Abstract
We report here the crystal structure of the minimal ligand-binding segment of the Staphylococcus aureus MSCRAMM, clumping factor A. This fibrinogen-binding segment contains two similarly folded domains. The fold observed is a new variant of the immunoglobulin motif that we have called DE-variant or the DEv-IgG fold. This subgroup includes the ligand-binding domain of the collagen-binding S.aureus MSCRAMM CNA, and many other structures previously classified as jelly rolls. Structure predictions suggest that the four fibrinogen-binding S.aureus MSCRAMMs identified so far would also contain the same DEv-IgG fold. A systematic docking search using the C-terminal region of the fibrinogen γ-chain as a probe suggested that a hydrophobic pocket formed between the two DEv-IgG domains of the clumping factor as the ligand-binding site. Mutagenic substitution of residues Tyr256, Pro336, Tyr338 and Lys389 in the clumping factor, which are proposed to contact the terminal residues 408AGDV411 of the γ-chain, resulted in proteins with no or markedly reduced affinity for fibrinogen.
Keywords: adhesins/clumping factor A/crystal structure/immunoglobulin fold/Staphylococcus aureus
Introduction
Fibrinogen (Fg) is a 340 kDa glycoprotein that is found at a concentration of ∼3 mg/ml in blood plasma and plays key roles in hemostasis and coagulation (Herrick et al., 1999). Fg is composed of six polypeptide chains, two Aα, two Bβ and two γ-chains, arranged in a symmetrical dimeric structure. The C-terminal residues of the γ-chain represent a biologically important part of Fg that interacts with the platelet integrin αIIβ3 during Fg-dependent platelet adherence and aggregation (Kloczewiak et al., 1984). This region of the γ-chain is also targeted by the pathogenic bacterium Staphylococcus aureus, resulting in Fg-dependent cell clumping and tissue adherence (Hawiger et al., 1982a,b). Clumping factor A (ClfA) was the first Fg γ-chain-binding S.aureus adhesin identified, and later the fibronectin-binding proteins A and B (FnbpA and B) of the bacterium were recognized as bi-functional proteins and found to bind the same C-terminal peptide segment in the γ-chain of Fg (Wann et al., 2000). Detailed characterization of the binding of these adhesins, which belonged to the family of MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) (Patti and Höök, 1994; Höök and Foster, 2000), to Fg have indicated that the C-terminal residues Ala408-Gly-Asp-Val411 of the γ-chain are critical in these interactions (Strong et al., 1982; McDevitt et al., 1994, 1997; Wann et al., 2000).
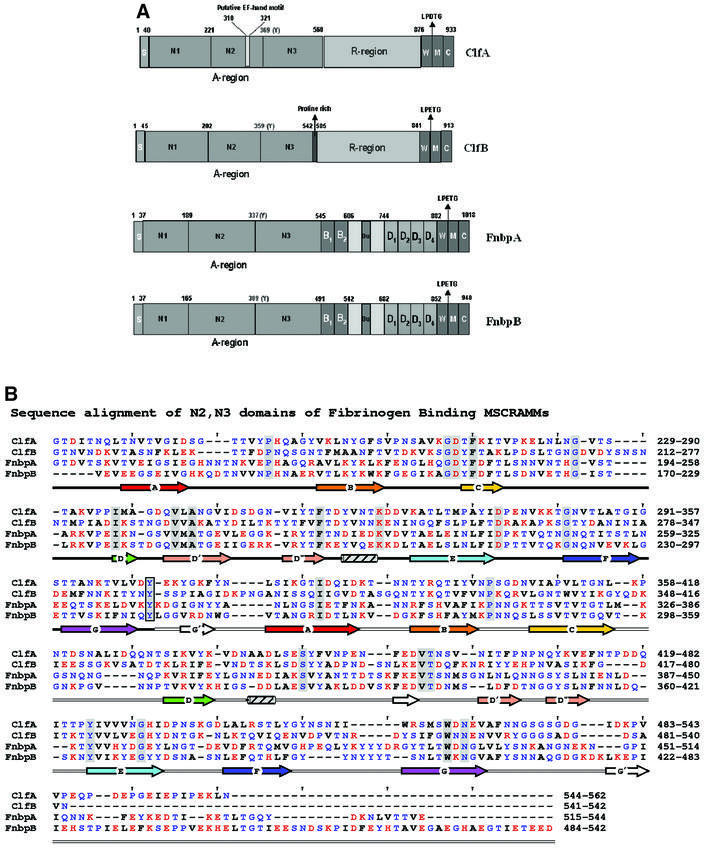
ClfA and the Fnbps have structural features that are common to other cell wall-anchored proteins expressed by Gram-positive bacteria, including ClfB, another S.aureus Fg-binding MSCRAMM that binds specifically to the α-chain (Figure 1A) (Patti and Höök, 1994; Nì Eidhin et al., 1998). These include an N-terminal signal sequence (S) and C-terminal features that are required for sorting the proteins to the cell wall [a proline-rich wall-spanning region (W), the wall-anchoring LPTXG motif, a hydrophobic transmembrane region (M) and a cytoplasmic tail of positively charged amino acid residues (C)]. ClfA and ClfB also contain a Ser-Asp repeat region (R region) in the C-terminal part of the protein, whereas the Fnbps contain C-terminal repeats (D repeats) that bind to fibronectin (Wann et al., 2000). The Fg-binding activity of these MSCRAMMs has been localized to the N-terminal A regions that are ∼500 amino acid residues long (Figure 1A) (McDevitt et al., 1995; Nì Eidhin et al., 1998; Wann et al., 2000). In the case of ClfA, the Fg-binding site has been further localized to residues 221–559. Furthermore, substitution of Glu526 and Val527 within the minimum Fg-binding truncate of ClfA [rClfA(221–559)] with Ala and Ser, respectively, abrogated the Fg-binding activity of this protein (Hartford et al., 2001).
Fig. 1. The Fg-binding MSCRAMMs of S.aureus: ClfA, ClfB, FnbpA and FnbpB. (A) The four Fg-binding MSCRAMMs of S.aureus identified so far have a common structural organization including a signal peptide(s) followed by the N-terminal ligand binding A region in which three subdomains, N1, N2 and N3, can be identified. The boundary between N2 and N3 is indicated by a conserved Tyr residue. At the C-terminus, the cell wall-binding region (W), the membrane-spanning domains (M) and the charged C-terminus (C) are present. ClfA has a unique putative EF-hand in the ligand binding A region and a ‘DS’ dipeptide repeat R region. ClfB is similar to ClfA and contains an additional proline-rich segment linking the A- and the R- regions. FnbpA and FnbpB contain the unique fibronectin-binding D repeats and B repeats of unknown function. (B) Sequence alignment of the N-terminal two-thirds of rClfA(221–559) with corresponding regions of ClfB, FnbpA and FnbpB. Red, blue and black letters represent charged, polar and hydrophobic residues, respectively. Identical residues are shaded and a conserved Tyr residue in the connector between the N2 and N3 domains is boxed. The secondary structural elements are colored in rainbow fashion similar to Figures 2, 4 and 5.
Analogous to αIIβ3 (Smith et al., 1994), the Fg-binding activity of ClfA is affected by the divalent cations Ca2+ and Mn2+. High concentrations of both cations inhibit Fg-dependent bacterial clumping and the binding of rClfA to isolated Fg. A putative cation-binding EF-hand-like motif (residues 310–321) has been proposed within the A region of ClfA (Figure 1A) (O’Connell et al., 1998).
In a previous study, we reported the crystallization of recombinant fragments of the ligand-binding A regions of ClfA and ClfB (Deivanayagam et al., 1999). These fragments, encompassing residues 221–559 and 199–542 of rClfA and rClfB, respectively, arose from the N-terminal degradation of the full-length Escherichia coli-expressed A regions. Subsequently, it was reported that the N-terminal region (residues 45–220) of native S.aureus-expressed ClfB was cleaved by the S.aureus metalloprotease aureolysin, generating small peptides that could not be detected by SDS–PAGE (McAleese et al., 2001).
In the present study, we report the crystal structure of the proteolytically stable minimum Fg-binding truncate of ClfA, rClfA(221–559) (Figure 1A). This protein consists of two domains of a new variant of the immunoglobulin (IgG) fold, which we called the DE-variant (DEv) IgG fold. Furthermore, using a combination of molecular modeling and site-directed mutagenesis, we tentatively localize the binding site in rClfA(221–559) for the C-terminal residues (Ala408-Gly-Asp-Val411) of the Fg γ-chain.
