Abstract
The U1 small nuclear ribonucleoprotein (U1 snRNP) binds to the pre-mRNA 5′ splice site (ss) at early stages of spliceosome assembly. Recruitment of U1 to a class of weak 5′ ss is promoted by binding of the protein TIA-1 to uridine-rich sequences immediately downstream from the 5′ ss. Here we describe a molecular dissection of the activities of TIA-1. RNA recognition motifs (RRMs) 2 and 3 are necessary and sufficient for binding to the pre-mRNA. The non- consensus RRM1 and the C-terminal glutamine-rich (Q) domain are required for association with U1 snRNP and to facilitate its recruitment to 5′ ss. Co-precipitation experiments revealed a specific and direct interaction involving the N-terminal region of the U1 protein U1-C and the Q-rich domain of TIA-1, an interaction enhanced by RRM1. The results argue that binding of TIA-1 in the vicinity of a 5′ ss helps to stabilize U1 snRNP recruitment, at least in part, via a direct interaction with U1-C, thus providing one molecular mechanism for the function of this splicing regulator.
Keywords: TIA-1/U1-C/U1 snRNP
Introduction
The excision of introns from mRNA precursors is important to generate translatable mRNAs in higher eukaryotes. The process is often regulated to generate alternatively spliced transcripts able to encode distinct proteins (Hastings and Krainer, 2001; Will and Lührmann, 2001; Modrek and Lee, 2002). The chemical process of intron removal occurs within the spliceosome, a complex of >100 polypeptides and five uridine-rich small nuclear ribonucleoproteins (U snRNPs) assembled on the pre-mRNA (Nilsen, 2002; Zhou and Reed, 2002).
U1 snRNP recognizes the 5′ splice site (ss) and is among the first factors to interact with the pre-mRNA to form complexes (complex E in mammalian extracts) that commit the pre-mRNA to the splicing pathway (Ruby and Abelson, 1988; Séraphin and Rosbash 1989; Michaud and Reed, 1991, 1993). Human U1 snRNP is composed of a 165 nucleotide RNA (U1 snRNA), seven different Sm proteins common to other snRNPs and three U1-specific polypeptides: U1-70K, U1-A and U1-C (Lührmann et al., 1990). The sequence of the 5′ end of U1 snRNA is complementary to the 5′ ss region (Rinke et al., 1984), and stem–loops I and II are bound directly by U1-70K and U1-A, respectively. A uridine-rich motif, the Sm site, is bound by the Sm proteins, most probably forming a ring-like structure around the Sm site (Hamm et al., 1987; Patton and Pederson 1988; Scherly et al., 1989, 1990; Lutz-Freyermuth et al., 1990; Kambach et al., 1999). U1-C does not interact directly with naked U1 snRNA, but depends on other U1 protein components for association with the snRNP. Interactions have been detected between the N-terminal 45 amino acids of U1-C, which include a zinc finger-like motif, and U1-70K, as well as with the Sm proteins B′/B (Nelissen et al., 1994). This region of U1-C has been shown to stimulate formation or stabilization of complex E (Will et al., 1996). In contrast, reconstituted snRNPs lacking U1-A, or lacking the binding sites for U1-A or U1-70K, can support splicing and complex E formation (Will et al., 1996). These results indicate that U1-C plays an important role in U1 snRNP function. Recent results indicate that U1-C recognizes the sequence of the 5′ ss, and argue that this RNA–protein recognition precedes base pairing with U1 snRNA (Du and Rosbash, 2002).
Comprehensive biochemical and mass spectrometric analysis of the composition of yeast U1 snRNP revealed the association of four polypeptides not observed in purified preparations of human U1 (Gottschalk et al., 1998). One of these, the protein Nam8p, interacts with sequences downstream from the 5′ ss and modulates U1 snRNP binding (Puig et al., 1999). A human homolog of Nam8p, the protein TIA-1, has been shown to bind to uridine-rich sequences downstream from 5′ ss and promote U1 snRNP binding (Del Gatto et al., 2000; Förch et al., 2000; Le Guiner et al., 2001b).
TIA-1 is composed of three RNA recognition motifs (RRMs) and a C-terminal Q-rich domain (Figure 1A) (Tian et al., 1991). Here we identify domains of TIA-1 required for pre-mRNA recognition, association with U1 snRNP and recruitment of U1 snRNP to 5′ ss. We report that the U1-C protein interacts directly with TIA-1 and, based on the results of functional domain analysis, we propose that this interaction is important for the recruitment of U1 snRNP by TIA-1.
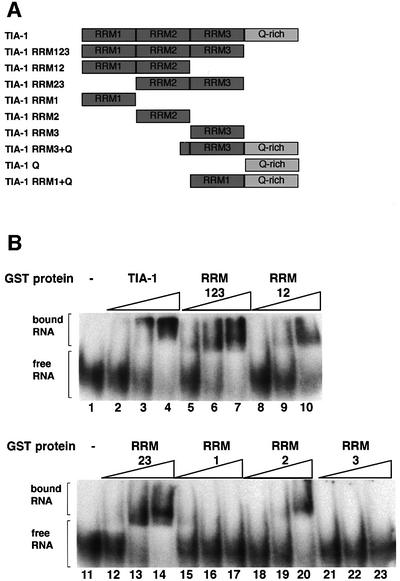
Fig. 1. Binding of TIA-1 and deletion mutants to the msl-2 5′ ss region. (A) Domain structure of TIA-1 and mutant derivatives. Schematic representation of the domains present in the different recombinant proteins. RRMs and the Q-rich domain are indicated. (B) Mobility shift assays. 32P-Labeled 5′ half msl-2 RNA was incubated with the indicated recombinant purified proteins at final concentrations of 10–8, 10–7 and 10–6 M, and the complexes resolved by native polyacrylamide gel electrophoresis. The positions of bound and unbound RNAs are indicated.
Results
TIA-1 was reported to cross-link to uridine-rich regions downstream of 5′ ss of Drosophila msl-2 and the human Fas receptor pre-mRNAs (Förch et al., 2000). To identify domains of TIA-1 required for pre-mRNA recognition, mobility shift assays were carried out using a battery of domain deletion mutants (Figure 1A) and in vitro transcribed RNAs containing the msl-2 5′ ss region. Binding of individual RRM domains was only detectable with RRM2 (Figure 1B, lanes 18–20). A protein including RRMs 2 and 3 bound the RNA with essentially the same affinity as the full-length protein (compare lanes 2–4 with 12–14, apparent KD 3 × 10–8 M), while a construct composed of RRMs 1 and 2 displayed the same affinity as RRM2 alone (compare lanes 8–10 with 18–20, apparent KD 3 × 10–7 M). The presence of the C-terminal Q-rich domain did not increase RNA binding (compare lanes 2–4 with 5–7), although it caused some retention of the complexes at the origin of the gel. Taken together, the binding data argue that pre-mRNA recognition is achieved by RRMs 2 and 3, which is consistent with previous results indicating that these domains can select uridine-rich sequences from a random pool of RNAs (Dember et al., 1996).
