Abstract
The HIV-1 transactivator protein, Tat, is an atypical transcriptional activator that functions through binding, not to DNA, but to a short leader RNA, TAR. Although details of its functional mechanism are still unknown, emerging findings suggest that Tat serves primarily to adapt co-activator complexes such as p300, PCAF and P-TEFb to the HIV-1 long terminal repeat. Hence, an understanding of how Tat interacts with these cofactors is crucial. It has recently been shown that acetylation at a single lysine, residue 50, regulated the association of Tat with PCAF. Here, we report that in the absence of Tat acetylation, PCAF binds to amino acids 20–40 within Tat. Interestingly, acetylation of Tat at Lys28 abrogates Tat–PCAF interaction. Acetylation at Lys50 creates a new site for binding to PCAF and dictates the formation of a ternary complex of Tat–PCAF–P-TEFb. Thus, differential lysine acetylation of Tat coordinates the interactions with its co-activators, cyclin T1 and PCAF. Our results may help in understanding the ordered recruitment of Tat co-activators to the HIV-1 promoter.
Keywords: cyclin T1/differential acetylation/HIV-1/PCAF/Tat
Introduction
Tat regulates processive transcription from the HIV-1 long terminal repeat (LTR). Expression from the HIV-1 LTR is activated 200- to 300-fold by Tat. Although the mechanism for Tat transactivation has been studied intensively, it remains incompletely understood. Several key observations have, however, been established. Tat is an atypical transactivator that functions by binding, not to DNA, but to a short nascent leader RNA, TAR (Berkhout et al., 1989; Dingwall et al., 1989). Tat controls an early step in transcriptional elongation (Xiao et al., 1997) that is sensitive to inhibitors of protein kinase (Marciniak and Sharp, 1991; Mancebo et al., 1997) and requires the C-terminal domain (CTD) of the large subunit of RNA polymerase II (RNAP II) (Chun and Jeang, 1996; Okamoto et al., 1996; Parada and Roeder, 1996; Yang et al., 1996). Tat binds a nuclear serine/threonine kinase, TAK (Tat-associated kinase) (Herrmann and Rice, 1993, 1995), which is identical to the CDK9 kinase subunit of human P-TEFb (Mancebo et al., 1997; Zhu et al., 1997), a positive-acting transcription elongation factor complex required for the expression of many cellular genes (Marshall and Price, 1992, 1995). CDK9 can phosphorylate RNAP II CTD, and CDK9 inhibitors, as well as a dominant-negative CDK9 mutant, can block Tat transactivation; these results verify the CDK9–P-TEFb complex as an important Tat co-activator (Mancebo et al., 1997; Yang et al., 1997; Zhu et al., 1997). The subsequent identification of cyclin T1 as a cyclin subunit of P-TEFb (Peng et al., 1998; Wei et al., 1998) has provided further insight into Tat transactivation. Currently, it is understood that cyclin T1 enhances Tat–TAR RNA interaction and that a Tat–cyclin T1/CDK9–TAR complex mediates phosphorylation of RNAP II CTD, dictating processive transcription from the LTR (Fujinaga et al., 1998; Garber et al., 1998; Bieniasz et al., 1999; Chen et al., 1999; Ivanov et al., 1999).
The packaging of genes into chromatin is increasingly recognized as being important in regulating transcription initiation and elongation (Struhl, 1998). Following integration into the host cell genome, the HIV-1 provirus is organized like cellular genes into chromatin. Integrated HIV-1 provirus has positioned nucleosomes in its 5′ LTR (Verdin et al., 1993; Sheridan et al., 1995). Inhibitors of histone deacetylases, trichostatin A (TSA) and trapoxin, potently induce HIV-1 transcription from chromatinized templates; and activation of the HIV-1 promoter by these agents is accompanied by the loss or rearrangement of a positioned nucleosome, nuc-1, near the viral mRNA start site (Van Lint et al., 1996a,b). Moreover, TSA strongly induces cell-free transcription of a chromatinized HIV-1 promoter (Sheridan et al., 1997). Collectively, these studies strongly suggest that chromatin is an important regulatory component of HIV-1 transcription. Relevent to the role of chromatin in HIV-1 gene expression, Tat has been shown to associate with histone acetyltransferases (HATs) p300/CBP, p300/CBP-associating factor (PCAF) and hGCN5 (Benkirane et al., 1998; Hottiger and Nabel, 1998; Marzio et al., 1998; Col et al., 2001). Tat-recruited HATs presumably acetylate histones in LTR-proximal nucleosomes to potentiate transcription (Benkirane et al., 1998; Marzio et al., 1998). Interestingly, we and others have found that Tat is also a substrate for acetylation by p300, PCAF and hGCN5 (Kiernan et al., 1999; Ott et al., 1999; Deng et al., 2000; Col et al., 2001), suggesting that acetyltransferases play multiple roles in HIV-1 transcription.
Tat is acetylated on two lysine residues: by PCAF on Lys28 (Kiernan et al., 1999) and by p300 and hGCN5 on Lys50 (Kiernan et al., 1999; Ott et al., 1999; Deng et al., 2000; Col et al., 2001). Acetylation of several non-histone proteins has been found to be important for their transcriptional activity (reviewed in Sterner and Berger, 2000). Indeed, we and others have also shown that acetylation regulates Tat activity (Kiernan et al., 1999; Ott et al., 1999; Deng et al., 2000; Col et al., 2001) and virus replication (Bres et al., 2002). Given that P-TEFb and PCAF/p300 activities are critical for Tat transcriptional activity, it is important to ask how acetylation at Lys28 and/or Lys50 may modulate these protein–protein interactions. A recent report has suggested that acetylation at Lys50 is critical for binding of Tat to the bromodomain of PCAF (Mujtaba et al., 2002). Here, we confirm that PCAF can bind to acetylated Lys50 and found that, more than just specifying Tat–PCAF interaction, acetylation at Lys50 also dictated the formation of a ternary Tat–PCAF–cyclin T1 complex. However, we additionally identified a second site (Tat amino acids 20–40) for PCAF binding found only in non-acetylated Tat protein. Acetylation of Tat at Lys28 abrogated the Tat–PCAF interaction. Thus, the differential acetylation of Tat at Lys 28 and Lys50 serves to coordinate its interaction with the co-activators, cyclin T1 and PCAF.