Results
Overall structure of rClfA(221–559)
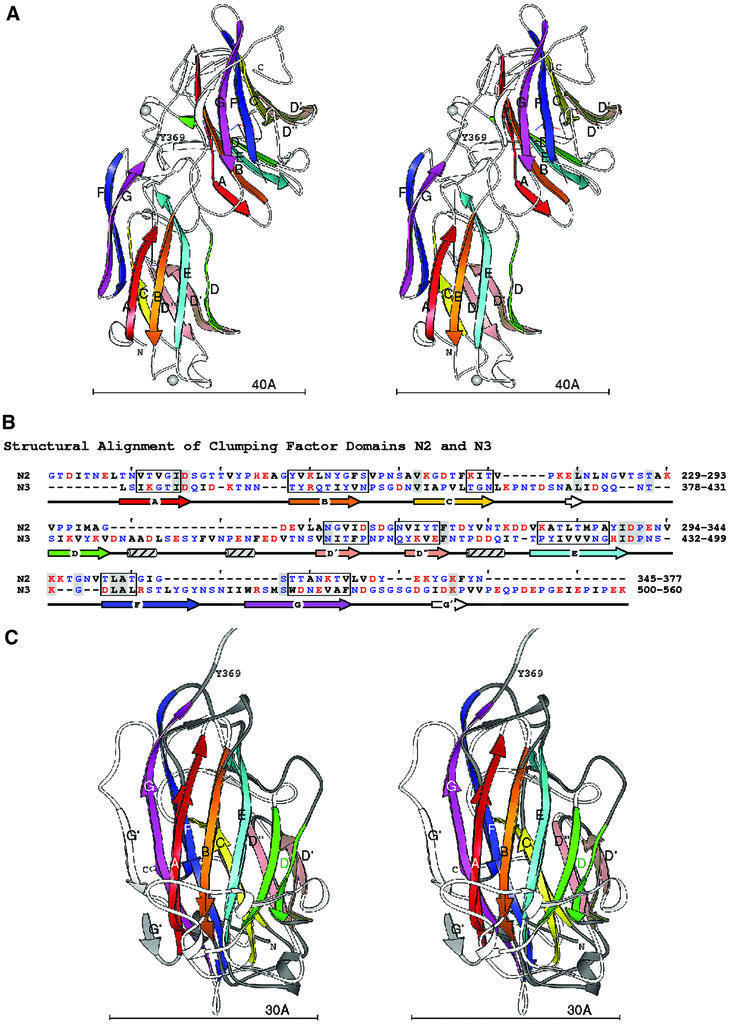
The structure of rClfA(221–559) is composed of two compact domains that we have named N2 and N3, respectively, each being dominated by anti-parallel β-strands (Figure 2A). The term N1 was assigned to the protease-sensitive N-terminal segment corresponding to residues 45–220 of the ClfA A region. The new N-terminal N2 domain contains a single-turn α-helix and two 310 helices, while the N3 domain contains three 310 helices. N2 represents the smaller domain, being composed of 140 residues (229–369), whereas the N3 domain encompasses 189 residues (370–559). No electron density was observed for the 20 N-terminal residues, which include 12 residues contributed by the vector His6 tag sequence and residues 221–228 of the rClfA(221–559) protein. Likewise, no electron density was observed for the two C-terminal residues, which originated from the expression vector. In addition, residues Gly532-Ser-Gly-Ser-Gly-Asp-Gly-Ile539 in the N3 domain did not have interpretable electron densities and were thus modeled into fragments of residual density over several cycles of refinement.
Fig. 2. Domain structure of rClfA(221–559). (A) Stereo ribbon diagram of the ClfA crystal structure. In the two independent domains, the β-strands A–G are colored in rainbow fashion. The N- and C-termini and the boundary between the domains indicated by Y369 are also labeled. Each domain has a coordinate axis defined by the relative orientation of the sheets. The long, black axis is the average sheet direction. The silver spheres represent the metal ions. (B) Structure-based sequence alignment of the ClfA N2 and N3 domains. The strands A–G are marked with arrows colored as in the ribbon diagrams. Ranges of residues where all Cα superpose within 1.5 Å are boxed. Hydrophobic residues are black, polar are blue and charged are red. Identical residues are highlighted with gray. (C) Superposition of the ClfA N2 and N3 domains. Stereo ribbon diagrams of the two domains are overlaid. β-strands A–G are labeled and colored as above. The coil regions of the first domain are dark gray, the coil linking the two domains is light gray and the coils of the second domain are shown in white. The average sheet direction is aligned with the y-axis, and the average direction between adjacent strands with the x-axis.
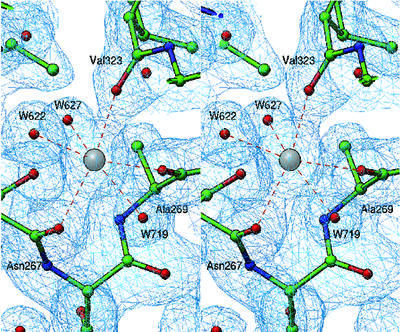
Although metal ions were not added to the buffers and precipitants used in the crystallization protocol, three metal ions (M1, M2 and M3) with octahedral coordination geometry are present in the rClfA(221–559) crystal structure (Figure 2A). The residues of the putative EF-hand-like motif (residues 310–321), which was previously identified within the ClfA primary structure (O’Connell et al., 1998), are not involved in coordinating any of these metal ions. Furthermore, the secondary structure of this region is composed of a β-turn followed by a β-strand and does not resemble the classical EF-hand motif (Kretsinger, 1996). The metal ions M1, M2 and M3 are principally coordinated through main-chain carbonyl oxygen atoms, with the exception of M3, which interacts with one side-chain carboxylate (Figure 3). Such metal ion coordination contrasts with that of other metal-binding motifs, where most of the coordination occurs via the side chains, specifically through the carboxylates (Kretsinger, 1996; Springer et al., 2000). In macromolecular crystal structures, the coordination number can vary from four to seven for Ca2+ and from four to six for Mg2+. The coordination number of six observed for M1, M2 and M3 in the rClfA(221–559) structure suggests that these metal ions might represent Mg2+. However, the average bond length of 2.37 Å observed at these coordination sites is suggestive of Ca2+ rather than Mg2+, where the average bond length would be expected to be 2.1 Å. The B-factors of these proteins are slightly higher than normal and indicate the possibility of a partial occupancy. Thus, the exact identity of the metal ions (Ca2+ or Mg2+) in the rClfA(221–559) structure could not be resolved from the refinement parameters.
Fig. 3. Structure of metal binding site M1. A representative 2fo – fc electron density map (rendered from a normalized map at 1.0σ level) displays octahedral coordination of the metal binding site M1, where three main-chain carbonyl oxygen atoms from Asn267, Ala269 and Val323 contribute together with three water molecules (W622, W627 and W719) to the coordination geometry.