U1 snRNP recruitment
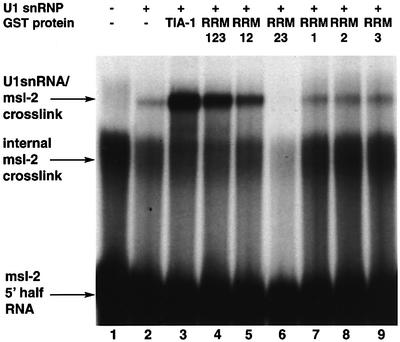
Next, the activity of TIA-1 deletion mutants in promoting the recruitment of U1 snRNP to the msl-2 5′ ss region was analyzed. This was monitored using psoralen-mediated cross-linking, which allows the detection of base-pairing interactions between the 5′ end of U1 snRNA and the 5′ ss of msl-2 (Wassarman and Steitz, 1992; Förch et al., 2000). Figure 2 shows that the presence of TIA-1 resulted in a significant increase in cross-linking of purified U1 snRNP to msl-2 5′ ss (compare lane 2 with 3). Neither the individual RRMs nor a protein containing RRMs 2 and 3 promoted cross-linking of U1 to msl-2 5′ ss (lanes 6–9). Importantly, a protein containing the three RRMs of TIA-1 significantly increased U1 cross-linking (lane 4), suggesting that the presence of RRM1 facilitates U1 snRNP recruitment (compare lane 4 with 6). Consistent with this, the presence of RRM1 in the RRM1–RRM2 fusion protein also provided some recruiting activity compared with RRM2 alone (compare lane 5 with 8). Finally, the activity of a protein containing the three RRM domains was lower than that of the full-length protein, arguing that the C-terminal Q-rich domain of TIA-1 contributes to U1 snRNP recruitment (compare lane 3 with 4). Similar results were obtained when cross-linking was analyzed in nuclear extracts instead of in assays using purified U1 snRNP (data not shown).
Fig. 2. Activity of TIA-1 derivatives in U1 snRNP recruitment. Psoralen-mediated cross-linking was carried out using 32P-labeled msl-2 5′ half RNA, 200 µg of purified U1 snRNP and 500 ng of GST–TIA-1 or deletion mutants, as indicated. After irradiation with 365 nm UV light, RNAs were purified and fractionated on polyacrylamide denaturing gels and the gels autoradiographed. The positions of free msl-2 5′ half RNA, an internal cross-link of this RNA and U1 snRNA–msl-2 adducts are indicated.
Taken together, the results of Figures 1 and 2 argue that RRMs 2 and 3 are important for pre-mRNA recognition, and that both RRM1 and the Q-rich domain contribute to the recruitment of U1 snRNP to the msl-2 5′ ss region.
TIA-1–U1 snRNP interaction
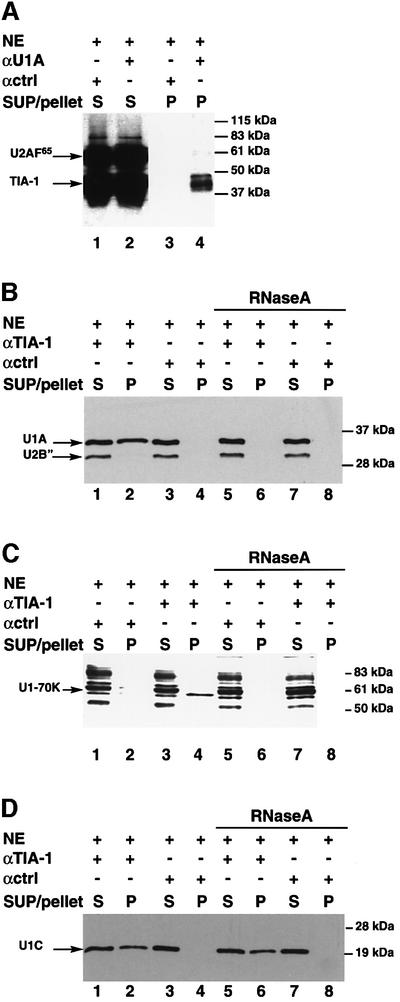
TIA-1 could promote U1 snRNP recruitment by establishing interactions with components of the snRNP. TIA-1, however, was not detected in purified preparations of U1 snRNP (Bringmann and Lührmann, 1986), suggesting that if such interactions exist, they are not as strong as those of stable components of the particle. To probe for relatively weak interactions between U1 snRNP and TIA-1, U1 snRNP was immunoprecipitated from HeLa nuclear extracts using an antibody against the U1-specific protein U1-A (Kambach and Mattaj, 1992), and the presence of TIA-1 in the precipitates was analyzed by western blot. As a control, the presence of the splicing factor U2AF65 was also tested. Low levels of TIA-1, but not of U2AF65, were detected reproducibly in precipitates of the snRNP, but not in precipitates using a control antibody (Figure 3A, compare lane 3 with 4). TIA-1 was also detectable in immunoprecipitates using antibodies against the other two U1-specific polypeptides, U1-70K and U1-C (see Supplementary data, available at The EMBO Journal Online).
Fig. 3. Association between TIA-1 and U1 snRNP. (A) Immuno precipitates of U1 snRNP contain TIA-1. Immunoprecipitates (P) of U1 snRNP present in HeLa cell nuclear extracts (NE) using anti-U1-A or control antibodies were analyzed by western blot for the presence of TIA-1 and U2AF65. Aliquots (10%) of the supernatants (S) were also analyzed in parallel. The positions of TIA-1, U2AF65 and molecular weight markers are indicated. (B) Immunoprecipitation of TIA-1 co-precipitates U1-A, but not U2-B′′. Anti-TIA-1 antibodies were used to immunoprecipitate U1 snRNP from NE or NE digested by RNase A prior to incubation. The presence of U1A and U2B′′ in the pellets was analyzed by western blot using an antibody that recognizes both U1-A and U2-B′′. Aliquots (5%) of the supernatants (SUP) were also analyzed in parallel. The positions of the U1-A and U2-B′′ proteins and molecular weight markers are indicated. (C) U1-70K is co-precipitated by TIA-1 antibodies. The experiment was performed as in (B) using U1-70K antibodies for western blot. (D) U1-C co-precipitates with TIA-1 in the presence or absence of treatment of the NE with RNase A. The assay was performed as described in (B) using anti-U1-C antibodies for western blot.
To validate further the association of TIA-1 with U1 snRNP, TIA-1 was immunoprecipitated with specific antibodies and the presence of U1 snRNP-specific proteins in the precipitates analyzed by western blot. Figure 3B and C shows that U1-A and U1-70K were detected in TIA-1 precipitates (compare lane 2 with 4). In contrast, the U2 snRNP-specific protein U2-B′′, which is recognized by the same antibody used to detect U1-A, was not detected in the precipitates, arguing that the association of TIA-1 with U1 snRNP is specific.
Both U1-A and U1-70K were absent when the extracts were treated with RNase A prior to immunoprecipitation (Figure 3B and C, compare lanes 1–4 with 5–8), suggesting that TIA-1 does not contact these proteins directly, and therefore that their presence in TIA-1 precipitates is most likely an indirect consequence of precipitation of intact U1 snRNP. Significantly, however, U1-C was detectable in TIA-1 immunoprecipitates in both the presence and the absence of RNase A treatment (Figure 3D), suggesting that the TIA-1–U1-C association is not dependent upon the integrity of the U1 particle.
