Abstract
The incorporation of selenocysteine into proteins is directed by specific UGA codons and mRNA secondary structures, designated SECIS elements. In bacteria, these elements are positioned within the reading frame of selenoprotein mRNAs immediately downstream of the triplet coding for selenocysteine, and they tether a complex of the selenocysteine-specific elongation factor SelB, GTP and selenocysteyl-tRNASec to the site of UGA decoding. A SECIS-like structure was identified in the 5′ non-translated region of the selAB transcript, encoding selenocysteine synthase and SelB. It specifically binds to SelB and the formation of a SelB·GTP·selenocysteyl-tRNASec complex on the SECIS-like element represses expression of the downstream gene. This effect is abolished by mutations preventing formation of the complex. The regulatory pattern observed correlated with the levels of sel gene products. As quaternary complex formation on the SECIS-like element did not influence the transcription rate and only slightly reduced the level of selAB mRNA, it was concluded that the structure is involved in regulating translation initiation efficiency, thereby coupling selenocysteine biosynthesis to the availability of the trace element selenium.
Keywords: elongation factor/mRNA-binding protein/RNA secondary structure/selenocysteine/translation
Introduction
The amino acid selenocysteine (Sec) has been identified as a component of selected proteins in bacteria, archaea and eukarya (Low and Berry, 1996; Stadtman, 1996; Rother et al., 2001). It is encoded by an in-frame UGA codon interrupting the reading frame of the respective selenoprotein gene and is co-translationally inserted into the nascent polypeptide chain. In Escherichia coli, there are three genes coding for selenium-containing formate dehydrogenases, which are expressed constitutively (fdoG) or under anaerobic conditions only (fdhF and fdnG) (Zinoni et al., 1986; Berg et al., 1991; Sawers et al., 1991). The biosynthesis of selenocysteine and its insertion into these proteins require the function of at least four gene products (see Figure 10). selC codes for tRNASec, which is charged with l-serine by seryl-tRNA synthetase (SerS) and serves as the adapter at which the seryl moiety is converted into the selenocysteyl derivative. The conversion reaction is catalyzed by selenocysteine synthase (the selA gene product) with the aid of monoselenophosphate as the selenium donor substrate, which is synthesized from selenide and ATP by selenophosphate synthetase (the selD gene product). SelB, finally, is a specialized translation factor taking over the function of elongation factor (EF) Tu in selenocysteine insertion. Its N-terminal three domains are homologous to EF-Tu, but it additionally exhibits a C-terminal extension of 272 amino acids, designated domain IV (for reviews, see Hüttenhofer and Böck, 1998; Böck, 2001; Böck et al., 2002).
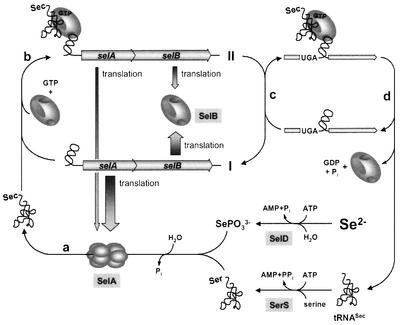
Fig. 10. Proposed model of the reactions involved in the regulation of selAB expression. Without the quaternary complex assembled at the SECIS-like element (state I), translation of selA and selB is derepressed. Biosynthesis of Sec-tRNASec (a), which is dependent on a sufficient supply of selenide, enables the formation of the regulatory complex on the 5′ mRNA structure of the selAB transcript (b), thereby decreasing the rate of translation of selA and, to a lesser extent, selB (state II). Competition between the SECIS-like element and the SECIS element of selenoprotein mRNAs (c) leads to the dissociation of the regulatory complex and to the formation of the SelB·GTP·Sec- tRNASec·SECIS complex, which catalyzes the decoding of UGA as selenocysteine. After A-site interaction and translocation of the ribosome (d), SelB and tRNASec are released and ready to re-enter the cycle.
SelB, in addition to interacting with guanosine nucleotides, binds two RNA ligands, namely Sec-tRNASec and the SECIS element of the mRNAs coding for selenoproteins. In bacteria, this mRNA structure is located immediately 3′ of the UGA codon that determines the position of selenocysteine incorporation in the nascent polypeptide chain. The binding order is random, but interaction with both RNA substrates leads to a stabilization of the resulting quaternary complex consisting of SelB, GTP, Sec-tRNASec and the SECIS element (Baron et al., 1993). Using rapid kinetic analysis, it was shown that the affinity of SelB for the SECIS element is maximized by Sec-tRNASec and decreases after its dissociation to allow the translation of codons 3′ of the UGA directing selenocysteine insertion (Thanbichler et al., 2000).
The interaction of SelB with the bacterial SECIS element has been additionally probed with a variety of techniques, like truncation of fdhF from the 3′ side (Zinoni et al., 1990), introduction of single or multiple mutations (Heider et al., 1992; Liu et al., 1998), gel retardation experiments (Baron et al., 1993; Kromayer et al., 1996), footprint analysis (Ringquist et al., 1994) and in vitro selection of RNA molecules (aptamers) that tightly bind to SelB (Klug et al., 1997, 1999). The main information gained and relevant for this communication was that: (i) the interaction involves a 17 kDa C-terminal domain of SelB (SelB472–614), designated domain IVb, and the apical part of the SECIS hairpin; (ii) the affinity of the interaction was lower between SelB and the SECIS element as compared with that between domain IVb and the SECIS element, but was equal when SelB was additionally complexed with Sec-tRNASec; (iii) finally, and intriguingly, some of the aptamers selected in vitro displayed excellent binding affinities but were not functional in the decoding process. Therefore, binding and function in the decoding process could be separated. In the present communication, we report the existence of a SECIS element present in the 5′ untranslated region of the mRNA for the bicistronic selAB operon. We show that this SECIS structure interacts with SelB and, in the presence of Sec-tRNASec, represses translation. By this novel mechanism, it adjusts the level of the gene products to the supply of the trace element selenium. Interestingly, this regulatory SECIS element is functional in the decoding process, although at reduced efficiency.
Results
The SECIS-like element
The structural genes coding for selenocysteine synthase (selA) and translation factor SelB (selB) in E.coli are organized in a bicistronic transcriptional unit and their reading frames overlap by four bases (Figure 1A). Previous analysis of the transcription of the selAB operon has shown that it is transcribed very weakly from a single start site at position –48 relative to the selA initiation codon (Sawers et al., 1991). When the 5′ non-translated RNA sequence was inspected for regulatory motifs, a putative RNA secondary structure was detected that strikingly resembled the SECIS element directing selenocysteine insertion (Figure 1B). Using mfold 3.1 (Mathews et al., 1999), its free energy was calculated to be –10.6 kcal/mol at 37°C. Since it is located outside the reading frame, it was designated SECIS-like. The putative minimal binding region of the SECIS-like element contains the essential structural and sequence elements that had been shown to be required for SECIS proper (Heider et al., 1992; Hüttenhofer et al., 1996; Liu et al., 1998, Li et al., 2000): the apical helix is closed by two G–C/C–G Watson–Crick base pairs and a preceding A–U/U–A base pair at the basal end and a C–G base pair at the distal end, and contains a bulged-out U residue at a position equivalent to +17 in the SECIS elements. The loop region contains the GU doublet that is supposed to make contact with the SECIS-binding pocket of SelB (Figure 1C).