Results
Physiological levels of PCAF and p300 govern Tat-mediated transactivation of the HIV-1 LTR
To date, several studies have proposed a role for PCAF and/or p300 in Tat activation of the HIV-1 LTR (Benkirane et al., 1998; Hottiger and Nabel, 1998; Marzio et al., 1998; Nakatani, 2002). However, most of the experimental evidence relied on artificial levels of overexpression of exogenously transfected PCAF and/or p300 plasmids. To confirm that physiological levels of PCAF and p300 indeed serve Tat activity, we employed small interfering RNAs (siRNAs) to inhibit endogenous expression in a HeLa cell line that contains an integrated HIV-1 LTR-β-gal reporter gene (HeLa P4).
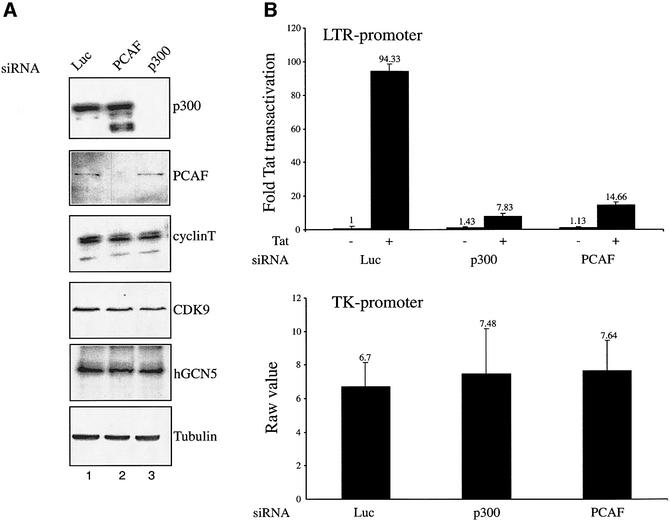
We first analyzed by western blotting the expression of cyclin T1, CDK9, hGCN5, p300 and PCAF in siRNA-transfected cells. As shown in Figure 1A, transfection of siRNA specific for p300 led to a dramatic decrease in p300 expression in these cells without affecting the control expression of PCAF, hGCN5, cyclin T1 or CDK9 (lane 3). Similarly, siRNA specific for PCAF repressed PCAF expression in transfected cells with no effect on expression of p300, hGCN5, cyclin T1 or CDK9 (lane 2). As a further control, siRNA specific for the luciferase gene, when transfected into HeLa P4 cells, did not affect the expression of p300, PCAF, hGCN5, cyclin T1, CDK9 or tubulin (lane 1).
Fig. 1. Endogenous p300 and PCAF are required for Tat-mediated transactivation of the HIV-1 LTR. (A) HeLa P4 cells containing the lacZ gene under the control of an integrated HIV-1 LTR were transfected with siRNA specific for p300, PCAF or luciferase. Western blotting analyses of p300, PCAF, hGCN5, cyclin T1, CDK9 and tubulin expression in HeLa P4 cells transfected with the indicated siRNA were performed. (B) Top panel: Tat-mediated transactivation of the LTR was analyzed 24 h post-transfection with 30 ng of a Tat expression plasmid. Fold Tat transactivation was calculated relative to transfection in the absence of Tat expression plasmid. Bottom panel: luciferase activity measured from an internal control plasmid encoding Renilla under the control of the TK promoter.
Next, Tat-mediated transactivation of the integrated HIV-1 LTR in the siRNA-transfected cells was assessed by β-galactosidase assay (Figure 1B). Tat activated β-gal expression in HeLa P4 containing luciferase-specific siRNA by 94-fold. This activation, however, was dramatically decreased by siRNA specific for either PCAF or p300 (6.4- and 12-fold reduction, respectively). As reported previously (Benkirane et al., 1998; Hottiger and Nabel, 1998; Marzio et al., 1998; Mujtaba et al., 2002), no effect was observed on the basal level of LTR-β-gal expression in the absence of Tat (Figure 1B, top panel) or on expression from a TK promoter (Figure 1B, bottom panel). These results support the view that physiological levels of PCAF and p300 are indeed critical for Tat activation of integrated HIV-1 LTR in HeLa cells. The residual transactivation detected may be due in part to acetylation of Tat by hGCN5 (Col et al., 2001), whose expression was not affected by any of the siRNAs used (Figure 1A).
PCAF interacts with amino acids 20–40 in non-acetylated Tat
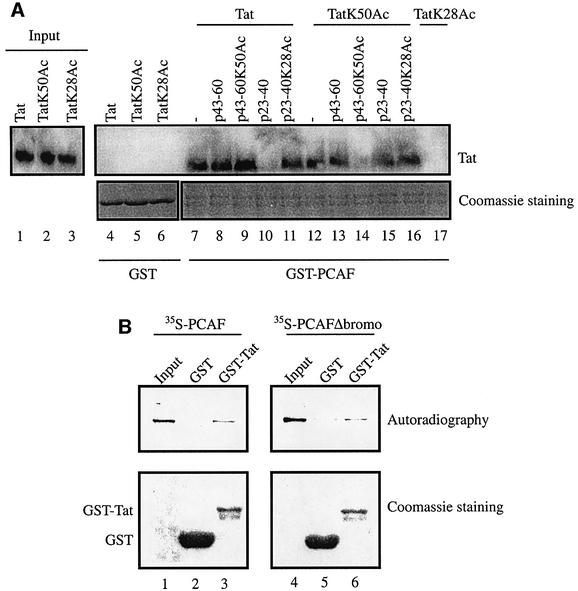
A recent study has accorded particular importance to PCAF binding of Tat (Mujtaba et al., 2002). It was proposed that PCAF binds Tat at acetylated Lys50 (Mujtaba et al., 2002). Interestingly, we had previously shown that unmodified, full-length HIV-1 Tat can bind directly to recombinant PCAF (Benkirane et al., 1998). To reconcile these results, we asked whether PCAF may differentially bind multiple regions of Tat, depending on the state of Tat acetylation. To test this hypothesis, bead-bound GST, GST–Tat1–72, GST–Tat1–40 or GST– Tat40–72 was equilibrated with purified recombinant Flag-tagged PCAF (rPCAF) for 1 h at 4°C. After extensive washing of pelleted beads in buffer containing 0.25 M KCl, the presence of rPCAF was assessed by western blotting using anti-Flag antibody. GST–Tat1–72 and GST–Tat1–40 bound rPCAF (Figure 2A, lanes 2 and 3). By contrast, no binding was observed when either GST or GST–Tat40–72 was used (Figure 2A, lanes 4 and 5). Thus, for unmodified Tat, PCAF bound to a Tat fragment that encompassed amino acids 1–40.