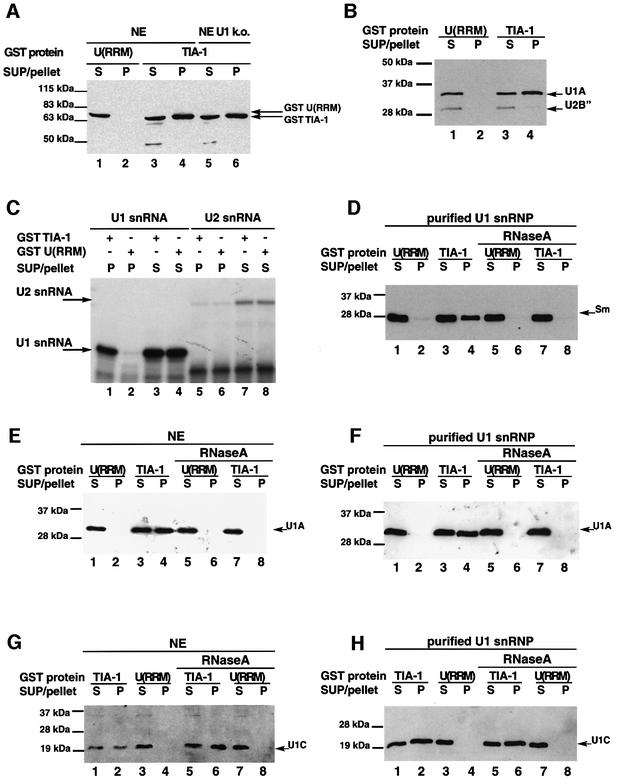
To document these observations further, we tested whether recombinant TIA-1 added to extracts also associates with U1 snRNP. GST–TIA-1 or a GST fusion containing the three RRM domains of U2AF65 [U(RRM)] were added to nuclear extracts and their association with U1 snRNP analyzed by probing for the presence of GST fusion proteins in immunoprecipitates of U1-A. Full-length U2AF65 was not used as a control in these experiments because the positively charged RS domain present at the N-terminus of the protein associates non-specifically with ribonucleoprotein complexes (data not shown). While GST–TIA-1 was detected in U1 precipitates, the control protein GST–U(RRM) was not (Figure 4A, compare lane 2 with 4). Association of GST–TIA-1 with U1 snRNP was also observed in extracts in which the 5′ end of U1 snRNA was degraded efficiently by RNase H in the presence of a complementary DNA oligo (lane 6), suggesting that the interaction was not a consequence of U1 assembly on pre-mRNAs present in the extract. Consistent with this idea, the levels of GST–TIA-1 in the precipitates were not increased by addition to the reaction of pre-mRNAs containing TIA-1-binding sites (data not shown).
Fig. 4. Recombinant TIA-1 associates with U1 snRNP. (A) Co-precipitation of GST–TIA-1 with U1 snRNP is independent of the integrity of the 5′ end of U1 snRNA. Anti-U1-A antibodies were used to immunoprecipitate U1 snRNP from HeLa nuclear extracts (NE) or from extracts in which the 5′ end of U1 snRNA was degraded using RNase H (NE U1 k.o.), in the presence of GST–TIA-1 or GST–U(RRM). The presence of recombinant proteins in the pellets was analyzed by western blot using anti-GST antibodies. Aliquots (5%) of the supernatants (SUP) were also analyzed in parallel. The positions of the GST fusion proteins and molecular weight markers are indicated. (B) Precipitation of U1 snRNP by GST–TIA-1. GST pull-down assays were carried out after addition of GST–TIA-1 or a control GST–U(RRM) protein to nuclear extracts, and the pellets analyzed by western blot using an antibody that recognizes U1-A and U2-B′′. Aliquots (5%) of the supernatants (SUP) were analyzed in parallel. The positions of U1-A, U2-B′′ and molecular weight markers are indicated. (C) Precipitation of U1 snRNA by GST–TIA-1. GST pull-down assays were carried out after addition of GST–TIA-1 or a control GST–U(RRM) protein to nuclear extracts, and RNAs present in the pellets were analyzed by primer extension using oligo nucleotides specific for U1 or U2 snRNAs. Aliquots (5%) of the supernatants (SUP) of the precipitations were also analyzed in parallel. The positions of the products of primer extension corresponding to U1 and U2 snRNAs are indicated. (D) RNase A-dependent interaction with Sm proteins from purified U1 snRNP. GST pull-down assays were carried out after incubation of GST–TIA-1 [or GST–U(RRM) as a control] with purified U1 snRNP. The presence of Sm proteins in the precipitates (position indicated by an arrow) was analyzed by western blot using Y12 antibodies. Treatment of the extracts with RNase A prior to the pull-down assays is indicated. (E) RNase A-dependent interaction with U1-A in nuclear extracts. Assays were performed as in (D) and the blots developed with anti-U1-A antibodies. (F) RNase A-dependent interaction with U1-A from purified U1 snRNP. Assays were performed as in (E) using purified U1 snRNP instead of nuclear extract. (G) RNase A-independent interaction with U1-C in nuclear extracts. Assays were carried out as in (D) and the blots developed with antibodies against U1-C. (H) RNase A-independent interaction with U1-C from purified U1 snRNP. Assays were performed as in (G) using using purified U1 snRNP instead of nuclear extract.
Reciprocal experiments were also consistent with a specific association between TIA-1 and U1 snRNP: GST–TIA-1 incubated with nuclear extracts was precipitated using glutathione–agarose beads, and the presence of U1 snRNP components in the precipitate analyzed. Western blot analysis using the antibody that recognizes both U1-A and the U2-specific protein U2-B′′ indicated that U1-A was detected in GST–TIA-1 precipitates, while U2-B′′ was barely detectable (Figure 4B, lane 4), consistent with the results of immunoprecipitation using TIA-1 antibodies (Figure 3B). Precipitation of the GST–U(RRM) fusion protein did not result in precipitation of either U1-A or U2-B′′ (lane 2). Primer extension analysis indicated that U1 snRNA, but not U2 snRNA, was enriched in GST–TIA-1 precipitates compared with GST–U(RRM) control precipitates (Figure 4C, compare lanes 1 and 2 with lanes 5 and 6). We conclude that recombinant TIA-1 establishes specific interactions with U1 snRNP components but not with other snRNPs such as U2.
The GST pull-down assay was also used to confirm the results of co-immunoprecipitation experiments regarding the nature of these interactions. Pull-down assays were carried out as described above, and the presence of different protein components of U1 snRNP in the precipitates analyzed by western blot. Once again, to distinguish between direct interactions and those detected as an indirect consequence of precipitation of the snRNP, the assays were carried out with and without treatment of the nuclear extracts with RNase A prior to the precipitation. To assess whether the interactions observed involved bridging factors between TIA-1 and U1 snRNP, both U1 from nuclear extracts and purified snRNP particles were tested in co-precipitation assays.
Sm proteins and U1A were precipitated by GST–TIA-1 in the absence of RNase A treatment, using both nuclear extracts and purified snRNP particles (Figure 4D–F, lanes 3 and 4; Supplementary data). Sm and U1-A proteins were absent, however, from the precipitates upon RNase A digestion (Figures 4D–F, lanes 7 and 8). These results argue that interactions between TIA-1 and Sm or U1-A take place in the context of U1 snRNP, but are not direct, as they depend upon the integrity of U1 snRNA. For reasons presently unclear, the protein U1-70K was undetectable in GST–TIA-1 precipitates, even in the absence or RNase A (see Supplementary data and Discussion).
In contrast, U1-C was present in GST–TIA-1 precipitates, even with RNase A treatment (Figure 4G and H, lanes 1 and 2, and 5 and 6), once more confirming that the association of TIA-1 and U1-C is likely to be based on direct protein–protein interactions.
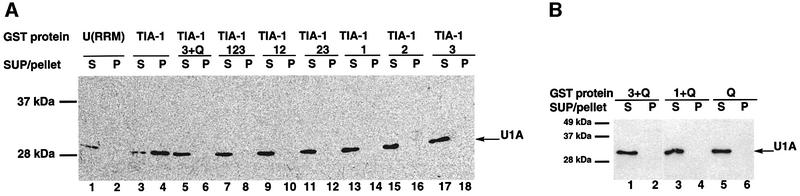
Finally, the GST pull-down assay was also used to identify the domains of TIA-1 involved in interaction with U1 snRNP. GST fusions of various TIA-1 deletion mutants were added to nuclear extracts, and co-precipitation of U1 snRNP was measured by western blot using antibodies against U1-A. The results of Figure 5A and B indicate that only full-length TIA-1 protein was able to precipitate detectable amounts of U1 snRNP.
Fig. 5. Functional domain analysis of the TIA-1–U1 snRNP association. (A) The integrity of TIA-1 is required for association with U1 snRNP. Recombinant GST–TIA-1 and deletion mutants were added to nuclear extracts, and the products of precipitation (P) with glutathione–agarose beads analyzed for the presence of U1-A by western blot. Aliquots of the supernatants (S) were analyzed in parallel. The positions of U1-A and molecular weight markers are indicated. (B) The Q-rich domain is not sufficient for association with U1 snRNP. The experiment was carried out as in (A) using GST fusions corresponding to the Q-rich domain alone or combinations of Q and individual RRMs, as indicated.