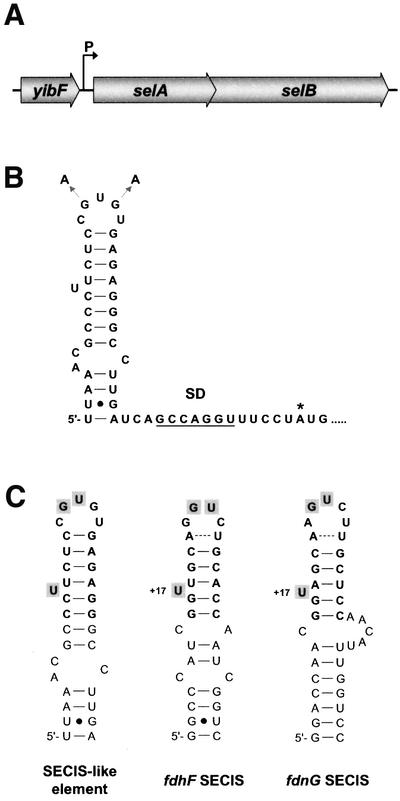
Fig. 1. (A) Organization of the selAB operon. (B) Fold of the stem–loop structure at the 5′ end of the selAB transcript as calculated by the mfold 3.1 program (Mathews et al., 1999). The transcrip tion initiation site as determined by Sawers et al. (1991), the Shine–Dalgarno motif (SD), the selA start codon (*) and the exchanges introduced to generate a mutant form of the SECIS-like element are indicated. (C) Comparison of the SECIS-like element with the SECIS structures of the fdhF and fdnG mRNAs. The minimal binding regions are depicted in bold. Bases supposed to make direct contact with SelB (Hüttenhofer et al., 1996) are shaded. The bulged-out U of the SECIS elements at position +17 relative to the selenocysteine-encoding UGA codon is indicated.
The SECIS-like structure is positioned at the immediate 5′ end of the mRNA and does not include the Shine–Dalgarno motif (Figure 1B).
The SECIS-like element binds SelB and forms a complex with SelB and Sec-tRNASec
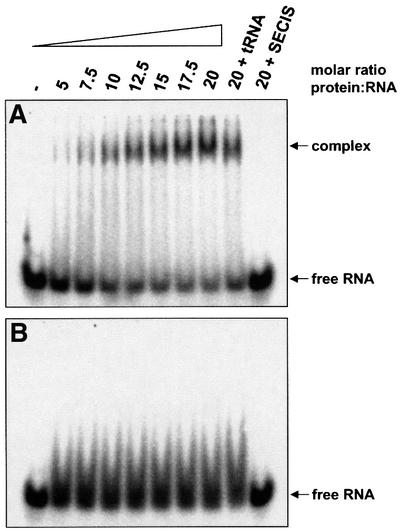
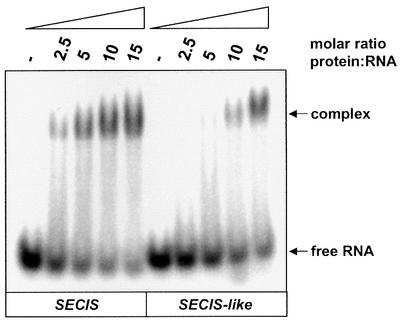
The striking similarities between the SECIS and the SECIS-like element prompted the analysis on whether the SECIS-like element is a binding target for translation factor SelB. A transcript was synthesized in vitro comprising the stem–loop structure at the 5′ end of the selAB mRNA (see Figure 1B). It was 32P-labeled at the 5′-terminus and used in gel retardation experiments (Figure 2). Figure 2A shows that the SECIS-like element yields a distinct shift signal at a 5-fold or higher molar excess of protein over the transcript despite the presence of a huge excess of unspecific competitor tRNA. The signal remains unchanged when additional competitor tRNA is added, but disappears completely if an equal amount of cognate SECIS RNA is included in the reactions. This indicates a specific interaction that involves the SECIS-binding site of SelB.
Fig. 2. Interaction of the SECIS-like element with wild-type SelB (A) and SelB472–614 (B). 32P-labeled SECIS-like element (80 nM) was incubated with increasing amounts of protein in the presence of 3.6 µM yeast tRNA. In the last two lanes, additional 3.6 µM unspecific (yeast tRNA) or specific (fdhF SECIS) competitor RNA was added to the reactions. After separation of the mixtures in a non-denaturing polyacrylamide gel, radioactivity was detected using a PhosphorImager.
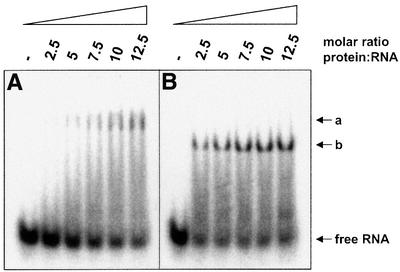
From previous experiments, it was known that a crucial requirement for SelB to function in the decoding of UGA is the capacity to undergo a sequence of conformational changes which ultimately result in the release of Sec-tRNASec at the ribosomal A-site and in the resolution of the SelB·SECIS complex (Thanbichler et al., 2000). These switches are reflected in the fact that SelB interacts with SECIS elements with a lower affinity and specificity, as does the C-terminal binding domain alone. To assess whether the SECIS-like element follows the same pattern, mobility shift experiments were conducted by probing the SECIS-like transcript with SelB472–614 (Figure 2B). Surprisingly, SelB472–614 is unable to form a stable complex with the SECIS-like element, whereas it tightly binds the fdnG (data not shown) and fdhF SECIS elements (Kromayer et al., 1996; Thanbichler et al., 2000). A weak interaction certainly takes place, as indicated by the tailing of the signal for the transcript, which is absent in the lane containing an excess of specific competitor RNA. There was no interaction detectable between the SECIS-like element and isolated SelB379–614, which comprises almost the entire domain IV of SelB (data not shown). These results are different from those obtained with the fdnG SECIS element, which binds to SelB379–614 with the same affinity as to full-length SelB (our unpublished results). The characteristics of the SECIS-like element are reminiscent of the binding behavior of some of the aptamers selected in vitro by Klug et al. (1997), which interact well with full-length SelB, but not with isolated SelB472–614. When Sec-tRNASec is included in the incubation mixture (Figure 3B), a strong retardation signal is already present at a 2.5-fold molar excess of protein, indicating a significant increase in the affinity of SelB for the SECIS-like element. Moreover, the mobility of the complex formed is enhanced, which is due to the additional negative charges introduced by binding of the tRNA. This resembles the binding behavior described previously for the fdhF SECIS element; namely, that a tighter and fast migrating complex is formed when both nucleic acid substrates are bound (Baron et al., 1993).
Fig. 3. Effect of Sec-tRNASec on complex formation between SelB and the SECIS-like element. 32P-labeled SECIS-like element (80 nM) was incubated with increasing concentrations of SelB in the presence of 3.6 µM yeast tRNA (A). To analyze the influence of Sec-tRNASec, 1.0 µM Sec-tRNASec and 100 µM GTP were additionally included in the reactions (B). GTP (100 µM) was added to the gel and electrophoresis buffer for separating the mixtures in both cases. Radioactivity was detected using a PhosphorImager. The positions of the ternary (a) and quaternary (b) complexes are indicated by arrows.