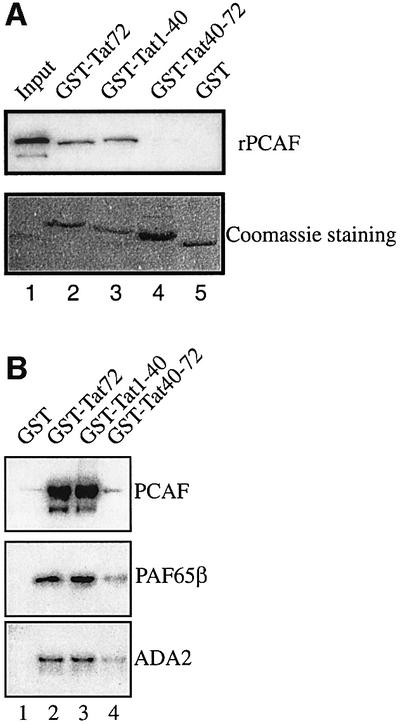
Fig. 2. Amino acids 1–40 within full-length non-acetylated Tat protein bind both free rPCAF and complex-associated PCAF. (A) Amino acids 1–40 of Tat bind PCAF. GST, GST–Tat1–72, GST–Tat1–40 or GST–Tat40–72 was incubated separately with recombinant, Flag-tagged PCAF (25 ng). The beads were extensively washed in buffer containing 0.25 M KCl and resuspended in Laemmli buffer. The presence of rPCAF was assessed by western blotting using anti-Flag M2 antibody (top panel). Input is shown in lane 1. Coomassie Blue stainings of GST and GST fusion proteins are shown (bottom panel). (B) Tat amino acids 1–40 bind complex-associated PCAF. GST, GST–Tat1–72, GST–Tat1–40 or GST–Tat40–72 was incubated with purified, nuclear, PCAF complex. The beads were extensively washed in buffer containing 0.25 M KCl and resuspended in Laemmli buffer. The presence of PCAF, PAF65β and ADA2 was assessed by western blotting.
Previous studies have shown that PCAF predominantly resides within a large multiprotein complex rather than as a free protein in cells (Ogryzko et al., 1998). To determine whether Tat could equally interact with PCAF complex, bead-bound GST, GST–Tat1–72, GST–Tat1–40 and GST–Tat40–72 were equilibrated with nuclear PCAF complex purified as described previously (Ogryzko et al., 1998). After extensive washing, binding of PCAF complex components to the beads was analyzed by western blotting (Figure 2B). GST–Tat1–72 and GST–Tat1–40 bound to PCAF as well as components of the PCAF complex, PAF65β and ADA2. Furthermore, PCAF complex binds to the same domain of Tat as recombinant PCAF.
Acetylation of Tat at lysines 28 and 50 regulates the Tat–PCAF interaction
We have previously shown that both p300 and PCAF acetylate Tat. p300 acetylates Lys50 within the TAR RNA-binding domain (Ott et al., 1999; Deng et al., 2000; Col et al., 2001), and PCAF acetylates Lys28 in the activation domain of Tat (Kiernan et al., 1999). To understand how Tat acetylation at either Lys28 (TatK28Ac) or Lys50 (TatK50Ac) could influence the Tat–PCAF complex (Figure 2B), we incubated bead-bound GST or GST– PCAF with unmodified Tat1–86, Tat(1–86)K28Ac or Tat(1–86)K50Ac. After incubation, beads were washed extensively in buffer containing 0.25 M KCl, and bound Tat was assessed by western blotting using anti-Tat antibody. GST–beads did not retain Tat, TatK28Ac or TatK50Ac (Figure 3A, lanes 4–6). By contrast, GST–PCAF retained Tat and TatK50Ac (lanes 7 and 12, respectively), but did not retain TatK28Ac (lane 17). Thus, Tat acetylated at Lys28 is unable to bind PCAF.
Fig. 3. Acetylation at Lys28 and Lys50 of Tat dictates the region of Tat interacting with PCAF. (A) GST (lanes 4–6) or GST–PCAF (lanes 7–17) beads were incubated with 100 ng of chemically synthesized Tat1–86 protein in non-acetylated (Tat), acetylated at Lys50 (TatK50Ac) or acetylated at Lys28 (TatK28Ac) forms for 1 h at 4°C. Additionally, GST–PCAF–beads were incubated separately with 1 µg of peptides corresponding to amino acids 43–60 (p43–60, lanes 8 and 13; p43–60K50Ac, lanes 9 and 14) and amino acids 23–40 (p23–40, lanes 10 and 15; p23–40K28Ac, lanes 11 and 16) for 1 h at 4°C prior to incubation with the indicated Tat proteins. After incubation, the beads were pelleted, extensively washed in buffer containing 0.25 M KCl and resuspended in Laemmli buffer. The presence of Tat was assessed by western blotting using anti-Tat antibody (top panel). Coomassie Blue stainings are shown (bottom panel). Lanes 1–3 correspond to the input materials. (B) GST or GST–Tat was incubated with in vitro translated, [35S]methionine/cysteine-labeled PCAF or PCAFΔbromo for 1 h at 4°C in binding buffer containing 0.25 M KCl. After incubation, the beads were pelleted and extensively washed in buffer containing 0.25 M KCl, and resuspended in Laemmli buffer. Bound materials were separated by SDS–PAGE and analyzed by Coomassie Blue staining and autoradiography. Lanes 1 and 4 correspond to the input materials.
