne A.F., Lee R.E., Lobanenkov V.V., Goodwin G.H. Molecular weight abnormalities of the CTCF transcription factor: CTCF migrates aberrantly in SDS–PAGE and the size of the expressed protein is affected by the UTRs and sequences within the coding region of the CTCF gene. Nucleic Acids Res. 1997;25:466–474. doi: 10.1093/nar/25.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iakoucheva L.M., Kimzey A.L., Masselon C.D., Smith R.D., Dunker A.K., Ackerman E.J. Aberrant mobility phenomena of the DNA repair protein XPA. Protein Sci. 2001;10:1353–1362. doi: 10.1110/ps.ps.40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stothard P. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 26.Moreau N.M., Champ M.M., Goupry S.M., Le Bizec B.J., Krempf M., Nguyen P.G., Dumon H.J., Martin L.J. Resistant starch modulates in vivo colonic butyrate uptake and its oxidation in rats with dextran sulfate sodium-induced colitis. J. Nutr. 2004;134:493–500. doi: 10.1093/jn/134.3.493. [DOI] [PubMed] [Google Scholar]
- 27.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 28.Brett D., Pospisil H., Valcarcel J., Reich J., Bork P. Alternative splicing and genome complexity. Nature Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- 29.Brinkman B.M.N. Splice variants as cancer biomarkers. Clin. Biochem. 2004;37:584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 30.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 31.Woodley L., Valcarcel J. Regulation of alternative pre-mRNA splicing. Brief Funct. Genomics Proteomics. 2002;1:266–277. doi: 10.1093/bfgp/1.3.266. [DOI] [PubMed] [Google Scholar]
- 32.Antona V., Cammarata G., De Gregorio L., Dragani T.A., Giallongo A., Feo S. The gene encoding the transcriptional repressor BERF-1 maps to a region of conserved synteny on mouse chromosome 16 and human chromosome 3 and a related pseudogene maps to mouse chromosome 8. Cytogenet. Cell Genet. 1998;83:90–92. doi: 10.1159/000015138. [DOI] [PubMed] [Google Scholar]