The novel DEv-IgG fold
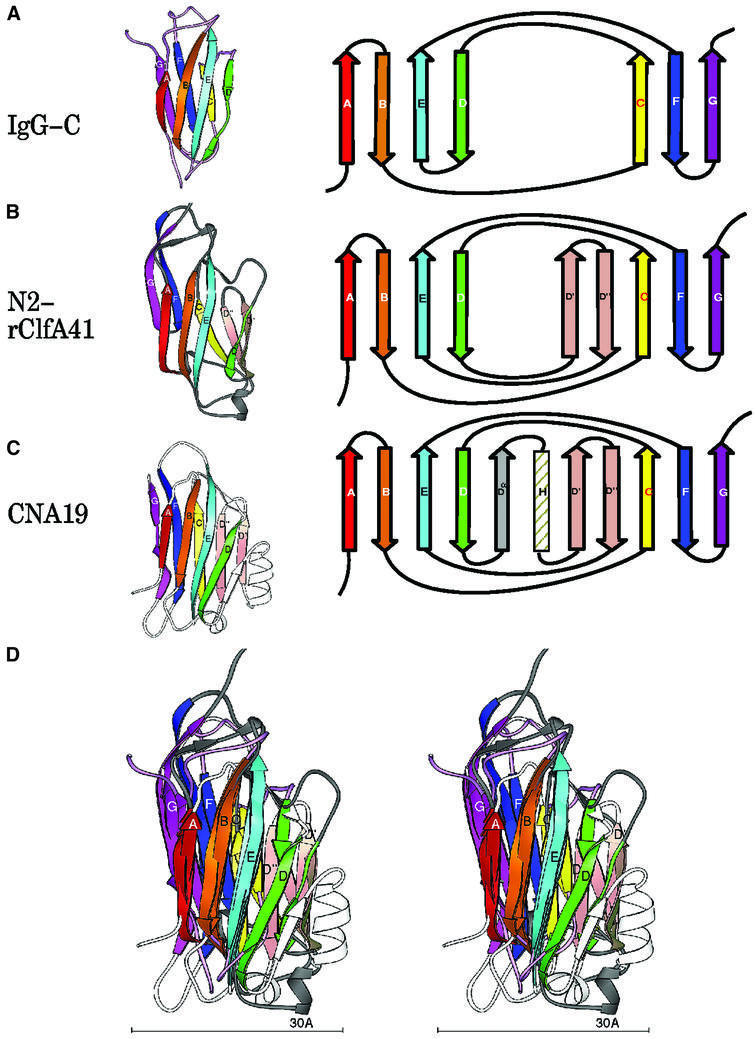
Primary sequence alignment of the N2 and N3 domains revealed 13% amino acid identity and 36% amino acid similarity over their entire lengths (Figure 2B). The secondary structural elements of these domains (boxed residues in Figure 2B) superpose with a root mean square deviation (r.m.s.d.) (on Cα) of 0.97 Å, highlighting the near identity of their structural motifs (Figure 2C). The topology of the N2 and N3 domains resembles that of the jelly roll barrel, yet differs in that the characteristic anti-parallel N- and C-terminal β-strands of the jelly roll fold (Branden and Tooze, 1999) are absent and are replaced by parallel β-strands. In this respect, the topology of the N2 and N3 domains more closely resembles that of the IgG fold (Figure 4A). In fact, we propose that the N2 and N3 structures represent a new variant of this fold (D-variant or DEv-type) that differs from the previously described V-, C-, H- and I-type folds of the immunoglobulin superfamily (Bork et al., 1994; Harpaz and Chothia, 1994). In the constant IgG fold (C-type), strands ABED and GFC are arranged at the front and back sides, respectively, of the fold (Figure 4A), whereas in its variants (V-, H- and I-type), additional structural elements are found between strands C and D. The structure of the N2 and N3 domains of rClfA(221–559) follows the topological arrangement of the IgG fold from strands A through D, with the prominent variation occurring between strands D and E, where two additional β-strands (D′ and D′′) are observed in each of the two domains in rClfA(221–559) (Figure 4B). Resuming from strand E through G, the topology of N2 and N3 again conforms to the IgG fold. Most of the variable region observed between strands D and E is on one side of the fold (on the right-hand side in Figure 4B), and does not perturb the core region of the IgG fold.
Fig. 4. Topology of the IgG and MSCRAMM domains. Ribbon diagrams of the labeled strands A–G colored in rainbow fashion as in Figure 2. The corresponding topology diagram of the structure is shown to the right. (A) IgG-C domain. (B) ClfA N2 domain [rClfA(221–559)]. (C) CNA19 domain (CNA169–318). (D) Superposition of IgG, rClfA(221–559)-N2 and CNA169–318.
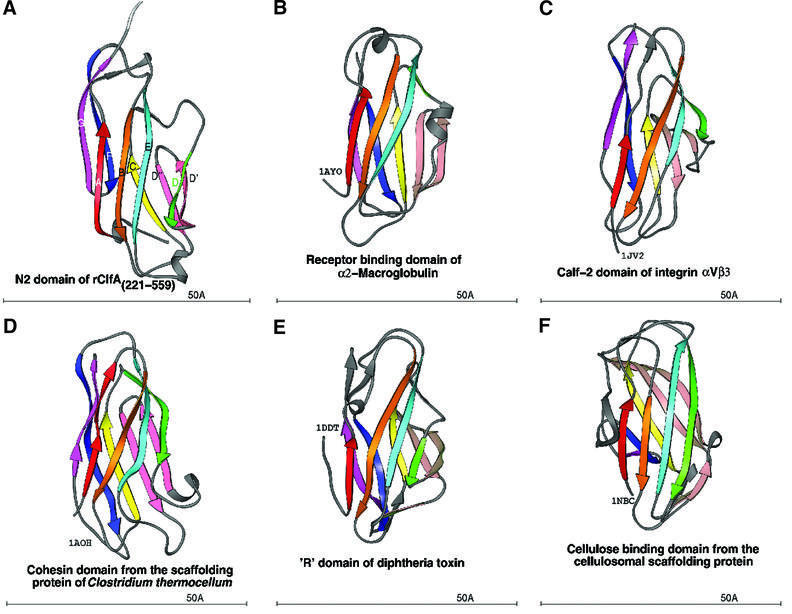
A search for similar structures in the Protein Data Bank (PDB) using the DALI server (Holm and Sander, 1993, 1994; Symersky et al., 1997) revealed that the DEv-IgG fold could be assigned to other crystal structures that were previously referred to as jelly roll barrels. Structures that we propose contain this new variant of the IgG fold include a truncate of the A region of the S.aureus colla gen-binding MSCRAMM CNA, CBD19 [CNA169–318)] (Symersky et al., 1997). The topology of the CNA(169–318) structure is identical to that of the N2 and N3 domains of rClfA(221–559) in the core regions. However, an additional β-strand (Dα) and a α-helix are found before strands D′ and D′′ in CNA(169–318) (Figure 4C and D). In addition, the DEv-IgG fold can be assigned to the receptor-binding domain of α2-macroglobulin (Jenner et al., 1998; 1AYO; Figure 5B), the Calf-1 and -2 domains of the extracellular region of the integrin αVβ3 (Xiong et al., 2001; 1JV2; Figure 5C), the cohesin domain from the scaffolding protein of Clostridium thermocellum (Tavares et al., 1997; 1AOH; Figure 5D), the ‘R’ domain of diphtheria toxin (Choe et al., 1992; 1DDT; Figure 5E) and the cellulose-binding domain from the cellulosomal scaffolding protein (Tormo et al., 1996; 1NBC; Figure 5F).
Fig. 5. Ribbon diagram of crystal structures with the DEv-IgG fold colored in rainbow fashion as in Figures 2 and 4. (A) N2 domain of ClfA. (B) Receptor-binding domain of α2-macroglobulin (PDB code: 1AYO, Z-score: 8.7, r.m.s.d.: 2.8, equiv. resid: 133). (C) Calf-2 domain of extracellular segment of the integrin αVβ3 (PDB Code: 1JV2, Z-score: 8.4, r.m.s.d.: 12.3, equiv. resid: 149). (D) Cohesin domain from the scaffolding protein of Clostridium thermocellum (PDB Code: 1AOH, Z-score: 7.6, r.m.s.d.: 3.4, equiv. resid: 123). (E) The R-domain of diphtheria toxin (PDB Code: 1DDT, Z-score: 7.2, r.m.s.d.: 4.2, equiv. resid: 129). (F) The cellulosomal scaffolding protein (PDB Code: 1NBC, Z-score: 7.0, r.m.s.d.: 3.0, equiv. resid: 109). Z-scores, r.m.s.d. values and equiv. resid. are from DALI.
The N2 and N3 DEv-IgG domains of rClfA(221–559) have a domain orientation resembling other proteins containing tandem IgG motifs with characteristic elbow, twist and swivel angles of 151, 56 and –112°, respectively (Deivanayagam et al., 2000). The N2 and N3 domains interact with each other and have a combined buried surface area of 925 Å2. A second crystal form (form II) of rClfA(221–559) was identified during the crystallization trials (see Materials and methods) and conformed to the P1 space group (Table I). This unit cell packing has two molecules in the asymmetric unit that superpose on each other with an r.m.s.d. (on Cα) of 0.56 Å and with the form I crystal structure with an r.m.s.d. (on Cα) of 0.53 Å. The near identity of the rClfA(221–559) structures derived from two different crystal forms of this protein makes it unlikely that the crystal packing affected the observed N2 and N3 domain interactions. This indicates that the orientation described for these two domains is fairly rigid.