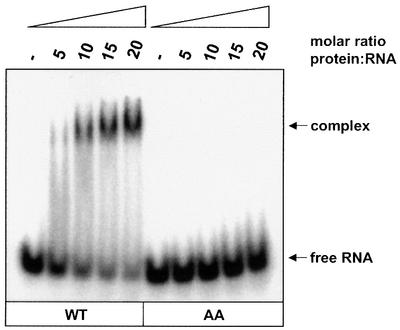
To test whether the interaction between SelB and the SECIS-like element is specific, we have introduced two base changes into the loop region of the hairpin (see Figure 1B). From previous experiments, it was known that the exchange of bases in the terminal loop of the fdhF SECIS element is detrimental to SelB binding and thus abolishes its function in the decoding process (Heider et al., 1992; M.Kromayer, unpublished results). Figure 4 indicates that this also holds for the binding capacity of the mutant SECIS-like element, as the ability to interact with SelB disappears.
Fig. 4. Specificity of the interaction between the SECIS-like element and SelB. 32P-labeled wild-type (WT) or mutant (AA) SECIS-like element (80 nM) (see Figure 1B) was mixed with increasing amounts of SelB in the presence of 3.6 µM yeast tRNA as unspecific competitor. Complexes formed were separated from unbound RNA by electrophoresis in a non-denaturing polyacrylamide gel and radioactivity was detected using a PhosphorImager.
To obtain qualitative information on the affinity of the interaction between SelB and the SECIS-like element, in vitro transcripts of the SECIS-like element and the fdnG SECIS element were titrated in parallel with increasing amounts of SelB. Complex formation, as analyzed by gel retardation, is depicted in Figure 5. It appears that the SECIS-like structure has an ∼3- to 4-fold lower affinity to the target protein. The KD value for the interaction of the fdnG SECIS with SelB has been determined to be 1.4 nM (Thanbichler et al., 2000). Therefore, the corresponding value for the binding of the SECIS-like element might be in the range of ∼10 nM.
Fig. 5. Affinity of the SECIS-like element for SelB. 32P-labeled fdnG SECIS or SECIS-like element (80 nM) was incubated with increasing amounts of SelB in the presence of 3.6 µM yeast tRNA as unspecific competitor. After separation in a non-denaturing polyacrylamide gel, unbound and complexed RNA was detected using a PhosphorImager.
The SECIS-like element functions in the regulation of selAB expression
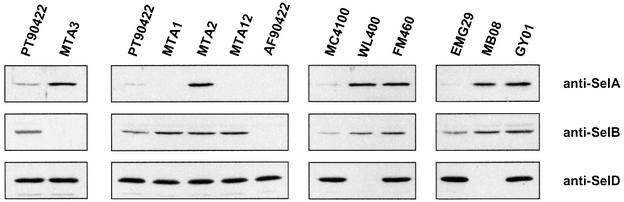
The characteristics of the SECIS-like element described above showed convincingly that it can form both a complex with SelB alone and a quaternary complex with SelB, Sec-tRNASec and presumably GTP, the latter interaction being significantly more stable. A speculation emanating from this is that the formation of the quaternary complex on the SECIS-like structure might affect the expression of the selAB mRNA. To follow this assumption, we have correlated the capacity of cells to form this complex with the amount of gene products for the sel genes. To this end, the wild-type strains PT90422, MC4100 and EMG29, and derivatives thereof carrying mutations in one or two of these genes, were grown to exponential phase, the cells were harvested and lysed, and the lysates analyzed for the quantity of sel gene products by immunoblotting (Figure 6). Intriguingly, a full correlation was detected: synthesis of the selA gene product was suppressed in a genetic background that renders the cells proficient for the formation of Sec-tRNASec and the concomitant production of SelB. In contrast, mutations in either selD, selC, selB or selA alleviate this suppression.
Fig. 6. Immunoblot analysis of the intracellular level of SelA, SelB and SelD in sel mutants derived from E.coli strains PT90422, MC4100 and EMG29. Cells of the strains PT90422 (wild type), MTA3 (ΔselB), MTA1 (ΔselA), MTA2 (ΔselC), MTA12 (ΔselA, ΔselC), AF90422 (ΔselAB), MC4100 (wild type), WL400 (ΔselD), FM460 (ΔselC), EMG29 (wild type), MB08 (selD165) and GY01 (selA160) were grown to an OD600 of 1.5, harvested and lysed in sample buffer. The lysates were applied to a 10% SDS–polyacrylamide gel. After separation of the proteins and transfer to a nylon membrane, immunoblot analysis was performed. Note that the point mutation carried by strain GY01 results in the synthesis of a stable but inactive form of SelA, whereas all other mutants do not synthesize stable products of the mutated genes.
The same regulatory pattern was observed for the selB gene product, although the effect was less pronounced. This correlates with previous results demonstrating that there is an internal translation initiation site upstream of the selB gene, which at least partly uncouples selB from selA mRNA translation and enables expression of selB when transcribed as a single gene under the control of the T7Φ10 promoter (Forchhammer et al., 1990). Physiologically, this is meaningful since SelB represents the key regulatory component, whose concentration should not fall below a critical level. The level of SelD remained constant in all strains tested and seems not to be regulated.
Analysis of regulation by the SECIS-like element employing lacZ reporter gene fusions
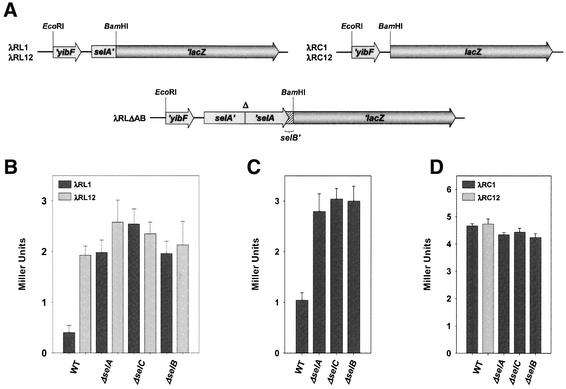
Since immunoblotting yields information on the steady-state level of gene products only and does not integrate the contribution of synthesis and degradation, the studies on the regulatory function of the SECIS-like element were extended by the use of lacZ reporter gene fusions. Both transcriptional and translational fusions were constructed (Figure 7A). To ensure the correct stoichiometry between regulatory factors and the mRNA, the fusion constructs were integrated into the λ phage and subsequently transferred in single copy to the λ attachment site of the chromosome. Assaying the level of β-galactosidase in cells lysogenized with λRL1, which carries the selA upstream region and a 5′ fragment of selA fused in-frame to lacZ, shows that the expression of the reporter gene is repressed in a wild-type genetic background, whereas it is increased ∼5-fold in strains with a mutation preventing formation of a functional quaternary complex (Figure 7B). The results obtained fit the regulation pattern as revealed by immunoblot analysis (see Figure 6) and indicate that binding of SelB·GTP·Sec-tRNASec to the SECIS-like element is crucial for the repression observed in the wild-type strain. This model is corroborated by the fact that only the derepressed level of β-galactosidase is measured in strains carrying λRL12, a phage whose insert is identical to that of λRL1 except for two mutations in the loop region of the SECIS-like element that have been shown to prevent binding of SelB (see Figure 4). The regulatory pattern of selB expression was followed using strains that were lysogenized with phage λRLΔAB carrying the selA upstream region, the selA open reading frame and a 5′ fragment of selB fused in-frame to lacZ. A deletion that has been shown not to have any polar effects on selB expression (data not shown) was introduced into the selA gene of the fusion construct to prevent complementation in a ΔselA genetic background. As depicted in Figure 7C, synthesis of β-galactosidase is repressed ∼3-fold in the wild type, whereas derepression occurs in strains deficient in quaternary complex formation. The effect is less distinct than the one measured for selA and again corresponds to the observations made by immunoblot analysis (Figure 6).