Direct interaction between TIA-1 and U1-C
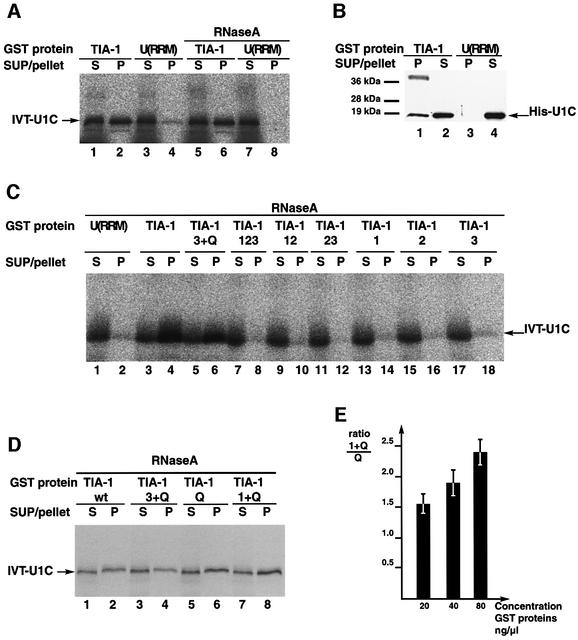
To test whether TIA-1 contacts U1-C directly, 35S-labeled U1-C was produced by in vitro translation in rabbit reticulocyte extracts, incubated with GST–TIA-1 and, after precipitation using glutathione–agarose beads, the presence of U1-C in the precipitates assessed by fractionation in SDS denaturing gels and autoradiography. U1-C was found to co-precipitate with TIA-1 (Figure 6A, lanes 1 and 2) but only marginally with the GST–U(RRM) control (lanes 3 and 4). The same result was obtained when the extracts were digested with RNase A after completion of in vitro translation (lanes 5 and 6, and 7 and 8).
Fig. 6. Direct interaction between TIA-1 and U1-C. (A) Interaction between GST–TIA-1 and in vitro translated U1-C. Assays were carried out as in Figure 5, using 35S-labeled U1-C translated in vitro instead of nuclear extracts or purified U1 snRNP. (B) Recombinant purified U1-C interacts with GST–TIA-1. The assay was carried out as in (A) using recombinant purified histidine-tagged U1-C instead of in vitro translated protein. The presence of U1-C (His-U1-C) in the precipitates was analyzed by western blot. (C) Functional domain analysis of TIA-1–U1-C interaction. Pull-down assays were carried out with RNase A treatment as in (A), using the GST fusion proteins indicated. (D) The presence of RRM1 enhances interaction with U1-C. Assays were as in (C) using the indicated recombinant purified proteins. (E) Quantification of the enhancement effect of RRM1 in U1-C interaction. The ratio between the amounts of U1-C precipitated by the RRM1 + Q and Q proteins is represented against the concentration of proteins used in the assay. Averages and standard deviations are indicated.
As a second test for direct interactions between TIA-1 and U1-C, pull-down assays were carried out using recombinant U1-C expressed in and purified from Escherichia coli. Consistent with the results obtained using in vitro translated protein, recombinant purified U1-C was co-precipitated with GST–TIA-1, but not with the control GST–U(RRM) (Figure 6B, compare lanes 1 and 2, and 3 and 4). Taken together, the results of Figure 6A and B argue for a direct protein–protein interaction between TIA-1 and U1-C.
Next we determined the domains of TIA-1 necessary for interaction with U1-C. Pull-down assays using individual domains and deletion mutants of TIA-1 indicated that the Q-rich domain was required for co-precipitation of recombinant purified U1-C, as no individual RRM or RRM combination was sufficient (Figure 6C). The Q-rich domain alone was found to be sufficient for interaction with U1-C (Figure 6D, lanes 5 and 6). Although RRM1 by itself had no capacity to co-precipitate U1-C (Figure 6C, lanes 13 and 14), and only marginally when combined with RRMs 2 and 3 (lanes 7–8), a fusion protein between RRM1 and the Q-rich domain showed enhanced interaction with U1-C compared with that of the Q-rich domain alone (Figure 6D, compare lanes 5 and 6 with 7 and 8). The effect of RRM1 was highly reproducible, was more evident at higher concentrations of protein (Figure 6E) and was not observed when other RRM motifs were fused to the Q-rich domain (Figure 6D, lanes 3 and 4).
We conclude that the Q-rich domain of TIA-1, particularly when combined with the RRM1 domain, contributes to the interaction with U1-C. As these domains were also important for association with U1 snRNP, and for its recruitment to 5′ ss regions, the results are consistent with the hypothesis that interaction with U1-C is important for the activity of TIA-1 in U1 snRNP recruitment.
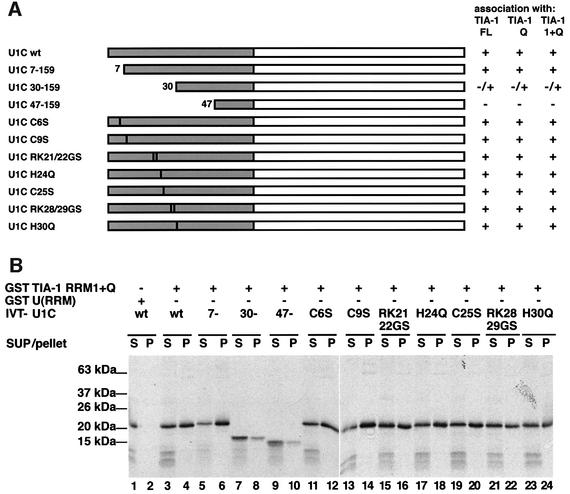
To define the region of U1-C that interacts with TIA-1, in vitro translated U1-C and mutant derivatives (kindly provided by G.Pruijn and W.van Venrooij) (Figure 7A) were tested in pull-down assays using GST–TIA-1, GST–Q and GST–RRM1–Q, with similar results (Figure 7B; data not shown). Deletion of the 30 or 47 N-terminal residues of U1-C significantly compromised interaction with TIA-1 (compare lanes 3 and 4 with lanes 7 and 8, and 9 and 10). As deletion of the seven N-terminal residues did not affect binding (lanes 5 and 6), we conclude that amino acids between positions 7 and 47 are import ant for interaction with TIA-1. Interestingly, this region includes the zinc finger motif and homodimerization domain of U1-C. Point mutations predicted to disrupt the zinc finger motif or known to prevent homodimerization, however, did not significantly compromise interaction with TIA-1 (lanes 11–24).
Fig. 7. Interaction with TIA-1 involves the N-terminal region of U1-C, but does not require the zinc finger structure or dimerization domains. (A) Schematic representation of deletion and point mutants of human U1-C. Amino acid numbers are indicated. Vertical black bars represent point mutations in the zinc finger or dimerization domains. The first letter corresponds to the mutated residue, the number to its position in the sequence and the second letter to the amino acid change. The white box represents the proline and methionine-rich C-terminal domain of U1-C. Co-precipitation with full-length (FL), glutamine-rich (Q) or glutamine-rich and RRM1 (1 + Q) domains of TIA-1 is indicated by +, – or −/+. (B) Interaction of GST–TIA-1 (RRM1 + Q) with U1-C and mutant derivatives. Assays were performed as in Figure 6A using the U1-C derivatives indicated in (A). Products of in vitro translation were treated with RNase A before pull-down assays were carried out.
Although technical difficulties in expressing C-terminal deletions of U1-C prevent us from being able to conclude that the contact surface is restricted to residues 7–47, we propose that these residues are important, directly or indirectly, for interaction with TIA-1, and that this interaction does not depend on the structure of the zinc finger or homodimerization domains of U1-C.