Table I. Crystallographic data for rClfA(221–559).
| Parameter | Form I | Form II |
|---|---|---|
| a (Å) | 38.70 | 67.07 |
| b (Å) | 80.54 | 79.20 |
| c (Å) | 113.89 | 49.84 |
| α (°) | 90.00 | 96.82 |
| β (°) | 90.00 | 104.22 |
| γ (°) | 90.00 | 100.32 |
| Space group | P212121 | P1 |
| Resolution (Å) | 1.9 | 2.9 |
| I/σI | 15.7 | 6.8 |
| Rsyma (%) | 4.4 | 9.0 |
| Vm (Å3 Da–1) | 2.3 | 2.4 |
| No. of measured reflections | 148 963 | 61 627 |
| Unique reflections | 28 604 | 23 440 |
| Redundancy | 5.2 | 2.6 |
| Completeness (%) | 99.0 | 97.0 |
| % of reflections > 3σ (last shell) | 82.7 (61.2) | 77.1 (43.4) |
| Completeness (%) (last shell) | 98.7 (95.2) | 97.7 (97.4) |
| No. of molecules in asymmetric unit | 1 | 2 |
| R-factor | 20.61 | 21.58 |
| Rfree | 25.67 | 30.97 |
| R.m.s.d. (bonds) | 0.005 | 0.009 |
| R.m.s.d. (angles) | 1.37 | 1.360 |
| Average B-factor of protein (Å2) | 25.71 | 27.95 |
| Average B-factor of main chain (Å2) | 25.12 | 26.84 |
| Average B-factor of side chain (Å2) | 26.36 | 27.95 |
| No. of metal ions | 3 | 0 |
| No. of solvent molecules | 375 | 72 |
aRsym = Σ|Ih – <Ih>|/ΣIh, where <Ih> is the average intensity over symmetry equivalents.
The A regions of S.aureus Fg-binding MSCRAMMs have a common mosaic structure containing DEv-IgG domains
To explore the structural relationship between the S.aureus Fg-binding MSCRAMMs, the primary sequence of the C-terminal two-thirds of the A regions of ClfA (residues 221–559), ClfB (residues 212–542), FnbpA (residues 194–544) and FnbpB (170–542) were aligned using MPSA (Blanchet et al., 2000). In addition, the secondary structural components of each of these sequences were predicted using PHD (Rost and Sander, 1993). The primary sequence alignments display 24% overall amino acid identity. Furthermore, the predicted secondary structural elements of these sequences align well with the core strands (ABCDD′D′′EFG) of the two DEv-IgG folds of rClfA(221–559) (Figure 1B). These observations strongly suggest that the two-domain organization and the DEv-IgG fold of rClfA(221–559) are also found in the corresponding regions of ClfB (residues 212–542), FnbpA (residues 194–544) and FnbpB (residues 170–542) (Figure 1A).
Primary and secondary structural alignment of the N-terminal third of these proteins, corresponding to the proposed N1 domain, revealed very little similarity to the N2 and N3 domains (data not shown). However, this region is also predicted to be composed largely of β-sheet structure and may, in fact, be composed of more then one subdomain. Through biophysical analysis of recombinant forms of ClfB, we recently proposed a model for the structural organization of the Fg-binding A region of this protein (Perkins et al., 2001). In this model, the A region is composed of at least three domains, N1, N2 and N3, where N1 represents the protease-sensitive N-terminal domain that is degraded by the S.aureus protease aureolysin (Figure 1A) (McAleese et al., 2001). Now, we extend this model further and propose that the full-length A regions of all four S.aureus Fg-binding MSCRAMMs possess a similar multi-domain architecture that is composed of at least three domains: N1, N2 and N3 (Figure 1A).
Docking of the C-terminal residues of the Fg γ-chain
The C-terminal region of the Fg γ-chain (residues 398–411) is perceived to be a flexible structure that can adopt multiple conformations, enabling it to bind with specificity to different receptors, including the platelet integrin αIIβ3, and the MSCRAMMs ClfA, FnbpA and FnbpB (Hawiger et al., 1982a,b; Mayo et al., 1990). Consistent with this flexibility, the electron density of this region has been reportedly poor in the crystal structures of intact Fg and the appropriate fragments thereof (Pratt et al., 1997; Spraggon et al., 1997). In an attempt to elucidate the crystal structure of the C-terminal residues of the γ-chain (residues 398–411), a novel carrier protein-driven crystallization method involving both hen egg lysozyme (HEL) and glutathione S-transferase (GST) was used (Donahue et al., 1994; Ware et al., 1999). While the crystal structures of the γ-chain residues obtained in each case were very similar, displaying a general Z shape, two sites of large structural difference were observed, His401-Leu402 and Lys406-Gln407, which may represent different conformational states of the C-terminal γ-chain residues. The main-chain atoms of the terminal residues Ala408-Gly-Asp-Val411 were nearly identical in these structures and superposed with an average r.m.s.d. (on Cα) of 0.94 Å. However, the side chains appeared to adopt varying conformations, indicating flexibility of the termini.
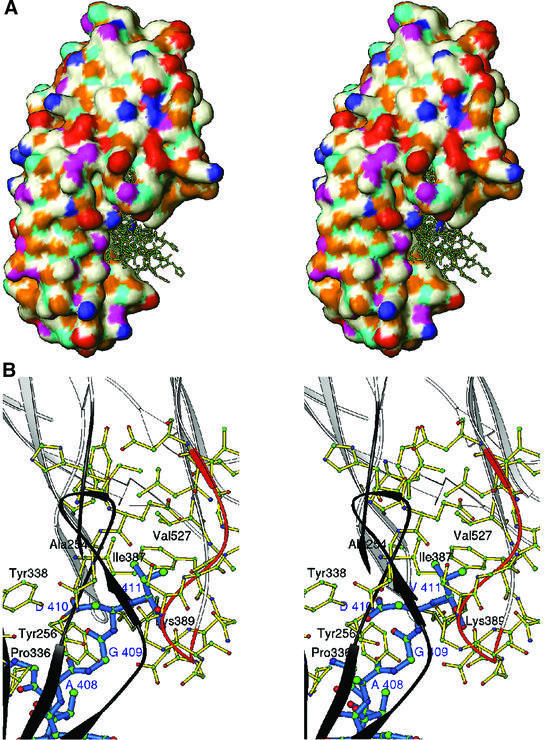
The γ-chain peptide crystal structures that were determined from the HEL and GST studies (1LSG and 1DUG, respectively) were used to model the rClfA(221–559)–γ-chain peptide complex. A brute force docking procedure (Materials and methods; Jiang and Kim, 1991) implemented in SoftDock (M.Carson, unpublished data) was used to dock the 15 residue γ-chain peptide onto rClfA(221–559). The top 20 solutions were examined and were further scored on the basis of the essential interaction of the γ-chain Ala408-Gly-Asp-Val411 residues with rClfA(221–559) (McDevitt et al., 1997; O’Connell et al., 1998). For seven of the top 20 solutions, the C-terminal γ-chain Ala408-Gly-Asp-Val411 residues were found to be extensively in contact with rClfA(221–559) residues. In all of these solutions, the Fg γ-chain peptide docked into a hydrophobic pocket formed at the interface between the N2 and N3 domains (Figure 6A). This hydrophobic pocket contains the Val527 residue that was previously shown to be important for the interaction of rClfA(221–559) with the γ-chain peptide (Figure 6B) (Hartford et al., 2001). Moreover, this Val residue is also present in FnbpB (Val464) and appears as a conserved substitution in FnbpA (Leu498). Both of these MSCRAMMs are known to interact with the C-terminal residues of the γ-chain (Figure 1B) (Wann et al., 2000). In our current model, the Val527 residue in the hydrophobic pocket of rClfA(221–559) interacts with the terminal Val411 residue of the γ-chain peptide. It is possible that replacement of Val527 with a Ser residue displaced the charge balance in this pocket, disrupting the hydrophobic interaction with Val411 in the γ-chain. Other residues of rClfA(221–559) that appear to interact with the Ala408-Gly-Asp-Val411 residues of the γ-chain in this model are Tyr256, Pro336 and Tyr338 in the N2 domain, and Ile387 and Lys389 in the N3 domain. Besides these interactions, several residues in the flexible loop region Gly532-Ser-Gly-Ser-Gly-Asp-Gly-Ile539 within rClfA(221–559) (colored orange in Figure 6B) also interact with the γ-chain peptide and form the backdrop for this suggestive binding site.