Fig. 7. Regulatory role of the SECIS-like element. (A) Reporter constructs employed to study regulation by the SECIS-like element. The restriction sites used to insert the respective fragment into plasmids pRS551 or pRS552 (Simons et al., 1987) are shown. The position of the deletion introduced into selA on the reporter construct harbored by phage λRLΔAB is indicated (Δ). λRL12 and λRC12 carry the mutated SECIS-like element (see Figure 1B), whereas all other phages contain the wild-type sequence. (B) β-galactosidase activities measured for strains PT90422 (WT), MTA1 (ΔselA), MTA2 (ΔselC) and MTA3 (ΔselB) after lysogenization with phages that carry selA′-′lacZ translational fusions encoding the wild-type (λRL1) or mutant (λRL12) SECIS-like element. (C) β-galactosidase activities determined for the same strains, when carrying the selAB′-′lacZ translational fusion harbored by λRLΔAB. (D) β-galactosidase activities exhibited by strains PT90422, MTA1, MTA2 and MTA3, when lysogenized with phages λRC1 and λRC12, which harbor transcriptional fusions encoding the wild-type and mutant SECIS-like element, respectively.
To ensure that the increase in β-galactosidase production that was measured in the mutant strains is a specific effect caused by the absence of a factor necessary to form the regulatory complex, complementation studies were performed. It was shown that the introduction of plasmids that lead to a moderate overproduction of SelA (with the SECIS-like element deleted from the upstream region of the plasmid-encoded selA gene) or tRNASec could completely override the derepression of reporter gene expression that was observed with strains MTA1::λRL1 (ΔselA) or MTA2::λRL1 (ΔselC), respectively, when they were carrying a control plasmid only (data not shown).
Effect on selAB mRNA synthesis and stability
There are several possible mechanisms that could underly the observed regulatory influence exerted by quaternary complex formation at the SECIS-like element, namely transcriptional attenuation, modulation of mRNA stability or regulation of translation initiation. Regulation of transcription by binding of proteins to their own message has emerged to be a general scheme for adjusting protein biosynthesis to the level of a gene product (Yanofsky, 2000). To analyze whether the SECIS-like element might mediate attenuation of selAB transcription, the level of β-galactosidase was assayed in strains lysogenized with phage λRC1, which carries the selA upstream region fused to the intact lacZ gene (Figure 7D). It appeared that the level of the reporter gene product was constant, independent of the genotype of the infected cells. The same specific β-galactosidase activity was obtained when the wild-type strain PT90422 was lysogenized with λRC12 instead of λRC1. This phage carries a similar construct with a mutated form of the SECIS-like element unable to interact with SelB (see Figure 4). These results clearly exclude a regulatory role for the SECIS-like element at the transcriptional level.
Next, it was investigated whether the stability of the selAB mRNA changes when the SECIS-like element is included in a quaternary complex. Many mRNAs have been shown to be protected against degradation by secondary structures positioned at their immediate 5′ ends, which are supposed to interfere with the initial endonucleolytic cleavage catalyzed by RNase E (Rauhut and Klug, 1999). Accordingly, binding of SelB·GTP· Sec-tRNASec might alter the conformation of the the SECIS-like element or the bordering region and thereby enhance the degradation of the selAB mRNA. To address this issue, the steady-state level and half-life of the selAB mRNA were analyzed using the S1 nuclease protection assay. Two labeled probes (Figure 8A) hybridizing with the 5′ and the 3′ end of the selA transcript, respectively, were synthesized and used to quantify the transcript levels. As depicted in Figure 8B, there was a reproducible, ∼1.5- to 2-fold increase in the steady-state amount of mRNA in the ΔselC mutant as compared with the wild-type strain. A similar effect was also observed in a ΔselA or ΔselB genetic background (data not shown). The ratios between the levels of the 3′ and the 5′ ends remained approximately constant at 60%. To investigate whether the half-life of the transcript changes under conditions of impeded quaternary complex formation, the decay profile of the selAB mRNA was determined for PT90422 (wild type) and MTA2 (ΔselC), which both have the wild-type operon. Figure 8C shows that the 5′ end of the transcript is rapidly degraded and reaches a basal level ∼6 min after inhibition of transcription. In the ΔselC genetic background, the mRNA is more stable (t1/2 = 67 s) than in the wild-type strain (t1/2 = 52 s), which explains the difference in the steady-state levels observed before. Very similar values were obtained with probe A2, which anneals to the 3′ region of selA (data not shown).
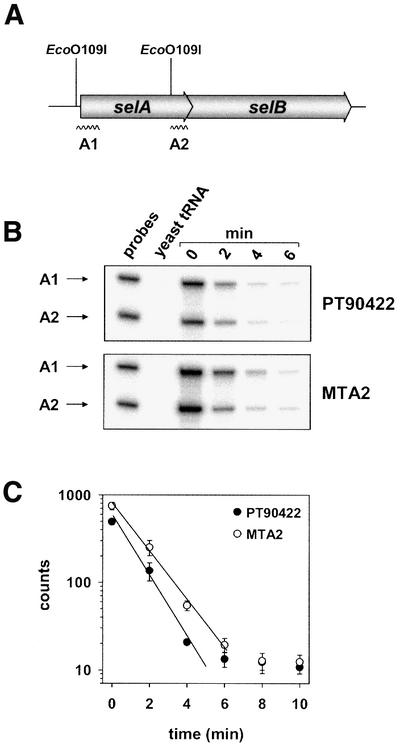
Fig. 8. Steady-state level and half-life of the selAB transcript. (A) Localization of probes A1 and A2 directed against the selA mRNA. The restriction sites used to generate the probes are indicated. (B) S1 nuclease protection assay. Seventy-five micrograms of yeast tRNA or total RNA that was extracted from cells of strains PT90422 (WT) and MTA2 (ΔselC) harvested before (0 min) or 2, 4 and 6 min after inhibition of transcription by addition of rifampicin were incubated with 10 000 c.p.m. of each probe. After treatment with S1 nuclease, the remaining probes were applied to an 8% urea–polyacrylamide gel, separated and visualized using a PhosphorImager. (C) Half-life of the selAB mRNA. The signals of probe A1 obtained in three independent experiments of the type described in Figure 8B were quantified using ImageQuant 2.1 (Molecular Dynamics), normalized to an internal standard and averaged. The straight lines fitted to the linear parts of the curves by regression analysis of the data correspond to half-lives of 67 s in strain MTA2 and 52 s in strain PT90422.