Discussion
Recent work has shown that the yeast protein Nam8p is a component of U1 snRNP that cross-links to a region of the pre-mRNA downstream from the 5′ ss, modulates U1 snRNP binding depending on the sequence present in that region and facilitates U1 snRNP assembly on weak 5′ ss or uncapped substrates (Gottschalk et al., 1998; Puig et al., 1999; Zhang and Rosbash, 1999). These activities resemble those of TIA-1 in the mammalian system (Del Gatto et al., 2000; Förch et al., 2000; Le Guiner et al., 2001b) and, together with the significant amino acid sequence homology between Nam8p and TIA-1, suggest that the two proteins are functional homologs. A difference between the two systems, however, is that only a limited fraction of the U1 snRNP pool in HeLa nuclear extracts is associated with TIA-1 (Figure 3A), while Nam8p is a stable component of yeast U1. This could be due to relatively weak interactions between TIA-1 and U1 when not assembled on a 5′ ss region. It is conceivable that a less tight association between TIA-1 and U1 snRNP serves to make their mutual stabilizing interactions on 5′ ss more sensitive to regulation, for example by independent modulation of the levels or activity of TIA-1. Alternatively, TIA-1 may associate with a subpopulation of U1 snRNP.
The observation that U1-70K was not detectable in precipitates of pull-down experiments using GST–TIA-1, which contained all other U1 components (see Supplementary data), opens up the possibility that U1 snRNP particles devoid of U1-70K can be recruited to certain 5′ ss by TIA-1. Interestingly, both TIA-1 and U1-70K can interact with the N-terminal region of U1-C (Nelissen et al., 1994; Figure 7). It cannot be ruled out, however, that U1-70K was not lost during the GST–TIA-1 pull-down procedure, which would be more consistent with the observation that TIA-1 can be found in precipitates of U1-70K and vice versa (Figure 3C; Supplementary data).
Several splicing regulators have been shown to modulate U1 snRNP binding to 5′ ss. This is in agreement with the idea that regulation often occurs at the earliest steps in spliceosome assembly that commit pre-mRNAs to the splicing pathway. Thus, the Drosophila regulator Sex-lethal has been shown to bind to uridine-rich sequences downstream from the 5′ ss of an intron at the 5′-untranslated region of msl-2 and prevent U1 snRNP binding by antagonizing TIA-1 activity (Förch et al., 2001).
The SR protein ASF/SF2 promotes U1 snRNP binding and modulates 5′ ss choice, most probably by equalizing differences in U1 binding affinity (Kohtz et al., 1994; Eperon et al., 2000). Protein–protein interactions between the arginine- and serine- (RS) rich domain of ASF/SF2 and a similar domain in U1-70K have been shown to be important for this activity (Wu and Maniatis, 1993; Kohtz et al., 1994; Cao and García-Blanco, 1998).
The Drosophila splicing regulator PSI is important for restriction of P element transposition to the germline. PSI protein is expressed only in the soma, where it is required for splicing inhibition of P element third intron (Siebel et al., 1994, 1995; Adams et al., 1997). PSI is part of a repressive complex that sequesters U1 snRNP at a cryptic, non-productive 5′ ss upstream from the bona fide 5′ ss (Siebel et al., 1992). PSI has been shown to bind directly to U1-70K, and this interaction is important for the regulatory function of PSI (Labourier et al., 2001). The region of PSI that contacts U1-70K has similarities to a sequence within KSRP, a protein that has been involved in alternative splicing of c-src exon N (Min et al., 1997). It is therefore possible that interaction with U1-70K is also at the basis of the regulatory activity of KSRP (Labourier et al., 2001).
The results of Figures 3–7 point to U1-C as one U1 snRNP component involved in direct protein–protein interactions with TIA-1. U1-C has been shown to be required for spliceosome assembly in yeast (Tang et al., 1997) and mammals (Heinrichs et al., 1990; Will et al., 1996). The protein can be cross-linked to the 5′ ss region and is responsible for the first recognition event of the 5′ ss sequence, most probably preceding and/or facilitating base pairing of U1 snRNA with the 5′ ss (Tang et al., 1997; Zhang and Rosbash, 1999; Du and Rosbash, 2002). As TIA-1 binds to the uridine-rich stretch immediately downstream from the 5′ ss (Förch et al., 2000), it is conceivable that the TIA-1–U1-C interaction can stabilize the association of both proteins (and consequently of U1 snRNP) with their adjacent binding sites (Figure 8). Cryo-electron microscopy studies (Stark et al., 2001) could be instrumental in validating or modifying this model.
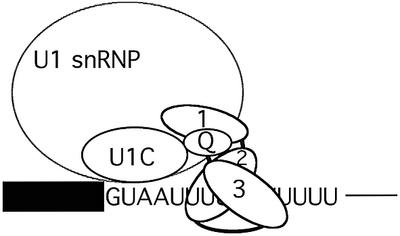
Fig. 8. A mechanism for TIA-1-mediated recruitment of U1 snRNP to weak 5′ ss followed by uridine-rich sequences. TIA-1 RRMs 2 and 3 recognize the uridine (U)-rich stretch, while the Q-rich domain contacts the U1-specific protein U1-C. U1-C may recognize the 5′ ss sequence (in this case represented by the non-consensus GUAAUU sequence found in msl-2 RNA) even before base-pairing interactions with U1 snRNA are established (Du and Rosbash, 2002). Thus, the U1-C–TIA-1 interaction can facilitate cooperative binding of the two proteins to their adjacent sites in the pre-mRNA and thereby U1 snRNP recruitment. RRM1 is important for TIA-1 activity and enhances the interaction of the Q-rich domain with U1-C. Additional activities of RRM1 are likely to contribute to U1 snRNP recruitment. The box represents the exon preceding the 5′ ss and the thin line indicates intronic sequences downstream from the TIA-1-binding site.
Interestingly, several other proteins have been shown to contact U1-C and be implicated in splicing regulation. The gene EWS encodes a transcription factor, and its translocation is often associated with progression of a variety of tumors. Translocation of the EWS locus to the gene encoding another transcription factor, FLI, or that encoding the nuclear receptor NOR1 gives rise to fusion proteins that have been shown to have, in addition to abnormal properties as transcriptional activators, the ability to alter 5′ ss selection (Knoop and Baker, 2000, 2001; Ohkura et al., 2002). Interestingly, both EWS/FLI and EWS/NOR1 interact with U1-C and, remarkably, EWS/NOR1 expression can complement loss-of-function mutants of snu23p, a component of yeast spliceosomes (Gottschalk et al., 1998; Knoop and Baker, 2000; Ohkura et al., 2002). These observations open up the possibility that the regulatory activities of these proteins are based on direct contacts with U1-C, as for TIA-1.
The interaction of TIA-1 with U1-C requires a region of the protein that includes the cystidine/histidine zinc finger-like motif (Nelissen et al., 1991), which is essential for interaction with U1 snRNP (Nelissen et al., 1994) and also for homodimerization (Gunnewiek et al., 1995). The fact that mutations that disrupt these structural elements do not compromise TIA-1 binding (Figure 7B) argue that distinct surfaces mediate U1 snRNP attachment and TIA-1 binding.
Both RRM1 and the Q-rich domain contribute to U1-C binding, association with U1 snRNP and snRNP recruitment to 5′ ss (Figures 2, 4 and 6). While Q-rich domains have often been involved in protein–protein interactions (McBride and Silver, 2001), the RRM fold is a characteristic RNA-binding module (Varani and Nagai, 1998). RRM1, however, does not contribute to binding to the pre-mRNA, as expected from the presence of negatively charged residues within a conserved octamer motif involved in direct contacts with RNA (Varani and Nagai, 1998). Protein–protein interactions mediated by RRM domains, however, have also been reported previously (Scherly et al., 1990; Kielkopf et al., 2001).