We next performed competition assays using Tat peptides corresponding to Tat amino acids 43–60 and 23–40, which were either non-acetylated or acetylated at Lys50 or Lys28, respectively. Tat peptides were used at 10-fold excess over full-length Tat1–86. Thus, GST– PCAF–beads were first incubated with the indicated peptide for 1 h at 4°C, followed by another 1 h incubation with the indicated Tat1–86. Bead-bound material was analyzed by SDS–PAGE and western blotting using anti-Tat serum. We observed that incubation of GST–PCAF with either Tat peptide 43–60 or 43–60K50Ac did not affect binding to Tat (lanes 8 and 9). Consistent with the results in Figure 2A, these findings indicate that PCAF–Tat interaction does not require amino acids 43–60 (Figure 3A, compare lanes 8 and 9 with 7). On the other hand, competition with Tat peptide 23–40 completely abrogated PCAF–Tat interaction (Figure 3A, lane 10). Interestingly, an excess of Tat peptide 23–40K28Ac failed to perturb PCAF–Tat binding (Figure 3A, lane 11). Thus, in the absence of Lys28 acetylation, Tat amino acids 23–40 can directly contact PCAF; however, acetylation at Lys28 eliminated this interaction. When the same competitions were repeated for TatK50Ac–PCAF binding, peptide 23–40 had no effect (Figure 3A, lane 15). This finding suggests that PCAF binds Tat acetylated at Lys50 and non-acetylated Tat differently.
The finding that p43–60K50Ac was not able to compete for Tat binding to PCAF (Figure 3A, compare lane 9 with 7) suggested that the interaction between non-acetylated Tat and PCAF may involve a region of PCAF distinct from its bromodomain. To investigate this hypothesis, PCAF and PCAF lacking its bromodomain (PCAFΔbromo) were in vitro translated in the presence of [35S]methionine/cysteine and incubated with either GST or GST–Tat for 1 h at 4°C in buffer containing 0.25 M KCl. The beads were extensively washed and bound materials were separated by SDS–PAGE and analyzed by Coomassie Blue staining and autoradiography. As shown in Figure 3B, GST–Tat (lanes 3 and 6), but not GST (lanes 2 and 5), was able to interact with both wild-type PCAF and PCAFΔbromo. Thus, the interaction between unmodified Tat and PCAF involves a domain of PCAF distinct from the bromodomain.
Tat acetylation regulates formation of a Tat–TAR–PCAF complex
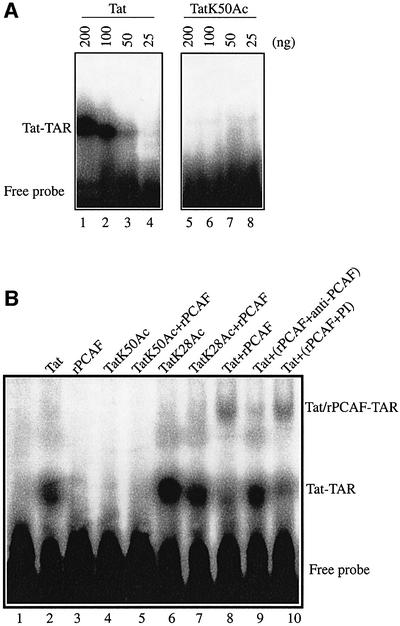
We and others have reported that acetylation of Tat at Lys50 leads to Tat dissociation from TAR RNA (Kiernan et al., 1999; Deng et al., 2000). Recently, others have shown that the bromodomain of PCAF binds specifically to a Lys50-acetylated Tat peptide corresponding to amino acids 46–55 (Mujtaba et al., 2002). The NMR structure of the PCAF bromodomain bound to Lys50-acetylated Tat peptide 46–55 showed that the peptide induced local conformational changes around its binding sites on the PCAF bromodomain (Mujtaba et al., 2002). Thus, Mujtaba et al. hypothesized that Tat–TAR RNA dissociation resulted from competition between PCAF and TAR RNA for binding to Lys50-acetylated Tat. We tested this hypothesis directly by performing electrophoretic mobility shift assay (EMSA) using 32P-labeled TAR RNA and chemically synthesized, full-length Tat which was either non-acetylated or acetylated at Lys50 (Figure 4A). In the setting of excess TAR RNA probe, the intensity of Tat–TAR RNA complex correlated with Tat addition in a dose-dependent manner from 25 to 200 ng (lanes 1–4). Informatively, no TatK50Ac–TAR RNA complex was observed even when 200 ng of Tat were added to the reaction (lanes 5–8). Thus, even in the absence of PCAF, Tat acetylated at Lys50 cannot bind TAR RNA.
Fig. 4. Tat acetylation regulates formation of a Tat–TAR–PCAF complex. (A) Acetylation of Tat at Lys50 is sufficient to abrogate binding to TAR RNA. 32P-labeled TAR RNA was incubated with increasing amounts of chemically synthesized Tat protein, which was either non-acetylated (lanes 1–4) or acetylated at Lys50, Tat K50Ac (lanes 5–8). Tat–TAR complexes were analyzed by non-denaturing acrylamide gel electrophoresis. (B) PCAF binds a Tat–TAR complex. 32P-labeled TAR RNA was incubated with 100 ng of chemically synthesized Tat protein either unmodified (lane 2), acetylated at Lys50, Tat K50Ac (lane 4), or acetylated at Lys28, TatK28Ac (lane 6), alone or with 10 ng of rPCAF (lanes 5, 7 and 8) or rPCAF that had been previously immunodepleted with anti-PCAF polyclonal antibody (lane 9) or control pre-immune serum (lane 10). Lane 1 corresponds to 32P-labeled TAR RNA alone and lane 3 corresponds to TAR RNA incubated with rPCAF.