Fig. 6. (A) Stereo surface plot showing seven solutions of the Fg γ-chain peptide docked into a hydrophobic pocket between N2 and N3 domains. The hydrophobic, polar, positive and negative residues are shown in white, magenta, blue and red, respectively. Cyan and gold represent hydrogen bond donors and acceptors, respectively. (B) Stereo ribbon diagram showing the interactions of residues 408AGDV411 (blue) of the γ-chain of Fg with residues from both the N2 (dark gray) and N3 (white) domains of rClfA(221–559). The ribbon of the glycine-rich region 532GSGSGDGI539 is colored in orange.
Mutational analysis of rClfA(221–559)
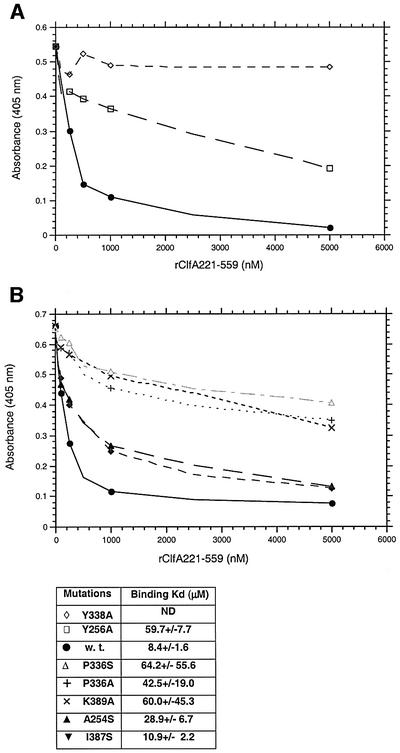
To test our docking model, we investigated the role of Ala254, Tyr256, Pro336, Tyr338, Ile387 and Lys389 residues that are present at the mouth of the hydrophobic pocket of rClfA(221–559). These residues were targeted by site-directed mutagenesis, being substituted by an Ala or Ser residue. The far-UV circular dichroism (CD) spectra of the mutant proteins were not detectably different from that of the wild-type rClfA(221–559) protein (data not shown), suggesting that the substitutions did not dramatically alter the conformation of the protein. The interaction of the mutant rClfA(221–559) proteins with immobilized intact Fg was analyzed in a direct ELISA-type binding assay (data not shown) and also in an inhibition ELISA-type assay in which the ability of the mutant proteins to inhibit the binding of soluble biotin-labeled wild-type rClfA(221–551) to immobilized Fg was determined (Figure 7). The Y338A mutant protein failed to bind to immobilized Fg in either assay, whereas the Y256A, K389A and P336S mutant proteins exhibited markedly reduced apparent affinity for immobilized Fg. In addition, the A254S and I387S mutant proteins had somewhat reduced apparent affinities. Fluorescence polarization was used to further characterize the binding of the wild-type and mutant ClfA(221–551) proteins to a fluorescein-labeled 17-amino-acid-long synthetic peptide corresponding to the C-terminus of the Fg γ-chain. The corresponding KD values were calculated from these assays (Figure 7). The Y338A mutant protein failed to bind to the γ-chain peptide in this assay, whereas the Y256A, K389A and P336S mutant proteins exhibited markedly reduced affinity for the labeled peptide (KD = 49–65 µM) compared with the wild-type rClfA(221–559) protein (KD = 8 µM). The KD values determined for the binding of the A254S and I387S protein to γ-chain peptide were 29 and 11 µM, respectively. Taken together, these results support the docking experiments and tentatively identify the hydrophobic pocket between the N2 and N3 domains as the binding site in ClfA for the C-terminal residues of the Fg γ-chain.
Fig. 7. (A) The relative affinities of rClfA221–559w.t. (closed circles), rClfA221–559Y256→A (open squares) and rClfA221–559Y338→A (open diamonds) for Fg were examined in an inhibition ELISA-type assay (Smith et al., 1993). Biotin-labeled rClfA221–559 was incubated with the indicated increasing concentration of unlabeled ClfA proteins in Fg-coated microtiter wells. The amount of labeled ClfA protein bound to the immobilized Fg was then quantitated. The Kd of each protein was calculated by determining the affinity of the proteins for a fluorescein-labeled 17-amino-acid-long synthetic peptide representing the C-terminus of the Fg γ-chain using fluorescence polarization. (B) The relative affinities of rClfA221–559A254→S (upright closed triangles), rClfA221–559I387→S (inverted closed triangles), rClfA221–559 K389→A (×), rClfA221–559P336→A (+), rClfA221–559P336→S (open upright triangles) for Fg were examined in the same way as above. Kd was also calculated as indicated.
Discussion
Nearly a century ago, Much (1908) first reported the clumping of S.aureus in the presence of blood plasma, which was later shown to be Fg dependent, indicating the mediation of Fg-binding surface proteins in the clumping phenomenon. The crystal structure of rClfA(221–559), which represents the minimum truncate of ClfA with Fg-binding activity, has now been determined. Recombinant rClfA(221–559) structure is composed of two discrete subdomains that have a very similar fold. This fold, which we have called DEv-IgG, represents a novel variation of the IgG fold. The topology of the DEv-IgG fold is very similar to that of the C-type IgG fold, but displays variation between the D and E strands on one side of the motif (right side in Figure 4B). From primary structure alignment and secondary structure prediction, it appears likely that the C-terminal two-thirds of the Fg-binding A regions of ClfB, FnbpA and FnbpB also contain two subdomains that display the DEv-IgG fold. In addition, we discovered that the minimum ligand-binding region of the collagen-binding MSCRAMM, CNA(151–318), which corresponds to the central part of the A region of this protein, also displays this fold (Symersky et al., 1997). Interestingly, we previously reported that the B repeats of CNA display the Inv-IgG fold, yet another variant of the immunoglobulin superfamily (Deivanayagam et al., 2000). Based on the crystal structure of rClfA(221–559), previous biophysical analysis of rClfB, and primary and secondary structure alignments of all four S.aureus Fg-binding MSCRAMMs, we propose that the A regions of these proteins contain at least three domains: N1, N2 and N3. N1 represents the N-terminal region, which, at least in the case of ClfA and ClfB, is known to be protease sensitive (Deivanayagam et al., 1999; McAleese et al., 2001). Thus, we propose that the A regions of these MSCRAMMs are mosaic proteins containing several immunoglobulin-like domains. The manifestation of MSCRAMMs through modules of IgG fold with varying functional characteristics (binding to different targets) suggests the evolution of these motifs from a common template. It is of interest to note that cell surface adhesins of Gram-negative bacteria, including invasin (Hamburger et al., 1999), FimC (Pellecchia et al., 1998) and FimH (Choudhury et al., 1999), contain domains of the traditional immunoglobulin superfamily. Furthermore, many eukaryotic cell adhesion molecules, such as ICAMs, VCAMs and MAdCAM, also contain these domains (Wang and Springer, 1998) and the recently solved αVβ3 integrin structure contain domains of the newly identified DEv-IgG fold. It is tantalizing to speculate that a structural organization encompassing the now extended superfamily of immunoglobulin folds is critical for the activity of these adhesion receptors.
In the crystal structure of rClfA(221–559), it was revealed that the putative EF-hand-like motif was composed of a β-turn followed by a β-strand, a structure that does not resemble a classical EF-hand. Therefore, this observation does not support the previously suggested models for ClfA–metal ion interactions (O’Connell et al., 1998). However, a role for the three metal ion-binding sites, identified in the crystal structure of rClfA(221–559), in ClfA–ligand interactions can not be ruled out at this point. Studies are in progress to further characterize these metal ions, their binding sites, and role in ClfA structure and function.