While RRM1 was essential for association of TIA-1 with U1 snRNP (Figure 4), and contributes significantly to U1 snRNP recruitment, even in the absence of the Q-rich domain (Figure 2), it failed to show by itself detectable binding to U1-C (Figure 6C). It promoted, however, binding to U1-C in the presence of the Q-rich domain (Figure 6E). It is possible that the U1-C-binding activity of RRM1 will be maximal only in the context of the complete protein, or upon recognition of the pre-mRNA by RRMs 2 and 3. Alternatively, RRM1 could contribute to U1 snRNP recruitment through other mechanisms in addition to facilitating contacts with U1-C (Figure 8). Finally, it is also possible that quantitative differences between activities for the various domain mutants are due to inherent differences in the stringency or detection levels of the assays used. In this context, it is worth noting that U1 snRNP binding involves multiple RNA–RNA, protein– RNA and protein–protein interactions, and that even the well-established base-pairing interactions between the 5′ end of U1 snRNA and the 5′ ss can be dispensable under certain experimental conditions (Du and Rosbash, 2001, 2002; Lund and Kjems, 2002).
Previous work has shown that TIA-1 modulates alternative splicing of the fibroblast growth factor (FGF) receptor 2 (Del Gatto et al., 2000). TIA-1 promotes inclusion of the K-SAM exon by binding downstream from the 5′ ss associated with the exon. The mechanism of TIA-1 function on this pre-mRNA can therefore be assimilated easily with that in msl-2: higher levels of TIA-1 activity will promote U1 snRNP assembly on exon K-SAM 5′ ss and therefore K-SAM exon inclusion. Additional cis-acting elements contribute to fine tuning of this splicing event (Del Gatto et al., 1997; Le Guiner et al., 2001a).
In summary, our results provide one molecular mechanism for how TIA-1 can promote U1 snRNP recruitment to 5′ ss by portraying a direct interaction between TIA-1 and U1-C as an important component of these effects.
Materials and methods
RNAs and recombinant proteins
In vitro transcription of msl-2 was as described (Gebauer et al., 1998; Förch et al., 2000). GST fusion proteins were expressed in and purified from E.coli as previously described (Förch et al., 2000). TIA-1 deletion mutants include: RRM1 (amino acids 1–92); RRM1 + 2 (amino acids 1–196); RRM1 + 2 + 3 (amino acids 1–273); RRM2 (amino acids 93–196); RRM2 + 3 (amino acids 93–273); RRM3 (amino acids 197–273); RRM3 + Q (amino acids 186–375, also known as T4T8); Q (amino acids 274–375); and RRM1 + Q (amino acids 1–92 + 274–375). These constructs were described previously (Dember et al., 1996), with the exception of Q and RRM1 + Q, which were generated by PCR, cloned in pGEX2T and confirmed by sequencing. GST–U(RRM) corresponds to GST–U2AF65 lacking the N-terminal 93 amino acids (Valcárcel et al., 1996). Recombinant histidine-tagged U1C was prepared as described previously (Gunnewiek et al., 1995). Templates for in vitro translation of U1-C were described previously (Nelissen et al., 1991; Gunnewiek et al., 1995). Deletion and point mutants of U1-C were U1-C 7-[amino acids 7–159]; U1-C 30-[amino acids 30–159]; U1-C 47-[amino acids 47–159]; C6S, substitution of cysteine at position 6 by serine; C9S, substitution of cysteine at position 9 by serine; RK21/22GS, substitution of arginine and lysine at positions 21 and 22 by glycine and serine, respectively; H24Q, substitution of histidine at position 24 by glutamine; C25S, substitution of cysteine at position 25 by serine; RK28/29GS, substitution of arginine and lysine at positions 28 and 29 by glycine and serine, respectively; and H30Q, substitution of histidine at position 30 by glutamine. 35S-Labeled U1-C and derivatives were generated in rat reticulocyte lysates using the T7 Quick transcription/translation system (Promega) according to the manufacturer’s instructions.
Nuclear extracts and purification of U1 snRNP
Nuclear extract was prepared as described previously by Dignam et al. (1983). Purification of U1 snRNP and purity criteria were as described previously by Lewis et al. (1996).
RNase H-mediated inactivation of U1 snRNP
Inactivation of U1 snRNA in HeLa nuclear extracts by RNase H in the presence of a DNA oligonucleotide complementary to the 5′ end of U1 snRNA was performed as described previously (Merendino et al., 1999).
RNA binding assays
A 2 fmol concentration of uniformly 32P-labeled msl-2 5′ half RNAs was incubated with 10–6, 10–7 or 10–8 M GST–TIA-1 or deletion mutants in a total volume of 15 µl of buffer D with 0.1 M KCl (Dignam et al., 1983) in the presence of tRNA (200 ng/µl) for 30 min on ice. After incubation, the samples were fractionated by electrophoresis on non-denaturing 6% polyacrylamide gels (acrylamide:bis-acrylamide ratio 60:1), dried and exposed to film.
Psoralen-mediated UV cross-linking
Radioactively labeled RNA (10 fmol) was incubated with 1 µl (200 ng/µl) of partially purified U1 snRNP in the presence of 13 µg/ml 4′ aminomethyl-4,5′,8-trimethylpsoralen, in the absence or presence of 500 ng of recombinant GST–TIA-1 or mutant derivatives in a total volume of 15 µl on ice. The samples were irradiated for 10 min with 365 nm UV light in a Stratalinker oven at 0°C. RNA was purified by digestion with proteinase K, phenol/chloroform extraction and ethanol precipitation, and analyzed by electrophoresis on a 6% denaturing polyacrylamide gel. As previously described (Förch et al., 2000), daylight was avoided in psoralen cross-linking experiments using msl-2 RNA.
Immunoprecipitation
Anti-U1A antibodies (856; Kambach and Mattaj, 1992), anti-TIA-1 (anti-2G9; Anderson et al., 1990), anti-U170k (16H3; Neugebauer et al., 1995) and anti-U1C (Dumortier et al., 1998) were bound to protein A–Sepharose beads and incubated in 15 µl of nuclear extracts complemented with buffer D to a total volume of 25 µl for 30 min on ice. After addition of 60 µl of IPP 150 buffer (10 mM Tris pH 8.0, 150 mM NaCl, 0.1% NP-40), the reaction was incubated for 2 h on a rotating wheel at 4°C. An 8 µl aliquot per sample was removed as loading control, and the beads were sedimented at 1000 r.p.m. and washed four times with ice-cold IPP 150. Loading control and pellet were fractionated by electrophoresis on a 10% SDS–polyacrylamide gel, blotted onto a nitrocellulose membrane (Schleicher & Schuell) and probed with antibodies against TIA-1 (anti-2G9; Anderson et al., 1990), U2AF65 (MC3; Gama-Carvalho et al., 1997), GST (B14; Santa Cruz), anti U1A/U2B′′ (9A9; Habets et al., 1989), anti-U1-70k (16H3) and anti-U1-C. Anti-mouse or anti-rabbit antibodies coupled to horseradish peroxidase (HRP) were used as secondary reagents, and visualized by chemoluminescence (ECL, Amersham).