We next checked the effect of Tat acetylation on the formation of a rPCAF–Tat–TAR RNA complex (Figure 4B). In EMSA using chemically synthesized, non-acetylated Tat protein and 32P-labeled TAR RNA, a band shift corresponding to a Tat–TAR complex was observed (lane 2). No complex was seen when rPCAF was incubated with [32P]TAR RNA, verifying that rPCAF does not directly interact with TAR (lane 3). No complex was observed between TatK50Ac and TAR RNA in either the presence or absence of rPCAF (lanes 4 and 5). While TatK28Ac readily formed a complex with TAR RNA (lane 6), this complex was not supershifted by incubation with rPCAF (lane 7) since acetylation at K28 abrogates the interaction between Tat and PCAF (Figure 3A, lane 17). Interestingly, rPCAF, when added with Tat and 32P-labeled TAR RNA, supershifted the Tat–TAR complex, indicating the formation of a Tat–TAR–PCAF ternary complex (lane 8), which could be inhibited by immunodepletion of rPCAF with anti-PCAF polyclonal antibodies (lane 9) but not pre-immune serum (lane 10). Collectively, these results confirm that acetylations of Tat at lysines 28 and 50 regulate the formation of PCAF–Tat–TAR RNA complex.
Formation of a Tat–PCAF–cyclin T1 complex is regulated by acetylation of Tat at Lys50
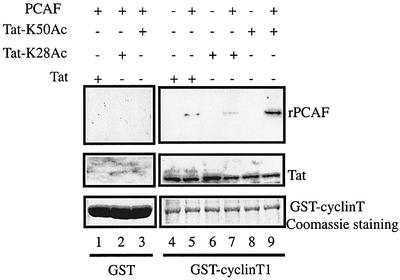
Cyclin T1, the regulatory subunit of P-TEFb, and PCAF contribute significantly to Tat transactivation of the HIV-1 LTR (reviewed in Jones, 1997; Jeang et al., 1999). Previously, we showed that acetylation of Tat at Lys28 enhanced the Tat–cyclin T1 interaction (Kiernan et al., 1999). Thus, we asked how Tat acetylation would further regulate interactions with cyclin T1 and PCAF (Figure 5). GST–cyclin T1–beads were incubated with Tat (lane 4), TatK28Ac (lane 6) or TatK50Ac (lane 8) alone or followed by subsequent incubation with rPCAF (lanes 5, 7 and 9). All beads were washed with buffer containing 0.25 M KCl, and bound material was analyzed by SDS–PAGE and western blotting using anti-Tat or anti-Flag antibodies. As shown in Figure 5, GST–cyclin T1 bound Tat, TatK28Ac and TatK50Ac (middle panel). No interaction was observed between cyclin T1 and rPCAF in the absence of Tat (data not shown). Interestingly, PCAF interacted weakly with the cyclin T1–Tat complex (lane 5) and very weakly with cyclin T1–TatK28Ac complex (lane 7), while it capably bound a TatK50Ac–cyclin T1 complex (lane 9). Thus, acetylation at Lys50 has two separate effects: it weakens Tat–TAR interaction (Figure 4) while it promotes Tat–PCAF–cyclin T1 complex formation (Figure 5).
Fig. 5. Formation of the cyclin T1–Tat–PCAF complex is regulated by acetylation of Tat Lys50. GST (lanes 1–3) or GST–cyclin 1 (lanes 4–9) beads were incubated separately with 100 ng of Tat (lanes 1 and 4), TatK28Ac (lanes 2 and 6) or TatK50Ac (lanes 3 and 8). GST–cyclin T1 was incubated first with either Tat (lane 5), TatK28Ac (lane 7) or TatK50Ac (lane 9) for 1 h at 4°C, washed, and further incubated with 25 ng of rPCAF for an additional hour at 4°C. After extensive washing of beads, the presence of Tat and rPCAF was assessed by western blotting using anti-Flag (top panel) or anti-Tat (middle panel) antibody. Coomassie Blue stainings are shown (bottom panel).
Discussion
The activity of transcription factors can be regulated by post-translational modifications such as phosphorylation, ubiquitylation and/or glycosylation (Beckett, 2001). These modifications can affect the cellular localization of transcription factors, as well as their interaction with partner proteins or DNA targets. Acetylation of transcription factors has also been shown to regulate activity by modulating interactions between partner proteins (reviewed in Roth et al., 2001). Acetylation can either enhance or repress transcription factor activity or, in the case of HMG I (Y), differential acetylation can independently have both positive and negative effects. Acetylation of HMG I (Y) at Lys71 by PCAF/GCN5 stabilized the INFβ enhanceosome, while acetylation at Lys65 by CBP destabilized the enhanceosome, leading to promoter shut-off (Munshi et al., 1998, 2001).
We and others have previously reported that the HIV-1 transactivator protein, Tat, is acetylated on two lysines (Kiernan et al., 1999; Ott et al., 1999; Deng et al., 2000; Col et al., 2001). Acetylation of Tat at Lys50 by p300 promoted the dissociation of Tat from TAR RNA (Kiernan et al., 1999; Deng et al., 2000), while acetylation of Tat at Lys28 enhanced Tat–P-TEFb interaction (Kiernan et al., 1999). Tat acetylations at K28 and K50 were recently shown to be important for HIV-1 replication (Bres et al., 2002). By knocking down the expression of p300 and PCAF using siRNAs specifically directed against these HATs, we show here that endogenous p300 and PCAF contribute significantly to Tat-mediated transactivation. This effect is probably due to a combination of their acetyltransferase activities directed towards the local chromatin environment, general factors involved in transcription as well as specific factor acetylation of Tat.
Recently, Mujtaba and co-workers reported that a Lys50-acetylated short Tat peptide was specifically bound by the bromodomain of PCAF. It was proposed that dissociation of Tat from TAR RNA resulted from competition between PCAF and TAR RNA for binding to Lys50-acetylated Tat (Mujtaba et al., 2002; reviewed in Nakatani, 2002). Here, we have further analyzed the interaction between PCAF using full-length Tat in its non-acetylated, Lys50-acetylated or Lys28-acetylated forms. Whereas the peptide used in the study by Mujtaba et al. contained only the short Lys50 PCAF-binding site, we found that, in the context of other regions in a full-length non-acetylated Tat protein, PCAF bound a second site within the cysteine-rich domain between amino acids 20 and 40. Unlike binding to the Lys50 site, PCAF binding to site 20–40 is negatively regulated by acetylation. Hence, we found that acetylation of Tat at Lys28 abrogated this Tat–PCAF binding. Interestingly, interaction between PCAF and the cysteine-rich domain of Tat does not involve its bromodomain.