The SECIS-like element functions as a selenocysteine insertion element
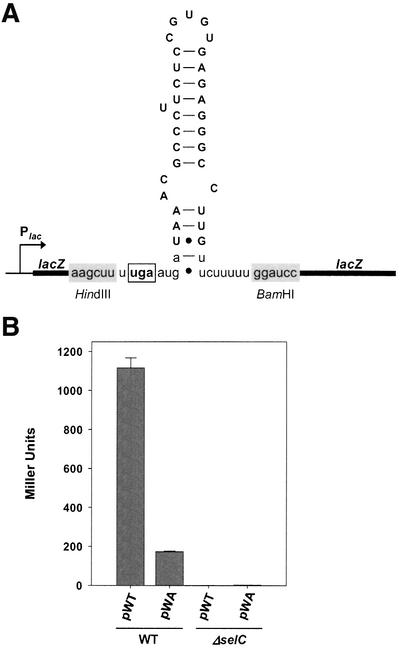
Having resolved the function of the SECIS-like structure as a novel type of regulatory element, it was tempting to assess whether it may still act as a genuine SECIS element, i.e. mediate the decoding of UGA with selenocysteine. To follow this possibility, a plasmid (pWA) was constructed that carries a lacZ gene with an in-frame UGA codon followed by the sequence encoding the SECIS-like element inserted into its 5′ region (Figure 9A). The distance between the UGA codon and the sequence of the SECIS-like element corresponding to the minimal binding region of SECIS elements was adjusted to 11 bp, which has been shown to be optimal for SelB activity (Heider et al., 1992; Chen et al., 1993; Liu et al., 1998). As a control, plasmid pWT was employed, which contains the fdhF SECIS in exactly the same position. The two plasmids were transferred into E.coli PT90422 (wild type) and MTA2 (ΔselC), and β-galactosidase activities were determined as a measure for read-through at the UGA codon (Figure 9B). The results clearly show that the SECIS-like element is able to act as an insertion sequence, but only at a reduced efficiency.
Fig. 9. Ability of the SECIS-like element to mediate SelB-dependent UGA read-through. (A) Schematic representation of the insert carried by plasmid pWA. The sequence of the insert replacing the fdhF SECIS cartridge of pWT is depicted as RNA in the predicted stem–loop conformation. Residues identical to those of the SECIS-like element are represented in upper case (B) β-galactosidase activities measured for strains PT90422 (WT) and MTA2 (ΔselC) transformed with pWT (fdhF SECIS) and pWA (SECIS-like element).
Discussion
Selenocysteine insertion sequences in bacterial selenoprotein mRNAs guide the ribosome to translate in-frame UGA codons with selenocysteine. They do so by the formation of a quaternary complex with translation factor SelB, Sec-tRNASec and GTP. Concomitant binding of the SECIS element and the charged tRNASec confers to SelB a conformation required to interact with the ribosome and to compete with release factor 2 (for a review, see Böck, 2001; Thanbichler and Böck, 2001). The ‘export’ of a SECIS element into the 5′ untranslated region of the mRNA and its effect on the expression of the selAB mRNA represent a novel mechanism of post-transcriptional control. The moderate affinity of the SECIS-like element for SelB alone and the increase in the tightness of the complex in the presence of Sec-tRNASec may be crucial for this function. In this way, the system measures the level of charging of tRNASec with selenocysteine and suppresses translation of selA, the gene for the main selenocysteine biosynthetic enzyme. What is the physiological need for this control? It may reside in the fact that selenocysteine is a highly toxic molecule because of the reactivitiy of its selenol group and that it should not be accumulated in the cell (even when attached to tRNA) beyond the level required for selenoprotein formation. Excess Sec-tRNASec might cause an increased intracellular level of free selenocysteine by spontaneous hydrolysis of the labile ester bond. It has been shown that selenocysteine can be recycled and serve as a selenium donor for the de novo synthesis of Sec-tRNASec (Lacourciere, 2002; Mihara et al., 2002). However, selenocysteine also readily infiltrates protein biosynthesis by substituting for cysteine (Müller et al., 1997), which in many cases leads to the inactivation of the affected enzyme due to the considerable difference in the redox potentials of the thiol and the selenol group. The fact that the expression of selA is regulated even though the level of the gene product is generally very low might argue for the toxicity of abundant selenocysteine formation. The regulatory function of the SECIS-like element also enables the cell to respond to the availability of the rare trace element selenium: at high supply the formation of the biosynthetic enzyme is suppressed and at limiting supply it is increased. Since no regulation has been detected for selD expression, this represents the only way to facilitate scavenging of the substrate under selenium-limiting conditions. Competition between the SECIS-like and the SECIS elements might additionally allow the cell to quickly adjust selenocysteine biosynthesis to sudden shifts in the intracellular concentration of selenoprotein mRNAs, as induced by abrupt changes between aerobic and anaerobic growth conditions. An increase in the cellular concentration of SECIS elements withdraws SelB·GTP·Sec-tRNASec from the regulatory structure and thereby induces the production of additional SelA molecules to meet the elevated need for selenocysteylated tRNASec. In contrast, reduced synthesis of selenoprotein mRNAs leads to a higher occupancy of the SECIS-like element and consequently to a decrease in the capacity to synthesize selenocysteine (Figure 10).
The finding that the SECIS-like element also functions as an ‘insertion sequence’ is intriguing, as its sequence strongly deviates from the SECIS consensus and exhibits exchanges that would cause a total loss of function when introduced into a genuine SECIS element as single mutations (Heider et al., 1992). By in vitro selection of aptamers able to bind SelB, it was possible to identify RNA molecules that showed an even higher affinity for SelB than the original SECIS element, although their sequences were only distantly related to that of the SECIS element (Klug et al., 1997). However, in contrast to the SECIS-like element, most of them were inactive in promoting UGA read-through at selenoprotein mRNAs. It was speculated that there exist multiple possibilities to fit RNA molecules into a binding site at the protein, although these molecules do not necessarily include all the features needed for biological activity. The SECIS-like element is a natural example for such a system: the overall binding properties are maintained, but the steric arrangement of determinants that are necessary to trigger the conformational change enabling SelB to interact with the ribosome is different, so that the ability to decode a UGA codon is drastically impaired. This reduction in function is reflected by the inability of the SECIS-like element to stably interact with domain IVb of SelB. Isolated domain IVb shows a very high stringency in the recognition of the SECIS element (Kromayer et al., 1996) and this may be the reason why the SECIS-like structure is partially excluded.
Although the finding that a translation factor complexed with a specific tRNA regulates gene expression is novel, the underlying mechanism bears some resemblance to the way translational control is achieved in the biosynthesis of threonyl-tRNA synthetase (Sacerdot et al., 1998) and several ribosomal proteins (reviewed by Zengel and Lindahl, 1994). These proteins repress translation by binding to the 5′ region of their own mRNA, which mimics the structure of the cognate RNA substrate, and thereby modulate the accessibility and quality of the translation initiation region (TIR). The exact mechanism enabling post-transcriptional control of selA and selB expression by the SECIS-like element in its complex with SelB and Sec-tRNASec is still open. Attenuation of transcription can be excluded as quaternary complex formation on the SECIS-like element showed no influence on the transcription rate. An ∼1.5-fold increase in the steady-state level of the selAB mRNA was observed in the absence of the functional complex, which can be explained by the elevated stability of the transcript observed under the same condition. A direct influence of quaternary complex formation at the SECIS-like element on mRNA stability is improbable as the capability to form the complex did not influence the synthesis of β-galactosidase in strains harboring a transcriptional fusion of the selA upstream region with lacZ. Rather, increased translation initiation rates in the absence of the complex might lead to a more frequent occupancy of the TIR and thereby to a more efficient masking of internal RNase target sites, as shown for several other systems (Baumeister et al., 1991; Yarchuk et al., 1991; Jain and Kleckner, 1993; Braun et al., 1998).