GST pull-down and western blot analyses
A 2 µg aliquot of recombinant GST–TIA-1 or mutant derivatives was incubated with 15 µl of nuclear extract, 3 µl of in vitro translated U1-C in RNase A-treated reticulocyte lysates, 2 µg of recombinant U1-C or 3 µg of purified U1 snRNP in a total volume of 25 µl, complemented with buffer D (20 mM HEPES pH 8.0, 20% glycerol, 0.2 mM EDTA, 100 mM KCl), for 30 min at 0°C. After addition of 60 µl of IPP 150 and 10 µl of glutatione–agarose beads (Sigma) pre-equilibrated in the same buffer, the reaction was incubated for 2 h on a rotating wheel at 4°C. A 4 µl aliquot of the reaction was removed for loading control and the pellet was washed four times with 400 µl of ice-cold IPP 150 buffer. After sedimentation by centrifugation at 1000 r.p.m. for 1 min, the pellet was resuspended in SDS loading buffer, fractionated together with the aliquot of supernatant by electrophoresis on a 10–15% SDS–polyacrylamide gel and transferred onto nitrocellulose (Schleicher & Schuell). The blots were probed with anti-TIA-1, anti-U1A, anti-U1A/U2B′′, anti-U1-70k, anti-U1C (Dumortier et al., 1998; Hoet et al., 1998), anti-Sm (Y12; Pisetsky and Lerner, 1982) or anti-GST antibodies. HRP-conjugated anti-mouse or anti-rabbit antibodies were used as secondary reagents, and visualized by enhanced chemoluminescence (ECL; Amersham).
Primer extension
GST pull-down assays were scaled up 2-fold and carried out as in the previous section. RNA was isolated from an aliquot from supernatant and pellet by digestion with proteinase K, phenol/chloroform extraction and precipitation with ethanol. Primer extension was carried out using 5′ end 32P-labeled antisense U1 snRNA- or U2 snRNA-specific oligonucleotides and AMV reverse transcriptase (Promega) for 1 h at 42°C. The products of primer extension were analyzed on a 10% denaturing acrylamide gel, dried and exposed to film.
Supplementary data
Supplementary data available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank N.Kedersha, P.Anderson, G.Pruijn, S.Muller, W.van Venrooij, R.Lührmann, D.Bilbao and I.Mattaj for reagents, D.Bilbao and other colleagues at the EMBL Gene Expression Programme for critical reading of the manuscript, and I.Mattaj for discussions. P.F. was supported by fellowships from the Gertrud Reemtsma and Boehringer-Ingelheim Stiftung, and O.P. by a fellowship from Fundación Ramón Areces. This work was supported in part by a grant from the Human Frontier Science Program Organization.
References
- Adams M.D., Tarng,R.S. and Rio,D.C. (1997) The alternative splicing factor PSI regulates P-element third intron splicing in vivo. Genes Dev., 11, 129–138. [DOI] [PubMed] [Google Scholar]
- Anderson P., Nagler-Anderson,C., O’Brien,C., Levine,H., Watkins,S., Slayter,H.S., Blue,M.L. and Schlossman,S.F. (1990) A monoclonal antibody reactive with a 15-kDa cytoplasmic granule-associated protein defines a subpopulation of CD8+ T lymphocytes. J. Immunol., 144, 574–582. [PubMed] [Google Scholar]
- Bringmann P. and Lührmann,R. (1986) Purification of the individual snRNPs U1, U2, U5 and U4/U6 from HeLa cells and characterization of their protein constituents. EMBO J., 5, 3509–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W. and Garcia-Blanco,M.A. (1998) A serine/arginine-rich domain in the human U1 70k protein is necessary and sufficient for ASF/SF2 binding. J. Biol. Chem., 273, 20629–20635. [DOI] [PubMed] [Google Scholar]
- DelGatto F., Plet,A., Gesnel,M.C., Fort,C. and Breathnach,R. (1997) Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol., 17, 5106–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelGatto-Konczak F., Bourgeois,C.F., Le Guiner,C., Kister,L., Gesnel,M.C., Stevenin,J. and Breathnach,R. (2000) The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol., 20, 6287–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dember L.M., Kim,N.D., Liu,K.Q. and Anderson,P. (1996) Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem., 271, 2783–2788. [DOI] [PubMed] [Google Scholar]
- Dignam J., Lebovitz,R. and Roeder,R. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H. and Rosbash,M. (2001) Yeast U1 snRNP–pre-mRNA complex formation without U1snRNA–pre-mRNA base pairing. RNA, 7, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H. and Rosbash,M. (2002) The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base-pairing. Nature, 419, 86–90. [DOI] [PubMed] [Google Scholar]
- Dumortier H., Klein Gunnewiek,J., Roussel,J.P., van Aarssen,Y., Briand,J.P., van Venrooij,W.J. and Muller,S. (1998) At least three linear regions but not the zinc-finger domain of U1C protein are exposed at the surface of the protein in solution and on the human spliceosomal U1 snRNP particle. Nucleic Acids Res., 26, 5486–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon I.C., Makarova,O.V., Mayeda,A., Munroe,S.H., Caceres,J.F., Hayward,D.G. and Krainer,A.R. (2000) Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol., 20, 8303–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förch P., Puig,O., Kedersha,N., Martinez,C., Granneman,S., Seraphin,B., Anderson,P. and Valcarcel,J. (2000) The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell, 6, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Förch P., Merendino,L., Martinez,C. and Valcarcel,J. (2001) Modulation of msl-2 5′ splice site recognition by Sex-lethal. RNA, 7, 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho M., Krauss,R., Chiang,L., Valcárcel,J., Green,M. and Carmo-Fonseca,M. (1997) Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol., 137, 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F., Merendino,L., Hentze,M. and Valcárcel,J. (1998) The Drosophila splicing regulator Sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. RNA, 4, 142–150. [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A. et al. (1998) A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA, 4, 374–393. [PMC free article] [PubMed] [Google Scholar]
- Gunnewiek J.M., van Aarssen,Y., Wassenaar,R., Legrain,P., van Venrooij,W.J. and Nelissen,R.L. (1995) Homodimerization of the human U1 snRNP-specific protein C. Nucleic Acids Res., 23, 4864–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets W.J., Hoet,M.H., De Jong,B.A., Van der Kemp,A. and Van Venrooij,W.J. (1989) Mapping of B cell epitopes on small nuclear ribo nucleoproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J. Immunol., 143, 2560–2566. [PubMed] [Google Scholar]
- Hamm J., Kazmaier,M. and Mattaj,I.W. (1987) In vitro assembly of U1 snRNPs. EMBO J., 6, 3479–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings M.L. and Krainer,A.R. (2001) Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol., 13, 302–309. [DOI] [PubMed] [Google Scholar]
- Heinrichs V., Bach,M. and Lührmann,R. (1990) U1-specific protein C is required for efficient complex formation of U1 snRNP with a 5′ splice site. Mol. Biol. Rep., 14, 165. [DOI] [PubMed] [Google Scholar]
- Hoet R.M., Raats,J.M., de Wildt,R., Dumortier,H., Muller,S., van den Hoogen,F. and van Venrooij,W.J. (1998) Human monoclonal autoantibody fragments from combinatorial antibody libraries directed to the U1snRNP associated U1C protein; epitope mapping, immunolocalization and V-gene usage. Mol. Immunol., 35, 1045–1055. [DOI] [PubMed] [Google Scholar]
- Kambach C. and Mattaj,I.W. (1992) Intracellular distribution of the U1A protein depends on active transport and nuclear binding to U1 snRNA. J. Cell Biol., 118, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C., Walke,S., Young,R., Avis,J.M., de la Fortelle,E., Raker,V.A., Lührmann,R., Li,J. and Nagai,K. (1999) Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell, 96, 375–387. [DOI] [PubMed] [Google Scholar]
- Kielkopf C.L., Rodionova,N.A., Green,M.R. and Burley,S.K. (2001) A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell, 106, 595–605. [DOI] [PubMed] [Google Scholar]