Brute force docking of the terminal residues (398–411) of the γ-chain into the rClfA(221–559) structure produced seven solutions that docked into the hydrophobic pocket formed between the N2 and N3 domains. In an earlier study, the replacement of Val527 (which we now have located inside this pocket) with Ser resulted in a loss of Fg-binding activity (Hartford et al., 2001). From the present model, it is apparent that such a substitution would disrupt the charge balance in the pocket and would thus be deleterious to the hydrophobic interactions with the terminal Val411 of the γ-chain. Besides the hydrophobic interactions described above, the Ala408-Asp-Gly-Val411 of the γ-chain in our proposed model also have charge-to-charge interactions with Tyr256 (N2) and Tyr338, and Lys389 (N3) present at the mouth of the pocket. Substitution of these residues with Ala resulted in mutant proteins with no detectable or a markedly reduced apparent affinity for Fg. Furthermore, sequence analyses of the N3 domain of ClfA from a number of S.aureus strains (Newman, 8324-5, COL, MRSA252, MSSA476, N315, Mu50, RN4256, Cowan I, MN8, Yeh, McNamara, P1 and BB) revealed that the amino acids in the hydrophobic pocket that we propose are involved in binding to the γ-chain of Fg are all conserved (data not shown). We have also observed that the independently expressed recombinant N2 and N3 domains fail to bind to Fg (data not shown), providing additional support for the hypothesis that both domains contribute to the formation of the Fg-binding site. The flexible Gly-rich sequence (Gly532-Ser-Gly-Ser-Gly-Asp-Gly-Ile539) perhaps provides the cushion for the flexible γ-chain peptide to permeate into the hydrophobic pocket (Figure 6B), and this region could potentially acquire a stable structure in the ClfA–γ-chain complex.
The S.aureus MSCRAMMs ClfA (McDevitt et al., 1997; O’Connell et al., 1998), FnbpA and FnbpB (Wann et al., 2000) bind to the C-terminal residues of the γ-chain, a region that is also involved in platelet aggregation (Farrell et al., 1992) and fibrin monomer cross-linking during the formation of a fibrin clot (Chen and Doolittle, 1969). Similarly, SdrG, a Fg-binding MSCRAMM from Staphylococcus epidermidis, binds to an N-terminal segment of the Fg β-chain that overlaps with the thrombin cleavage site (Davis et al., 2001). The fact that these staphylococcal MSCRAMMs bind to the terminal amino acids of various Fg chains that are involved in physiological processes is an intriguing observation. Whether staphylococci target these flexible regions of Fg to inhibit its physiological function, or simply because these regions are easily accessible, is a question that remains to be answered.
Materials and methods
Purification of recombinant proteins
For crystallization, the rClfA(221–599) protein, which is equivalent to the proteolytic product ClfA37 (Deivanayagam et al.,1999), was expressed in E.coli and purified by immobilized metal chelate affinity chromatography, followed by anion-exchange and gel-filtration chromatography, as described previously (O’Connell et al., 1998; Deivanayagam et al., 1999). The purity and molecular mass of the rClfA(221–559) protein were analyzed by SDS–PAGE and MALDI mass spectroscopy, respectively. For the Fg-binding assays, the rClfA proteins were expressed and purified as above, except that the final gel-filtration purification step was not performed.
Crystallization and data collection
Form I. A droplet made from 2 µl of rClfA(221–559) at 20 mg/ml and 2 µl of 1.2 M sodium citrate, 100 mM succinic acid at pH 5.6 was equilibrated against a reservoir containing 1 ml of 1.2 M sodium citrate, 100 mM succinic acid at pH 5.6. After 1 week, the crystals grew to 0.4 × 0.3 × 0.15 mm. These crystals were ‘flash’ frozen without any addition of cryo-protectants and were subjected to X-rays generated on the X-9B beamline at the Synchrotron light source in the Brookhaven National Laboratories (BNL). Diffraction data to 1.9 Å resolution were collected on a MAR CCD detector at a distance of 170 mm with a 0.5° oscillation sweeping the reciprocal lattice with 262 frames (131°) at 0.91676 Å wavelength (λ). Data were integrated and scaled using DENZO and SCALEPACK (Otwinowski, 1993), and the data statistics are reported in Table II.
Table II. Heavy atom derivative statistics for rClfA(221–559).
| Heavy atom compound | Resolution (Å) | Rsyma (%) | Unique reflections | Solve |
Phases |
||
|---|---|---|---|---|---|---|---|
| Rcentric | R.M.S. (FH)/R.M.S.(E) | Rcullisb | Phasing powerc | ||||
| (CH3)3PbOOCCH3 | 2.45 | 6.2 | 13 162 | 0.57 | 1.35 | 0.567 | 1.98 |
| K2PtCl4 | 3.2 | 16.3 | 7177 | 0.64 | 1.22 | 0.657 | 1.15 |
| PCMBS | 3.0 | 7.2 | 7450 | 0.56 | 1.42 | 0.628 | 1.45 |
aRsym = Σ|Ih – <Ih>|/ΣIh, where <Ih> is the average intensity over symmetry equivalents.
bRcullis = Σ||FPH ± FP| – FHcalc|/Σ|FPH – FP|
cPhasing power = Σ|FH|/Σ||FPHobs| – |FPHcalc||
Form II. High concentrations of Ca2+ or Mn2+ interfere with the rClfA(221–559)–γ-chain interactions and hence rClfA(221–559) was dialyzed into a solution containing 20 mM Tris pH 8.0, 100 mM NaCl and 5 mM EDTA, as opposed to 1 mM EDTA reported in the form I crystallization condition. The 17 residue synthetic peptide 395GEGQQHHLGGAK QAGDV411 imitating the C-terminal residues of the γ-chain of Fg was mixed at 1:20 molar ratio and incubated at 4°C overnight with purified rClfA(221–559) (0.1 mg/ml) with occasional gentle stirring. The above solution was then concentrated under 40 p.s.i. argon gas pressure using the Amicon micro-ultrafiltration system with a YM10 membrane. The concentrated protein–peptide mixture was used to set up hanging drop crystallization trials. Large diamond-shaped single crystals measuring 0.3 × 0.3 × 0.1 mm were grown from a droplet made of 3 µl of the protein–peptide mixture (14 mg/ml) and 3 µl of the well solution containing 200 mM ammonium acetate, 100 mM sodium citrate dihydrate pH 5.6 and 26% (w/v) PEG4000. Crystals were flash frozen in the presence of 15% glycerol and diffraction data were collected at BNL (X-9B) on a MAR CCD detector at a distance of 220 mm with a 1.0° oscillation sweeping the reciprocal lattice with 276 frames (276°) at 0.98 Å wavelength (λ). Data were integrated and scaled using DENZO and SCALEPACK and the data statistics are reported in Table I. The form II crystals diffracted poorly (2.9 Å resolution) compared with the form I crystals (1.9 Å resolution).
Structure determination and refinements
Form I. The structure was solved by the MIR (multiple isomorphous replacement) method. Three derivatives, trimethyl lead acetate (TMLA), potassium platinum tetrachloride (PPT) and p-chloromercuri-benzenesulfonic acid (PCMPS), were identified by difference Patterson analysis using the program XTALVIEW (McRee, 1993). The details of the heavy atom derivative statistics are presented in Table II. The PPT and PCMPS derivatives had one site each, whereas TMLA had two. Refinement of heavy atom derivatives and subsequent phase calculations were performed using the program PHASES (Furey and Swaminathan, 1997) and SOLVE (Terwilliger and Berendzen, 1999). An initial 3.5 Å electron density map clearly defined the solvent and revealed well-connected main-chain densities. The initial Cα trace was made using the baton mode in O (Jones et al., 1991). Side chains were incorporated into the model and further refinements were carried out with ARP/WARP (Perrakis et al., 1999) and CNS-Solve-1.0 (Adams et al., 1997). All reflections (≥2σ) from 100 to 1.9 Å were used in refinement, with 10% partitioned in a test set for monitoring the refinement process (Brünger, 1992). The initial R-factor/Rfree of the fully built model with side chains was 33.9%/41.0%. The molecular model building was performed with the help of the graphics program O, in conjunction with Oops (Kleywegt and Jones, 1997). Solvent molecules were added based on geometry and electron density, and model convergence was achieved after several cycles of model building and refinement. Maximum likelihood refinement and bulk solvent correction were performed with CNS-Solve in the last few cycles, resulting in 375 solvent molecules and three metal ions in the asymmetric unit with the final R-factor/Rfree being 20.6%/25.7%. The average B-factor for the protein is 25.71 Å2. The model has 99.4% of the residues in the allowed regions of the Ramachandran plot, two residues in the generously allowed regions and one of these residues (Asp330) appears in a loop region where the electron density is not clearly defined.