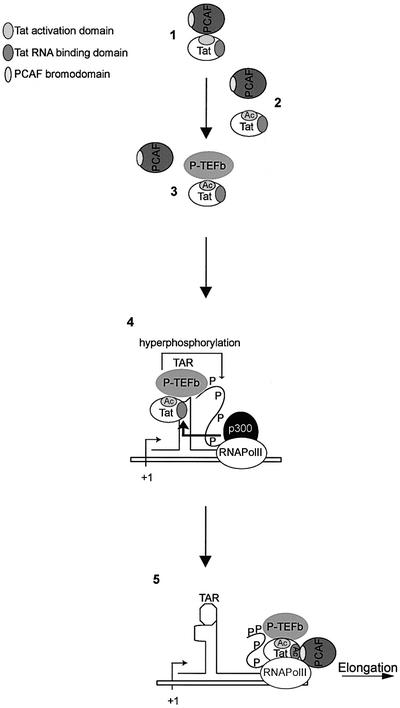
Acetylation of Tat by p300 and PCAF may help in understanding the order of Tat-mediated recruitment of its co-activators to the HIV-1 promoter. The observations that, in the absence of Tat acetylation, cyclin T1 and PCAF bind to the same region of Tat (cysteine-rich domain) and that acetylation of Tat at Lys28 enhances its interaction with P-TEFb (Kiernan et al., 1999) suggest that non-acetylated Tat binds first to PCAF. Shown in Figure 6 is a proposed model for the role of Tat acetylations in transactivation of transcription that incorporates data described here and elsewhere. Following its acetylation at Lys28 by PCAF, Tat dissociates from PCAF and then binds to cyclin T1 (steps 1–3). Tat/P-TEFb binds with high affinity to TAR RNA, facilitating hyperphosphorylation of the RNAPolII CTD. The interaction between RNAPolII and p300 (Cho et al., 1998) may help to bring p300 within the vicinity of Tat. Subsequently, Tat can be acetylated at Lys50 by p300 (step 4). The p300-mediated acetylation of Tat at Lys50 leads to two consequences. First, it induces P-TEFb/Tat dissociation from TAR RNA (Kiernan et al., 1999) and, secondly, it creates a new binding site for PCAF in the TAR RNA-binding domain of Tat, which leads to the formation of a P-TEFb–Tat–PCAF complex. This complex remains associated with the elongation complex during transcriptional elongation, possibly stabilized by interactions between Tat–P/TEFb and RNAPolII CTD through a histidine-rich region of cyclin T1 (Taube et al., 2002) and Tat and RNAPolII holoenzyme (Cujec et al., 1997) (step 5). hGCN5 also acetylates Tat on K50 and K51, leading to increased transactivation of the LTR (Col et al., 2001). Whether hGCN5-mediated acetylation induces the same effects as p300 or leads to other consequences important in transactivation remains to be determined.
Fig. 6. Proposed model for the regulation of Tat transcriptional activity by p300 and PCAF. (1) PCAF interacts with Tat amino acids 20–40. (2) PCAF acetylates Tat at Lys28 and dissociates from Tat. (3) P-TEFb associates with Tat. (4) P-TEFb–Tat complex binds TAR RNA, and p300 acetylates Tat at Lys50. (5) Tat dissociates from TAR RNA and PCAF interacts with K50-acetylated TAR RNA-binding domain of Tat. Tat–P-TEFb–PCAF complex associates with the transcription elongation complex.
An issue that remains unresolved is the mechanism that governs the temporal and TAR-dependent regulation of the two independent Tat acetylations. To date, very little is known about the regulation of HAT activity. HAT activity of both p300 and ATF-2 has been shown to be regulated by phosphorylation (Ait-Si-Ali et al., 1998; Kawasaki et al., 2000). It is possible, therefore, that the specific Tat acetylation by p300 could be regulated by one or more of the kinase activities known to associate with Tat. Another possibility is that recruitment of Tat/P-TEFb to the HIV-1 promoter through TAR RNA will bring Tat into the vicinity of p300, which in turn acetylates Tat at Lys50, leading to its dissociation from TAR RNA and its subsequent binding to the bromodomain of PCAF. The P-TEFb–Tat–PCAF complex may then associate with the transcription elongation complex. Further work will be needed to understand the regulation of acetylation of Tat and, indeed, other substrates.
Materials and methods
Cell culture and transfection
HeLa and HeLa P4 cells that contain the lacZ gene under the control of an integrated HIV-1 LTR (Clavel and Charneau, 1994) were propagated in DMEM with 10% FBS and transfected according to the manufacturer’s instructions using either oligofectamine or lipofectamine (Invitrogen; Gibco) as indicated in the figure legends. β-galactosidase activity was measured 48 h after transfection according to the manufacturer’s protocol (Roche).
siRNAs
RNA oligonucleotides corresponding to PCAF (forward: 5′-UCG CCGUGAAGAAAGCGCATT-3′; reverse: 5′-UGCGCUUUCUUCAC GGCGATT-3′), p300 (forward: 5′-CAGAGCAGUCCUGGAUUAGTT-3′; reverse: 5′-CUAAUCCAGGACUGCUCUGTT-3′) and luciferase (forward: 5′-CGUACGCGGAAUACUUCGATT-3′; reverse: 5′-UCG AAGUAUUCCGCGUACGTT-3′) were synthesized (Eurogentec, Belgium). HeLa P4 cells that contain the lacZ gene under the control of the HIV-1 LTR (Clavel and Charneau, 1994) were transfected with 100 ng of siRNA using oligofectamine (Invitrogen). Twenty-four hours after transfection, cells were retransfected with Tat-flag expression vector using lipofectamine (Invitrogen). Forty-eight hours after transfection of Tat-flag plasmid, expression levels of PCAF, p300, hGCN5, cyclin T1, CDK9 and tubulin were analyzed by western blotting, and β-gal activity was analyzed according to the manufacturer’s protocol (Roche Molecular Biochemicals).