- Knoop L.L. and Baker,S.J. (2000) The splicing factor U1C represses EWS/FLI-mediated transactivation. J. Biol. Chem., 275, 24865–24871. [DOI] [PubMed] [Google Scholar]
- Knoop L.L. and Baker,S.J. (2001) EWS/FLI alters 5′-splice site selection. J. Biol. Chem., 276, 22317–22322. [DOI] [PubMed] [Google Scholar]
- Kohtz J., Jamison,S., Will,C., Zuo,P., Lührmann,R., García-Blanco,M. and Manley,J. (1994) RS domain mediated interactions between ASF/SF2 and U1 snRNP: a mechanism for 5′ splice site recognition in mammalian mRNA precursors. Nature, 368, 119–124. [DOI] [PubMed] [Google Scholar]
- Labourier E., Adams,M.D. and Rio,D.C. (2001) Modulation of P-element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70K protein. Mol. Cell, 8, 363–373. [DOI] [PubMed] [Google Scholar]
- Le Guiner C., Plet,A., Galiana,D., Gesnel,M.C., Del Gatto-Konczak,F. and Breathnach,R. (2001a) Polypyrimidine tract-binding protein represses splicing of a fibroblast growth factor receptor-2 gene alternative exon through exon sequences. J. Biol. Chem., 276, 43677–43687. [DOI] [PubMed] [Google Scholar]
- Le Guiner C., Lejeune,F., Galiana,D., Kister,L., Breathnach,R., Stevenin,J. and Del Gatto-Konczak,F. (2001b) TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem., 276, 40638–40646. [DOI] [PubMed] [Google Scholar]
- Lewis J.D., Izaurralde,E., Jarmolowski,A., McGuigan,C. and Mattaj,I.W. (1996) A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev., 10, 1683–1698. [DOI] [PubMed] [Google Scholar]
- Lührmann R., Kastner,B. and Bach,M. (1990) Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta, 1087, 265–292. [DOI] [PubMed] [Google Scholar]
- Lund M. and Kjems,J. (2002). Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA, 8, 166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz-Freyermuth C., Query,C.C. and Keene,J.D. (1990) Quantitative determination that one of two potential RNA-binding domains of the A protein component of the U1 small nuclear ribonucleoprotein complex binds with high affinity to stem–loop II of U1 RNA. Proc. Natl Acad. Sci. USA, 87, 6393–6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride A.E. and Silver,P.A. (2001) State of the arg: protein methylation at arginine comes of age. Cell, 106, 5–8. [DOI] [PubMed] [Google Scholar]
- Merendino L., Guth,S., Bilbao,D., Martinez,C. and Valcarcel,J. (1999) Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature, 402, 838–841. [DOI] [PubMed] [Google Scholar]
- Michaud S. and Reed,R. (1991) An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev., 5, 2534–2546. [DOI] [PubMed] [Google Scholar]
- Michaud S. and Reed,R. (1993) A functional association between the 5′ and 3′ splice sites is established in the earliest prespliceosome complex (E) in mammals. Genes Dev., 7, 1008–1020. [DOI] [PubMed] [Google Scholar]
- Min H., Turck,C.W., Nikolic,J.M. and Black,D.L. (1997) A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev., 11, 1023–1036. [DOI] [PubMed] [Google Scholar]
- Modrek B. and Lee,C. (2002) A genomic view of alternative splicing. Nat. Genet., 30, 13–19. [DOI] [PubMed] [Google Scholar]
- Nelissen R.L., Heinrichs,V., Habets,W.J., Simons,F., Lührmann,R. and van Venrooij,W.J. (1991) Zinc finger-like structure in U1-specific protein C is essential for specific binding to U1 snRNP. Nucleic Acids Res., 19, 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen R.L., Will,C.L., van Venrooij,W.J. and Lührmann,R. (1994) The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J., 13, 4113–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer K.M., Stolk,J.A. and Roth,M.B. (1995) A conserved epitope on a subset of SR proteins defines a larger family of pre-mRNA splicing factors. J. Cell Biol., 129, 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T.W. (2002) The spliceosome: no assembly required? Mol. Cell, 9, 8–9. [DOI] [PubMed] [Google Scholar]
- Ohkura N., Yaguchi,H., Tsukada,T. and Yamaguchi,K. (2002) The EWS/NOR1 fusion gene product gains a novel activity affecting pre-mRNA splicing. J. Biol. Chem., 277, 535–543. [DOI] [PubMed] [Google Scholar]
- Patton J.R. and Pederson,T. (1988) The Mr 70 000 protein of the U1 small nuclear ribonucleoprotein particle binds to the 5′ stem–loop of U1 RNA and interacts with Sm domain proteins. Proc. Natl Acad. Sci. USA, 85, 747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisetsky D.S. and Lerner,E.A. (1982) Idiotypic analysis of a monoclonal anti-Sm antibody. J. Immunol., 129, 1489–1492. [PubMed] [Google Scholar]
- Puig O., Gottschalk,A., Fabrizio,P. and Seraphin,B. (1999) Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev., 13, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke J., Appel,B., Blocker,H., Frank,R. and Lührmann,R. (1984) The 5′-terminal sequence of U1 RNA complementary to the consensus 5′ splice site of hnRNA is single-stranded in intact U1 snRNP particles. Nucleic Acids Res., 12, 4111–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby S.W. and Abelson,J. (1988) An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science, 242, 1028–1035. [DOI] [PubMed] [Google Scholar]
- Scherly D., Boelens,W., van Venrooij,W.J., Dathan,N.A., Hamm,J. and Mattaj,I.W. (1989) Identification of the RNA binding segment of human U1 A protein and definition of its binding site on U1 snRNA. EMBO J., 8, 4163–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherly D., Boelens,W., Dathan,N.A., van Venrooij,W.J. and Mattaj,I.W. (1990) Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B′′ and their cognate RNAs. Nature, 345, 502–506. [DOI] [PubMed] [Google Scholar]
- Séraphin B. and Rosbash,M. (1989) Identification of functional U1 snRNA–pre-mRNA complexes committed to spliceosome assembly and splicing. Cell, 59, 349–358. [DOI] [PubMed] [Google Scholar]
- Siebel C., Fresco,L. and Rio,D. (1992) The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes Dev., 6, 1386–1401. [DOI] [PubMed] [Google Scholar]
- Siebel C.W., Kanaar,R. and Rio,D.C. (1994) Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes Dev., 8, 1713–1725. [DOI] [PubMed] [Google Scholar]
- Siebel C.W., Admon,A. and Rio,D.C. (1995) Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes Dev., 9, 269–283. [DOI] [PubMed] [Google Scholar]
- Stark H., Dube,P., Lührmann,R. and Kastner,B. (2001) Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature, 409, 539–542. [DOI] [PubMed] [Google Scholar]
- Tang J., Abovich,N., Fleming,M.L., Seraphin,B. and Rosbash,M. (1997) Identification and characterization of a yeast homolog of U1 snRNP-specific protein C. EMBO J., 16, 4082–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q., Streuli,M., Saito,H., Schlossman,S.F. and Anderson,P. (1991) A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell, 67, 629–639. [DOI] [PubMed] [Google Scholar]
- Valcárcel J., Gaur,R., Singh,R. and Green,M. (1996) Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science, 273, 1706–1709. [DOI] [PubMed] [Google Scholar]
- Varani G. and Nagai,K. (1998) RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys Biomol. Struct., 27, 407–445. [DOI] [PubMed] [Google Scholar]
- Wassarman D.A. and Steitz. J.A. (1992). Interactions of small nuclear RNA’s with precursor messenger RNA during in vitro splicing. Science, 257, 1918–1925. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol., 13, 290–301. [DOI] [PubMed] [Google Scholar]
- Will C.L., Rumpler,S., Klein Gunnewiek,J., van Venrooij,W.J. and Lührmann,R. (1996) In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res., 24, 4614–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. and Maniatis,T. (1993) Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell, 75, 1061–1070. [DOI] [PubMed] [Google Scholar]
- Zhang D. and Rosbash,M. (1999) Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev., 13, 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Licklider,L.J., Gygi,S.P. and Reed,R. (2002) Comprehensive proteomic analysis of the human spliceosome. Nature, 419, 182–185. [DOI] [PubMed] [Google Scholar]