Form II. The crystal structure of form II was solved by the molecular replacement method, where the crystal structure of form I was used as the initial model. Self-rotation function calculated using 15–4 Å resolution diffraction data revealed one strong peak (60% of the origin peak) at ω = 210° and φ = 162 ° in the κ = 180° section, indicating the presence of a non-crystallographic two-fold. A good rotation solution was obtained using the program AMoRe (Navaza, 1994) and positional searches initiated using the best rotations produced two strong translational peaks. After fixing the orientation and position of one molecule, the rotation and translation values for the second molecule were obtained. Each model was subjected to rigid body refinement using AMoRe and expressed an R-factor of 35.4% and correlation coefficient of 0.54. These two molecules were further refined in CNS using the simulated annealing protocol where their R-factor/Rfree fell from 42.8%/42.3% to 26.9%/34.0%. Several cycles of model building were carried out using program O in conjunction with Oops, and the last few cycles were subjected to maximum likelihood refinement and bulk solvent correction incorporated in CNS-Solve-1.0. The addition of 5 mM EDTA chelated the metal ions in this structure, and with 90 solvent molecules in the asymmetric unit the model had a final R-factor/Rfree of 21.58%/30.97% (resolution 100–3.2 Å). This model had an average B-factor of 27.95 Å2, which is comparable to that of form I, and has all the residues within the allowed regions of the Ramachandran plot. On the difference Fourier maps, residual density was observed in a region of contact between the two asymmetrically related molecules that could accommodate three polyalanine residues. However, after several cycles of model building and refinements, the improved phases did not extend the residual density to account for the 17 residue peptide.
Orientation and superposition of domains
The orientation of all the figures and superposition presented in this paper were performed using the common-coordinate system, as described previously (Deivanayagam et al., 2000). All the figures in this paper were made using the program RIBBONS (Carson, 1997).
Buried surface area
The analytical molecular surface area was calculated using the MSP program (Connolly, 1993). The coordinates of the two domains, D1 and D2, were combined to form D1D2 and the surface area determined for each set. The combined buried surface was approximated with the areas: D1 + D2 – D1D2.
Docking
The docking proceedure holds a target molecule stationary, while a full six degrees of freedom search is performed with the probe molecule. For each discrete rotational orientation (typically ∼10 000), a molecular dot surface is calculated and classified per atom type. Each molecule is partitioned into cubes classified as solvent, surface or internal, and then compared cube by cube for each possible translation. The scoring function rewards geometric and chemical complementarity between two surfaces, while penalizing internal volume overlap. The geometric complementarity requires the surface normals to be oriented toward each other, within an angular tolerance. The chemical complementarity is based on the atomic classification as shown in Figure 6A. A simple ±1 sum is accumulated pairwise over the surface dots inside each cube; for example, a positive charge interacting with hydrophobic is –1; a positive charge interacting with a hydrogen-bond acceptor is +1. Details are given in Jiang and Kim (1991).
Site-directed mutagenesis
A two-step, PCR-based, strand overlap extension (SOE) site-directed mutagenesis (Ho et al., 1989) procedure was used to specifically substitute the targeted residues in rClfA(221–559). The final PCR products generated were gel purified, digested with BamHI and HindIII, and ligated into the expression vector pQE30, which had been digested with the same enzymes. The DNA sequence of each of the mutations was verified by automated nucleotide sequencing at the University of Texas, Houston, core facility.
CD spectroscopy
All the protein samples were dialyzed into 1 mM Tris–HCl pH 7.4. Far-UV CD data were collected and the secondary structure composition was estimated as described previously (Hartford et al., 2001).
Fg-binding assays
The binding of the rClfA proteins to immobilized intact Fg (Enzyme Research Laboratories) was analyzed in an inhibition ELISA-like assay as described previously (Smith et al., 1994). The binding of the rClfA proteins to the fluorescein-labeled C-terminal γ-chain peptide was quantitated by fluorescence polarization, and this method, together with peptide synthesis and labeling procedures, have been described previously (O’Connell et al., 1998).
PDB accession code
Coordinates of the rClfA(221–559) structure have been deposited in the Protein Data Bank (accession code IN67).
Acknowledgments
Acknowledgements
We would like to thank Sita Danthuluri, Shital Patel and Karthe Ponnuraj for their invaluable technical help in setting up crystallization, and Krishna Murthy and Dwight Moore for useful suggestions. This study was supported in part by NIH grant 1AI20624.
References
- Adams P.D., Pannu,N.S., Read,R.J. and Brünger,A.T. (1997) Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement Proc. Natl Acad. Sci. USA, 94, 5018–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet C., Combet,C., Geourjon,C. and Deleage,G. (2000) MPSA: integrated system for multiple protein sequence analysis with client/server capabilities. Bioinformatics, 16, 286–287. [DOI] [PubMed] [Google Scholar]
- Bork P., Holm,L. and Sander,C. (1994) The immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set, which is close to that containing the variable domains. J. Mol. Biol., 242, 309–320. [DOI] [PubMed] [Google Scholar]
- Branden C. and Tooze,J. (1999) Introduction to Protein Structure, 2nd edn. Garland Publishing, New York, NY.
- Brünger A.T. (1992) Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–475. [DOI] [PubMed] [Google Scholar]
- Carson M. (1997) RIBBONS. Methods Enzymol., 277, 493–505. [PubMed] [Google Scholar]
- Chen R. and Doolittle,R.F. (1969) Synthetic peptide probes and the location of fibrin polymerization sites. Protides Biol. Fluids, 28, 311–316. [Google Scholar]
- Choe S., Bennet,M.J., Fujii,G., Curmi,P.M.G., Kantardjieff,K.A., Coller,R.J. and Eisenberg,D. (1992) The crystal structure of diphtheria toxin. Nature, 357, 216–222. [DOI] [PubMed] [Google Scholar]
- Choudhury D., Thompson,A., Stojanoff,V., Langermann,S., Pinker,J., Hultgren,S.J. and Knight,S.D. (1999) X-ray structure of the FimC–FimH chaperone–adhesin complex from uropathogenic Escherichia coli. Science, 285, 1061–1066. [DOI] [PubMed] [Google Scholar]
- Connolly M.L. (1993) The molecular surface package. J. Mol. Graph., 11, 139–141. [DOI] [PubMed] [Google Scholar]
- Davis S.L., Gurusiddappa,S., McCrea,K.W., Perkins,S. and Höök,M. (2001) SdrG, a fibrinogen-binding bacterial adhesin of the MSCRAMM subfamily of the Staphylococcus epidermis, targets the thrombin cleavage site in the β chain. J. Biol. Chem., 276, 27799–27805. [DOI] [PubMed] [Google Scholar]
- Deivanayagam C.C., Perkins,S., Danthuluri,S., Owens,R.T., Bice,T., Nanavathy,T., Foster,T.J., Höök,M. and Narayana,S.V.L. (1999) Crystallization of ClfA and ClfB fragments: the fibrinogen-binding surface proteins of Staphylococcus aureus. Acta Crystallogr. D, 55, 554–556. [DOI] [PubMed] [Google Scholar]
- Deivanayagam C.C., Rich,R.L., Carson,M., Owens,R.T., Danthuluri,S., Bice,T., Höök,M. and Narayana,S.V.L. (2000) Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure, 8, 67–78. [DOI] [PubMed] [Google Scholar]
- Donahue J.P., Patel,H., Anderson,W.F. and Hawiger,J. (1994) Three-dimensional structure of the platelet integrin recognition segment of the fibrinogen γ chain obtained by carrier driven protein crystallization. Biochemistry, 91, 12178–12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell D.H., Thiagarajan,P., Chung,D.W. and Davie,E.W. (1992) Role of fibrinogen α and γ chain sites in platelet aggregation. Proc. Natl Acad. Sci. USA, 89, 10729–10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey W. and Swaminathan,W. (1997) PHASES-95: a program package for processing and analyzing diffraction data from macromolecules. Methods Enzymol., 277, 590–620. [DOI] [PubMed] [Google Scholar]
- Hamburger Z.A., Brown,M.S., Isberg,R.R. and Bjorkman,P.J. (1999) Crystal structure of invasin: a bacterial integrin binding protein. Science, 286, 291–295. [DOI] [PubMed] [Google Scholar]
- Harpaz Y. and Chothia,C. (1994) The immunoglobulin fold: structural classification, sequence patters and common core. J. Mol. Biol., 238, 528–539.8176743 [Google Scholar]
- Hartford O.M., Wann,E.R., Höök,M. and Foster,T.J. (2001) Identification of residues in the Staphylococcus aureus fibrinogen binding MSCRAMM clumping factor A (ClfA) that are important for ligand-binding. J. Biol. Chem., 276, 2466–2473. [DOI] [PubMed] [Google Scholar]
- Hawiger J.S., Timmons,S., Strong,D.D., Cottrell,B.A., Riley,M. and Doolittle,R.F. (1982a) Identification of a region of human fibrinogen interacting with staphylococcal clumping factor. Biochemistry, 21, 1407–1413. [DOI] [PubMed] [Google Scholar]
- Hawiger J., Timmons,S., Kloczewiak,M., Strong,D.D. and Doolittle,R.F. (1982b) γ and α chains of human fibrinogen interacting with staphylococcal clumping factor. Proc. Natl Acad. Sci. USA, 79, 2068–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick S., Blanc-Brude,O., Gray,A. and Laurent,G. (1999) Fibrinogen. Int. J. Biochem. Cell Biol., 31, 741–746. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt,M.D., Hornton,R.M., Pullen,J.K. and Pease L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by distant alignment matrices. J. Mol. Biol., 233, 123–128. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1994) Searching protein structure database has come of age. Proteins, 19, 165–173. [DOI] [PubMed] [Google Scholar]
- Höök M. and Foster,T.J. (2000) Staphylococcal surface proteins. In Fischetti et al. (eds), Gram Positive Pathogens, Vol. 40. American Society for Microbiology, Washington, DC, pp. 386–392.