It is unlikely that this small influence on the mRNA level fully accounts for the 5-fold stimulation of LacZ production observed in strains carrying the selA′-′lacZ translational fusions. Therefore, the main function of quaternary complex formation on the SECIS-like element is presumably the regulation of translation initiation efficiency. In the presence of tRNAfMet, the ribosomal 30S subunit covers ∼54 bases around the initiation codon of an mRNA, ranging from position –35 to +19 (Hüttenhofer and Noller, 1994). The regulatory structure encompasses bases –48 to –16 of the selAB transcript and, therefore, overlaps with the binding site of the initiating ribosome. Either the stem–loop has to be melted completely to allow translation initiation to occur or it can be adopted by the ribosomal mRNA track, as shown for the mRNA structures mediating translational control of thrS expression (Sacerdot et al., 1998). In both situ ations, loading of the SECIS-like element with SelB·GTP·Sec-tRNASec might pose additional thermodynamic or steric constraints on the TIR–30S subunit interaction, leading to a decreased efficiency of initiation complex formation.
Strain MTA3, which lacks selB, did not show higher levels of β-galactosidase than strains with mutations in other sel genes, when lysogenized with λRL1. Consequently, association of the SECIS-like element with SelB alone does not seem to have any repressive effect on selA translation, which is reasonable as the structure is likely to be permanently occupied by SelB under the conditions prevalent in the cell. The distinct regulatory function of the quaternary complex can be explained by its increased stability, which might impede the displacement of SelB from its mRNA-binding site. However, it cannot be excluded that the formation of the translation initiation complex is sterically hindered by SelB and that this effect depends on the interaction of SelB with the bulky tRNA ligand or on a particular conformational state of SelB that is adopted in the quaternary complex only. Association of SelB·GTP·Sec-tRNASec with the SECIS-like element leads to the formation of a complex that is able to functionally interact with the ribosome and to decode UGA as selenocysteine. An interesting and unprecedented mechanism might, therefore, involve binding of a ribosome to the quaternary complex by A-site interaction, which may block translation initiation. Further experimentation, which is beyond the scope of this communication, is required to solve this issue.
Interestingly, there are some parallels between the SECIS-like element of the selAB mRNA and the SECIS element of eukaryotic cells. Both structures are located in an untranslated region and both of them are able to mediate the decoding of UGA as selenocysteine by functionally interacting with components of the selenocysteine incorporation machinery, when positioned in a functional context. Since SECIS elements are thought to be responsible for the hierarchy of selenoprotein formation in higher eukaryotes (Low et al., 2000), both structures additionally serve as regulatory elements that adjust the level of gene expression to the supply of the trace element selenium.
Materials and methods
Materials
[γ-32P]ATP (6000 Ci/mmol) was purchased from NEN Life Science Products (Boston, MA). Oligonucleotides were synthesized by MWG Biotech (Ebersberg, Germany). Chromosomal DNA from E.coli MC4100 and T7 RNA polymerase, purified according to the method of Wyatt et al. (1991), were a generous gift from S.Leonhartsberger. Nucleotides, tRNA from baker’s yeast and S1 nuclease were obtained from Roche Biochemicals (Mannheim, Germany). All other enzymes were from MBI Fermentas (St Leon-Roth, Germany) and New England Biolabs (Frankfurt, Germany).
Bacterial strains, phages, plasmids and standard procedures
The bacterial strains, phages and plasmids used in this study are listed in Table I. Generally, E.coli DH5α (F–, gyrA96, recA1, relA1, endA1, thi-1, hsdR17, glnV44, deoR, Δ(lacZYA-argF)U169, [Φ80dΔ(lacZ)M15]) (Woodcock et al., 1989) was used for the propagation of plasmids. Standard procedures were performed as described by Ausubel et al. (1997) and Sambrook et al. (1989). Owing to length limitations, details on the construction of plasmids, strains and recombinant phages are given in the Supplementary data available at The EMBO Journal Online.
Table I. Relevant bacterial strains, phages and plasmids used in this study.
| Genotype | Reference | |
|---|---|---|
| Strains |
|
|
| PT90422 | F–, araD139, Δ(argF-lac)U169, ptsF25, deoC1, relA1, flbB5301, rpsE+, strA1 | P.Tormay, unpublished |
| MTA1 | PT90422, ΔselA | This study |
| MTA2 | PT90422, ΔselC | This study |
| MTA3 | PT90422, ΔselB | This study |
| MTA12 | PT90422, ΔselA, ΔselC | This study |
| AF90422 | PT90422, Δ(selAB) | A.Friebel, unpublished |
| MC4100 | F–, araD193, Δ(argF-lac)U169, ptsF25, relA1, deoC1, flbB5301, rpsL150, λ– | Casadaban and Cohen (1979) |
| FM460 | MC4100, ΔselC400::neo | Sawers et al. (1991) |
| WL400 | MC4100, ΔselD204::cat | Leinfelder et al. (1990) |
| EMG29 (NCIB 10241) | F–, proC23, trp-30, his-51, lac-28, rpsL101, λ+ | NCIB |
| GY01 | EMG29, selA160 | Mandrand-Berthelot et al. (1978) |
| MB08 |
EMG29, selD165 |
Graham (1980) |
| Phages |
|
|
| λRS45 | lac′ZYA, imm21, ind+ | Simons et al. (1987) |
| λRL | λRS45, KnR, lac′ZYA | This study |
| λRL1 | λRS45, KnR, Φ(selA′-′lacZ), lacYA | This study |
| λRL12 | λRS45, KnR, Φ(selA′-′lacZ), lacYA, mutant SECIS-like element | This study |
| λRLΔAB | λRS45, KnR, Φ(selAΔ(490–936)B′-′lacZ), lacYA | This study |
| λRC | λRS45, KnR, lacZYA | This study |
| λRC1 | λRS45, KnR, selA upstream region, lacZYA | This study |
| λRC12 |
λRS45, KnR, selA upstream region, lacZYA, mutant SECIS-like element |
This study |
| Plasmids |
|
|
| pWT | ColE1 ori, ApR, (lacZ-fdhF)42, lacYA | Heider et al. (1992) |
| pWA | pWT derivative, SECIS-like element instead of fdhF SECIS | This study |
| pUAB | ColE1 ori, ApR, selAB | This study |
Growth conditions and β-galactosidase activity assay
To measure the efficiency of UGA read-through, the test strains, carrying either pWT or pWA, were grown aerobically in TP medium (Heider et al., 1992) at 37°C. Strains lysogenized with λRS45 derivatives were cultivated aerobically at 37°C in TP2 medium, which contains 1% peptone, 0.6% glucose, 100 mM potassium phosphate pH 7.0, 1 mM MgCl2, 0.1 mM CaCl2, 1 µM sodium selenite, and trace elements (Neidhardt et al., 1974). When reaching the mid-exponential phase (OD600 ∼ 1.5), the cells were chilled on ice and used for the determination of β-galactosidase activities as described by Miller (1972). The results of at least three independent measurements, which were each performed in triplicate, were averaged and corrected for the background values, which were measured with strain PT90422 lysogenized with λRL or λRC, respectively.