Synthesis, purification and characterization of Tat proteins and peptides
Tat peptides corresponding to amino acids 23–40 (p23–40) and 43–60 (p43–60) either non-acetylated or acetylated on Lys28 (p23–40K28Ac) or Lys50 (p43–60K50Ac) were chemically synthesized and purified to >95% homogeneity by mass spectral analysis purification procedures (Synt:em, France). Tat Bru (1–86) non-acetylated or acetylated at Lys50 (TatK50Ac) or acetylated at Lys28 (TatK28Ac) were assembled according to the method of Barany and Merrifield (1979) on 4-hydroxymethyl-phenoxy-methyl-copolystyrene–1% divinylbenzene preloaded resin (HMP) (0.65 mmol) (Perkin Elmer, Applied Biosystem Inc., Forster City, CA) on an automated synthesizer (ABI 433A; Perkin Elmer) as described previously (Peloponese et al., 1999). Purification was carried out with a Beckman HPLC apparatus on a Merck C8 reverse-phase column (10 × 250 mm) as described previously (Peloponese et al., 1999). For Tat acetylated forms, a cartridge containing N-α-Fmoc-N-ε-acetyl-l-lysine (Novabiochem) was used. HPLC analysis was performed as described previously (Peloponese et al., 1999). Electrospray mass spectrometry was carried out with a single quad PE-SCIEX API 150ex (Perkin Elmer). Amino acid analyses were performed on a 6300 Beckman analyzer.
Fusion protein affinity chromatography
PCAF, cyclin T1, HIV-1 Tat wild type and mutants were expressed as GST fusion proteins in BL21 (Pharmacia) as described previously (Benkirane et al., 1998). Twenty microliters of the various protein-bound resins were immobilized at 0.2 µg/µl onto glutathione–Sepharose beads for 1 h at 4°C. The resins were packed into columns, which were washed with buffer (20 mM HEPES–KOH pH 7.9, 120 mM KCl, 1 mM MgCl2, 17% glycerol, 2 mM DTT) containing 0.25 M KCl. Recombinant Flag-tagged PCAF was produced and purified from a baculovirus overexpression system (Ogryzko et al., 1996). PCAF complex was purified as described previously (Ogryzko et al., 1998). Components of the complex were confirmed by SDS–PAGE, followed by Coomassie Blue staining.
Western blot analysis
Column eluates or HeLa P4 extracts were resolved by SDS–PAGE. Proteins were transferred to PVDF membrane by semi-dry electroblotting (Millipore, Bedford) for 1 h at 400 mA. Membranes were incubated with the primary antibody overnight at 4°C, washed and incubated with the appropriate secondary antibody (Amersham) for 1 h. Proteins were visualized by chemiluminescence (Amersham) according to the manufacturer’s protocol. Primary antibodies used were anti-Flag (M2; Sigma), monoclonal anti-tubulin (Sigma), anti-GCN5 (N-18; Santa Cruz), anti-p300 (N-15; Santa Cruz), anti-PCAF polyclonal antibody (Ogryzko et al., 1998), anti-cyclin T1 (gift from D.Price), anti-CDK9 (gift from D.Price) and anti-Tat (K.-T.Jeang, unpublished data).
EMSA
TAR RNA was synthesized using T7 RNA polymerase from HindIII-digested pT7TAR by in vitro transcription (Promega) containing [γ-32P]UTP (Amersham). After treatment with RQ DNase, labeled TAR RNA was purified on a G-50 Sephadex column (Boehringer). Gel mobility shift reactions were performed as described previously (Wei et al., 1998), except that the reactions were performed for 15 min at room temperature.
References
- Ait-Si-Ali S. et al. (1998) Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature, 396, 184–186. [DOI] [PubMed] [Google Scholar]
- Barany G. and Merrifield,R.B. (1979) The Peptide. In Gross,E. and Meinhofer,J. (eds), Solid phase peptide synthesis, Vol. 2. Academic Press, New York, NY, pp. 1–284.
- Beckett D. (2001) Regulated assembly of transcription factors and control of transcription initiation. J. Mol. Biol., 314, 335–352. [DOI] [PubMed] [Google Scholar]
- Benkirane M., Chun,R.F., Xiao,H., Ogryzko,V.V., Howard,B.H., Nakatani,Y. and Jeang,K.T. (1998) Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J. Biol. Chem., 273, 24898–24905. [DOI] [PubMed] [Google Scholar]
- Berkhout B., Silverman,R.H. and Jeang,K.T. (1989) Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell, 59, 273–282. [DOI] [PubMed] [Google Scholar]
- Bieniasz P.D., Grdina,T.A., Bogerd,H.P. and Cullen,B.R. (1999) Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl Acad. Sci. USA, 96, 7791–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bres V., Kiernan,R.E., Emiliani,S. and Benkirane,M. (2002) Tat acetyl-acceptor lysines are important for human immunodeficiency virus type-1 replication. J. Biol. Chem., 277, 22215–22221. [DOI] [PubMed] [Google Scholar]
- Chen D., Fong,Y. and Zhou,Q. (1999) Specific interaction of Tat with the human but not rodent P-TEFb complex mediates the species-specific Tat activation of HIV-1 transcription. Proc. Natl Acad. Sci. USA, 96, 2728–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Orphanides,G., Sun,X., Yang,X.J., Ogryzko,V., Lees,E., Nakatani,Y. and Reinberg,D. (1998) A human RNA polymerase II complex containing factors that modify chromatin structure. Mol. Cell. Biol., 18, 5355–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun R.F. and Jeang,K.T. (1996) Requirements for RNA polymerase II carboxyl-terminal domain for activated transcription of human retroviruses human T-cell lymphotropic virus I and HIV-1. J. Biol. Chem., 271, 27888–27894. [DOI] [PubMed] [Google Scholar]
- Clavel F. and Charneau,P. (1994) Fusion from without directed by human immunodeficiency virus particles. J. Virol., 68, 1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Col E., Caron,C., Seigneurin-Berny,D., Gracia,J., Favier,A. and Khochbin,S. (2001) The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator, Tat. J. Biol. Chem., 276, 28179–28184. [DOI] [PubMed] [Google Scholar]
- Cujec T.P., Cho,H., Maldonado,E., Meyer,J., Reinberg,D. and Peterlin,B.M. (1997) The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol. Cell. Biol., 17, 1817–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L. et al. (2000) Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones. Virology, 277, 278–295. [DOI] [PubMed] [Google Scholar]