- Jenner L., Husted,L., Thirup,S., Sottrup-Jensen,L. and Nyborg,J. (1998) Crystal structure of the receptor-binding domain of α2-macroglobulin. Structure, 6, 595–604. [DOI] [PubMed] [Google Scholar]
- Jiang F. and Kim,S.H. (1991) Soft docking: matching of molecular surface cubes. J. Mol. Biol., 219, 79–102. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. D, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1997) Model rebuilding and refinement practice Methods Enzymol., 276, 208–330. [DOI] [PubMed] [Google Scholar]
- Kloczewiak M., Timmons,S., Lukas,T.J. and Hawiger,J. (1984) Platelet receptor recognition site on human fibrinogen. Synthesis and structure–function relationship of peptides corresponding to the carboxy-terminal segment of the γ chain. Biochemistry, 23, 1767–1774. [DOI] [PubMed] [Google Scholar]
- Kretsinger R.H. (1996) EF hands reach out. Nat. Struct. Biol., 3, 12–15. [DOI] [PubMed] [Google Scholar]
- Mayo K.H., Bruke,C., Lindon,J.N. and Kloczewiak,M. (1990) 1-H NMR sequential assignments and secondary structure analysis of human fibrinogen γ chain C-terminal residues 385–411. Biochemistry, 29, 3277–3286. [DOI] [PubMed] [Google Scholar]
- McAleese M.F., Walsh,E.J. Sieprawska,M. Potempa,J. and Foster,T.J. (2001) Loss of clumping factor B fibrinogen binding activity by Staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease J. Biol. Chem., 276, 29969–29987. [DOI] [PubMed] [Google Scholar]
- McDevitt D., Francois,P., Vaudaux,P. and Foster,T.J. (1994) Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol., 11, 237–248. [DOI] [PubMed] [Google Scholar]
- McDevitt D., Francois,P., Vaudaux,P. and Foster,T.J. (1995) Identification of the ligand-binding domain of the surface-located fibrinogen receptor (clumping factor) of Staphylococcus aureus. Mol. Microbiol., 16, 895–907. [DOI] [PubMed] [Google Scholar]
- McDevitt D., Nanavathy,T., House-Pompeo,K., Bell,E., Turner,N., McIntire,L., Foster,T.J. and Höök,M. (1997) Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur. J. Biochem., 247, 416–424. [DOI] [PubMed] [Google Scholar]
- McRee D.E. (1993) Practical Protein Crystallography. Academic Press, San Diego, CA.
- Much H. (1908) Uber eine vorstufe des fibrinfer mentes in kulturen von Staphylokokkus aureus. Biochem Z., 14, 143–144. [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nì Eidhin D., Perkins,S., Francois,P., Vaudaux,P., Höök,M. and Foster,T.J. (1998) Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol., 30, 245–257. [DOI] [PubMed] [Google Scholar]
- O’Connell D.P., Nanavathy,T., McDevitt,D., Gurusiddappa,S., Höök,M. and Foster,T.J. (1998) The fibrinogen-binding MSCRAMM (clumping factor) of Staphylococcus aureus has a Ca2+-dependent inhibitory site. J. Biol. Chem., 273, 6821–6829. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. (1993) DENZO, a Film Processing Program for Macromolecular Crystallography. Yale University, New Haven, CT.
- Patti J. M and Höök,M. (1994) Microbial adhesins recognizing extracellular matrix macromolecules Curr. Opin. Cell Biol., 6, 752–758. [DOI] [PubMed] [Google Scholar]
- Pellecchia M., Güntert,P., Glockshuber,R. and Wüthrich,K. (1998) NMR solution of the periplasmic chaperone FimC. Nat. Struct. Biol., 5, 885–890. [DOI] [PubMed] [Google Scholar]
- Perkins S., Walsh,E.J., Deivanayagam C.C.S., Narayana,S.V.L., Foster,T.J. and Höök,M. (2001) Structural organization of the fibrinogen-binding region of the clumping factor B MSCRAMM of Staphylococcus aureus. J. Biol. Chem., 276, 44721–44728. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R.J. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Pratt K.P., Côtè,H.C.F., Chung,D.W., Stenkamp,R.E. and Davie,E.W. (1997) The primary fibrin polymerization pocket: three-dimensional structure of a 30 kDa C-terminal γ chain fragment complexed with the peptide Gly-Pro-Arg-Pro. Proc. Natl Acad. Sci. USA, 94, 7176–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B. and Sander,C. (1993) Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol., 232, 584–599. [DOI] [PubMed] [Google Scholar]
- Smith J.W., Piotrowicz,R.S. and Mathis,D. (1994) A mechanism for divalent cation regulation of β3-integrins J. Biol. Chem., 269, 960–967. [PubMed] [Google Scholar]
- Spraggon G., Everse, E and Doolittle,R.F. (1997) Crystal structures of fragment D from human fibrinogen and its cross linked counterpart from fibrin. Nature, 389, 455–462. [DOI] [PubMed] [Google Scholar]
- Springer T.A., Jing,H. and Takagi,J. (2000) A novel Ca2+ binding β hairpin loop better resembles integrin sequence motifs than the EF hand. Cell, 102, 272–277. [DOI] [PubMed] [Google Scholar]
- Strong D.D., Laudano,A.P., Hawiger,J. and Doolittle,R.F. (1982) Isolation, characterization and synthesis of peptides from human fibrinogen that block the staphylococcal clumping reaction and construction of a synthetic clumping particle. Biochemistry, 21, 1414–1420. [DOI] [PubMed] [Google Scholar]
- Symersky J. et al. (1997) Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat. Struct. Biol., 4, 833–838. [DOI] [PubMed] [Google Scholar]
- Tavares G.A., Beguin,P. and Alzari,P.M. (1997) The crystal structure of type I cohesin domain at 1.7 Å resolution. J. Mol. Biol., 273, 701–713. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated structure solution for MIR and MAD. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo J., Lamed,R., Chirino,A.J., Morag,E., Bayer,E.A., Shoham, Y and Steitz,T.A. (1996) Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J., 15, 5739–5751. [PMC free article] [PubMed] [Google Scholar]
- Wang J.-H. and Springer,T. (1998) Structural specialization of immunoglobulin superfamily members for adhesion to integrins and viruses. Immunol. Rev., 163, 197–215. [DOI] [PubMed] [Google Scholar]
- Wann E.R., Gurusiddappa,S. and Höök,M. (2000) The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem., 275, 13683–13781. [DOI] [PubMed] [Google Scholar]
- Ware S., Donahue,J.P. Hawiger,J. and Anderson,W.F. (1999) Structure of the fibrinogen γ chain integrin binding and factor XIIIa cross-linking sites obtained through carrier driven protein crystallization. Protein Sci., 8, 2663–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.-P., Stehle,T., Diefenbach,B., Zhang,R., Dunker,R., Scott,D.L., Joachimiak,A., Goodman,S.L. and Arnaout,M.A. (2001) Crystal structure of the extracellular segment of the integrin αVβ3. Science, 294, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]