Immunoblot analysis
Cells were grown aerobically in TP2 medium to the mid-exponential phase, harvested by centrifugation, resuspended in SDS sample buffer (Ausubel et al., 1997) to an OD600 of 10 and lysed by incubation for 15 min at 95°C. The proteins were separated in a 10% SDS–polyacrylamide gel (Laemmli, 1970), transferred to a nitrocellulose membrane (BioTrace NT; Pall Corporation, Ann Arbor, MI) and probed with rabbit antisera raised against SelA, SelB or SelD. Immunocomplexes were detected using horseradish peroxidase–protein A conjugate (Bio-Rad Laboratories, Munich, Germany) and the ECL system from Roche Biochemicals (Penzberg, Germany).
Purification of proteins
Wild-type SelB from E.coli was purified as described previously (Thanbichler et al., 2000). SelB472–614 fused with an N-terminal His6 affinity peptide was prepared according to Kromayer et al. (1996).
Purification and aminoacylation of tRNASec
tRNASec was purified according to the method of Leinfelder et al. (1990). Aminoacylation of tRNASec with serine and conversion of the seryl into a selenocysteyl residue were carried out as described previously (Thanbichler et al., 2000).
Mobility shift assay
The fdnG stem–loop was prepared as described previously (Thanbichler et al., 2000). The fdhF stem–loop and the SECIS-like elements were prepared by in vitro transcription (Milligan et al., 1987; Wyatt et al., 1991) using oligo T7-top (5′-TAATACGACTCACTATAG-3′) and oligos fdhF-sl (5′-GACCGATTGGTGCAGACCTGCAACCGATGG GCCTATAGTGAGTCGTATTA-3′), SLE-wt (5′-TCAAGGCCCTCT CACACGGAGAAGGGCGTTTAACATAACCTATAGTGAGTCGTA TTA-3′) or SLE-aa (5′-TCAAGGCCCTCTCATATGGAGAAGG GCGTTTAACATAACCTATAGTGAGTCGTATTA-3′) to assemble the partially double-stranded templates. Quantification of the transcripts was performed as described elsewhere (Thanbichler et al., 2000). The transcripts were purified, folded and assayed for their interaction with SelB as described previously (Thanbichler and Böck, 2002). Radioactivity was detected by exposure of the dried gels on a phosphor screen (Molecular Dynamics, Sunnyvale, CA). After scanning the screen with a PhosphorImager (Storm 860 Laser Scanner; Molecular Dynamics), the signals were visualized using ImageQuant 2.1 (Molecular Dynamics).
S1 nuclease protection assay
To determine the steady-state concentration and half-life of the selAB mRNA, cells were grown in TP2 medium at 37°C under aerobic conditions. At an OD600 of 1.0, a 10 ml sample was withdrawn, immediately mixed with 1.25 ml of ice-cold 5% phenol (in ethanol) and stored on ice. Subsequently, rifampicin (240 µg/ml) was added to the cultures and additional samples were withdrawn at regular intervals. The cells were pelleted by centrifugation at 4°C and frozen in liquid nitrogen. Total RNA was extracted by the acid hot phenol method (Aiba et al., 1981). Oligos S1-selA1 (5′-GCTCGCCCAAGGTTGGTATGCAGCAC-3′) and S1-selA2 (5′-CGACCAATCACCGGCACTGGCAATTC-3′) were labeled at the 5′ end using [γ-32P]ATP and T4 polynucleotide kinase. Probes annealing with the 5′ and 3′ end of the selA transcript, respectively, were synthesized by PCR in reactions containing either of the two labeled oligos and EcoO109I-treated plasmid pUAB as template and gel purified. RNA was co-precipitated with 10 000 c.p.m. of each probe, washed with 80% ethanol and dissolved in 20 µl of hybridization buffer (80% formamide, 0.5 M NaCl, 1 mM EDTA, 40 mM PIPES pH 7.0). The solution was heated for 5 min at 95°C, immediately transferred to a 58°C water bath and incubated for 12 h. After addition of 200 µl of S1 digestion buffer (300 mM NaCl, 2 mM ZnSO4, 30 mM sodium acetate pH 5.5, 1 U/µl S1 nuclease) and incubation for 45 min at 37°C, the reaction was stopped by addition of 5.5 µl of 10% SDS and 2 µl of 10 mg/ml yeast tRNA. The nucleic acids were precipitated, dissolved in formamide loading dye and separated on an 8% urea–polyacrylamide gel. The signals were detected as described above.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank A.Malamoussi for her assistance in constructing the reporter gene fusions, and A.Resch and M.McMurray for helpful discussions. This work was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
References
- Aiba H., Adhya,S. and de Crombrugghe,B. (1981) Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem., 256, 11905–11910. [PubMed] [Google Scholar]
- Ausubel F., Brent,R., Kingston,R., Moore,D., Seidmann,J., Smith,J. and Struhl,K. (eds) (1997) Current Protocols in Molecular Biology. J.Wiley & Sons, New York, NY.
- Baron C., Heider,J. and Böck,A. (1993) Interaction of translation factor SELB with the formate dehydrogenase H selenopolypeptide mRNA. Proc. Natl Acad. Sci. USA, 90, 4181–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister R., Flache,P., Melefors,O., von Gabain,A. and Hillen,W. (1991) Lack of a 5′ non-coding region in Tn1721 encoded tetR mRNA is associated with a low efficiency of translation and a short half-life in Escherichia coli. Nucleic Acids Res., 19, 4595–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg B.L., Li,J., Heider,J. and Stewart,V. (1991) Nitrate-inducible formate dehydrogenase in Escherichia coli K-12. J. Biol. Chem., 266, 22380–22385. [PubMed] [Google Scholar]
- Böck A. (2001) Selenium metabolism in bacteria. In Hatfield,D.L. (ed.), Selenium: Its Molecular Biology and Role in Human Health. Kluwer Academic, Dordrecht, The Netherlands, pp. 7–22.
- Böck A., Thanbichler,M., Rother,M. and Resch,A. (2002) Selenocysteine. In Ibba,M., Francklyn,C. and Cusack,S. (eds), Aminoacyl-tRNA Synthetases. Landes Bioscience, Georgetown, TX.
- Braun B., Le Derout,J. and Régnier,P. (1998) Ribosomes inhibit an RNase E cleavage which induces the decay of the rpsO mRNA of Escherichia coli. EMBO J., 17, 4790–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M.J. and Cohen,S.N. (1979) Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl Acad. Sci. USA, 76, 4530–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.T., Fang,L. and Inouye,M. (1993) Effect of the relative position of the UGA codon to the unique secondary structure in the fdhF mRNA on its decoding by selenocysteinyl tRNA in Escherichia coli. J. Biol. Chem., 268, 23128–23131. [PubMed] [Google Scholar]
- Forchhammer K., Rücknagel,K. and Böck,A. (1990) Purification and biochemical characterization of SELB, a translation factor involved in selenoprotein synthesis. J. Biol. Chem., 265, 9346–9350. [PubMed] [Google Scholar]
- Graham A. (1980) The synthesis of formate dehydrogenase and nitrate reductase proteins in various fdh and chl mutants of Escherichia coli. FEMS Microbiol. Lett., 7, 145–151. [Google Scholar]
- Heider J., Baron,C. and Böck,A. (1992) Coding from the distance: dissection of the mRNA determinants required for the incorporation of selenocysteine into proteins. EMBO J., 11, 3759–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A. and Böck,A. (1998) RNA structures involved in selenoprotein synthesis. In Grunberg-Manago,M. and Simons,R.W. (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 603–639.
- Hüttenhofer A. and Noller,H. (1994) Footprinting mRNA–ribosome complexes with chemical probes. EMBO J., 13, 3892–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A., Westhof,E. and Böck,A. (1996) Solution structure of mRNA hairpins promoting selenocysteine incorporation in Escherichia coli and their base-specific interaction with special elongation factor SELB. RNA, 2, 354–366. [PMC free article] [PubMed] [Google Scholar]
- Jain C. and Kleckner,N. (1993) S10 mRNA stability and steady state levels in Escherichia coli: indirect effects of translation and role of rne function. Mol. Microbiol., 9, 233–247. [DOI] [PubMed] [Google Scholar]
- Klug S.J., Hüttenhofer,A., Kromayer,M. and Famulok,M. (1997) In vitro and in vivo characterization of novel mRNA motifs that bind special elongation factor SelB. Proc. Natl Acad. Sci. USA, 94, 6676–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug S.J., Hüttenhofer,A. and Famulok,M. (1999) In vitro selection of RNA aptamers that bind special elongation factor SelB, a protein with multiple RNA-binding sites, reveals one major interaction domain at the carboxyl terminus. RNA, 5, 1180–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kromayer M., Wilting,R., Tormay,P. and Böck,A. (1996) Domain structure of the prokaryotic selenocysteine-specific elongation factor SelB. J. Mol. Biol., 262, 413–420. [DOI] [PubMed] [Google Scholar]
- Lacourciere G. (2002) Selenium is mobilized in vivo from free selenocysteine and is incorporated specifically into formate dehydrogenase H and tRNA nucleosides. J. Bacteriol., 184, 1940–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Leinfelder W., Forchhammer,K., Veprek,B., Zehelein,E. and Böck,A. (1990) In vitro synthesis of selenocysteinyl-tRNAUCA from seryl-tRNAUCA: involvement and characterization of the selD gene product. Proc. Natl Acad. Sci. USA, 87, 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Reches,M. and Engelberg-Kulka,H. (2000) The bulged nucleotide in the Escherichia coli minimal selenocysteine insertion sequence participates in interaction with SelB: a genetic approach. J. Bacteriol., 182, 6302–6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Reches,M., Groisman,I. and Engelberg-Kulka,H. (1998) The nature of the minimal ‘selenocysteine insertion sequence’ (SECIS) in Escherichia coli. Nucleic Acids Res., 26, 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low S. and Berry,M. (1996) Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem. Sci., 21, 203–208. [PubMed] [Google Scholar]
- Low S.C., Grundner-Culemann,E., Harney,J.W. and Berry,M.J. (2000) SECIS–SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J., 19, 6882–6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrand-Berthelot M., Wee,M.Y.K. and Haddock,B.A. (1978) An improved method for the identification and characterization of mutants of Escherichia coli deficient in formate dehydrogenase activity. FEMS Microbiol. Lett., 4, 37–40. [Google Scholar]
- Mathews D., Sabina,J., Zuker,M. and Turner,D. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- Mihara H., Kato,S., Lacourciere,G., Stadtman,T., Kennedy,R., Kurihara,T., Tokumoto,U., Takahashi,Y. and Esaki,N. (2002) The iscS gene is essential for the biosynthesis of 2-selenouridine in tRNA and the selenocysteine-containing formate dehydrogenase H. Proc. Natl Acad. Sci. USA, 99, 6679–6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S., Heider,J. and Böck,A. (1997) The path of unspecific incorporation of selenium in Escherichia coli. Arch. Microbiol., 168, 421–427. [DOI] [PubMed] [Google Scholar]
- Neidhardt F., Bloch,P. and Smith,D. (1974) Culture medium for enterobacteria. J. Bacteriol., 119, 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut R. and Klug,G. (1999) mRNA degradation in bacteria. FEMS Microbiol. Rev., 23, 353–370. [DOI] [PubMed] [Google Scholar]
- Ringquist S., Schneider,D., Gibson,T., Baron,C., Böck,A. and Gold,L. (1994) Recognition of the mRNA selenocysteine insertion sequence by the specialized translational elongation factor SELB. Genes Dev., 8, 376–385. [DOI] [PubMed] [Google Scholar]
- Rother M., Resch,A., Wilting,R. and Böck,A. (2001) Selenoprotein synthesis in archaea. Biofactors, 14, 75–83. [DOI] [PubMed] [Google Scholar]
- Sacerdot C., Caillet,J., Graffe,M., Eyermann,F., Ehresmann,B., Ehresmann,C., Springer,M. and Romby,P. (1998) The Escherichia coli threonyl-tRNA synthetase gene contains a split ribosomal binding site interrupted by a hairpin structure that is essential for autoregulation. Mol. Microbiol., 29, 1077–1090. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E. and Maniatis,T. (eds) (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sawers G., Heider,J., Zehelein,E. and Böck,A. (1991) Expression and operon structure of the sel genes of Escherichia coli and identification of a third selenium-containing formate dehydrogenase isoenzyme. J. Bacteriol., 173, 4983–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R., Houman,F. and Kleckner,N. (1987) Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene, 53, 85–96. [DOI] [PubMed] [Google Scholar]
- Stadtman T. (1996) Selenocysteine. Annu. Rev. Biochem., 65, 83–100. [DOI] [PubMed] [Google Scholar]
- Thanbichler M. and Böck,A. (2001) Functional analysis of prokaryotic SELB proteins. Biofactors, 14, 53–59. [DOI] [PubMed] [Google Scholar]
- Thanbichler M. and Böck,A. (2002) Selenoprotein biosynthesis: purification and assay of components involved in selenocysteine biosynthesis and insertion in Escherichia coli. Methods Enzymol., 347, 3–16. [DOI] [PubMed] [Google Scholar]
- Thanbichler M., Böck,A. and Goody,R.S. (2000) Kinetics of the interaction of translation factor SelB from Escherichia coli with guanosine nucleotides and selenocysteine insertion sequence RNA. J. Biol. Chem., 275, 20458–20466. [DOI] [PubMed] [Google Scholar]
- Woodcock D., Crowther,P., Doherty,J., Jefferson,S., DeCruz,E., Noyer-Weidner,M., Smith,S., Michael,M. and Graham,M. (1989) Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res., 17, 3469–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt J.R., Chastain,M. and Puglisi,J.D. (1991) Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques, 11, 764–769. [PubMed] [Google Scholar]
- Yanofsky C. (2000) Transcription attenuation: once viewed as a novel regulatory strategy. J. Bacteriol., 182, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarchuk O., Iost,I. and Dreyfus,M. (1991) The relation between translation and mRNA degradation in the lacZ gene. Biochimie, 73, 1533–1541. [DOI] [PubMed] [Google Scholar]
- Zengel J. and Lindahl,L. (1994) Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol., 47, 331–370. [DOI] [PubMed] [Google Scholar]
- Zinoni F., Birkmann,A., Stadtman,T.C. and Böck,A. (1986) Nucleotide sequence and expression of the selenocysteine-containing polypeptide of formate dehyrogenase (formate-hydrogen-lyase-linked) from Escherichia coli. Proc. Natl Acad. Sci. USA, 83, 4650–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoni F., Heider,J. and Böck,A. (1990) Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc. Natl Acad. Sci. USA, 87, 4660–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]