- Dingwall C. et al. (1989) Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl Acad. Sci. USA, 86, 6925–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K., Cujec,T.P., Peng,J., Garriga,J., Price,D.H., Grana,X. and Peterlin,B.M. (1998) The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1 and Tat. J. Virol., 72, 7154–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M.E., Wei,P., KewalRamani,V.N., Mayall,T.P., Herrmann,C.H., Rice,A.P., Littman,D.R. and Jones,K.A. (1998) The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev., 12, 3512–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann C.H. and Rice,A.P. (1993) Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology, 197, 601–608. [DOI] [PubMed] [Google Scholar]
- Herrmann C.H. and Rice,A.P. (1995) Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol., 69, 1612–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger M.O. and Nabel,G.J. (1998) Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J. Virol., 72, 8252–8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D., Kwak,Y.T., Nee,E., Guo,J., Garcia-Martinez,L.F. and Gaynor,R.B. (1999) Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for tat-activation. J. Mol. Biol., 288, 41–56. [DOI] [PubMed] [Google Scholar]
- Jeang K.T., Xiao,H. and Rich,E.A. (1999) Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem., 274, 28837–28840. [DOI] [PubMed] [Google Scholar]
- Jones K.A. (1997) Taking a new TAK on tat transactivation. Genes Dev., 11, 2593–2599. [DOI] [PubMed] [Google Scholar]
- Kawasaki H., Schiltz,L., Chiu,R., Itakura,K., Taira,K., Nakatani,Y. and Yokoyama,K.K. (2000) ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature, 405, 195–200. [DOI] [PubMed] [Google Scholar]
- Kiernan R.E. et al. (1999) HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J., 18, 6106–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo H.S. et al. (1997) P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev., 11, 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak R.A. and Sharp,P.A. (1991) HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J., 10, 4189–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall N.F. and Price,D.H. (1992) Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol. Cell. Biol., 12, 2078–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall N.F. and Price,D.H. (1995) Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem., 270, 12335–12338. [DOI] [PubMed] [Google Scholar]
- Marzio G., Tyagi,M., Gutierrez,M.I. and Giacca,M. (1998) HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc. Natl Acad. Sci. USA, 95, 13519–13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujtaba S., He,Y., Zeng,L., Farooq,A., Carlson,J.E., Ott,M., Verdin,E. and Zhou,M.M. (2002) Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol. Cell, 9, 575–586. [DOI] [PubMed] [Google Scholar]
- Munshi N., Merika,M., Yie,J., Senger,K., Chen,G. and Thanos,D. (1998) Acetylation of HMG I(Y) by CBP turns off IFNβ expression by disrupting the enhanceosome. Mol. Cell, 2, 457–467. [DOI] [PubMed] [Google Scholar]
- Munshi N., Agalioti,T., Lomvardas,S., Merika,M., Chen,G. and Thanos,D. (2001) Coordination of a transcriptional switch by HMGI(Y) acetylation. Science, 293, 1133–1136. [DOI] [PubMed] [Google Scholar]
- Nakatani Y. (2002) HIV-1 transcription. Activation mediated by acetylation of Tat. Structure (Camb), 10, 443–444. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Kotani,T., Zhang,X., Schiltz,R.L., Howard,T., Yang,X.J., Howard,B.H., Qin,J. and Nakatani,Y. (1998) Histone-like TAFs within the PCAF histone acetylase complex. Cell, 94, 35–44. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Sheline,C.T., Corden,J.L., Jones,K.A. and Peterlin,B.M. (1996) Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc. Natl Acad. Sci. USA, 93, 11575–11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M., Schnolzer,M., Garnica,J., Fischle,W., Emiliani,S., Rackwitz,H.R. and Verdin,E. (1999) Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr. Biol., 9, 1489–1492. [DOI] [PubMed] [Google Scholar]
- Parada C.A. and Roeder,R.G. (1996) Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature, 384, 375–378. [DOI] [PubMed] [Google Scholar]
- Peloponese J.M. Jr, Collette,Y., Gregoire,C., Bailly,C., Campese,D., Meurs,E.F., Olive,D. and Loret,E.P. (1999) Full peptide synthesis, purification and characterization of six Tat variants. Differences observed between HIV-1 isolates from Africa and other continents. J. Biol. Chem., 274, 11473–11478. [DOI] [PubMed] [Google Scholar]
- Peng J., Zhu,Y., Milton,J.T. and Price,D.H. (1998) Identification of multiple cyclin subunits of human P-TEFb. Genes Dev., 12, 755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S.Y., Denu,J.M. and Allis,C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Sheridan P.L., Sheline,C.T., Cannon,K., Voz,M.L., Pazin,M.J., Kadonaga,J.T. and Jones,K.A. (1995) Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev., 9, 2090–2104. [DOI] [PubMed] [Google Scholar]
- Sheridan P.L., Mayall,T.P., Verdin,E. and Jones,K.A. (1997) Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev., 11, 3327–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E. and Berger,S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Taube R., Lin,X., Irwin,D., Fujinaga,K. and Peterlin,B.M. (2002) Interaction between P-TEFb and the C-terminal domain of RNA polymerase II activates transcriptional elongation from sites upstream or downstream of target genes. Mol. Cell. Biol., 22, 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint C., Emiliani,S., Ott,M. and Verdin,E. (1996a) Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J., 15, 1112–1120. [PMC free article] [PubMed] [Google Scholar]
- Van Lint C., Emiliani,S. and Verdin,E. (1996b) The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr., 5, 245–253. [PMC free article] [PubMed] [Google Scholar]
- Verdin E., Paras,P.,Jr and Van Lint,C. (1993) Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J., 12, 3249–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P., Garber,M.E., Fang,S.M., Fischer,W.H. and Jones,K.A. (1998) A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell, 92, 451–462. [DOI] [PubMed] [Google Scholar]
- Xiao H., Lis,J.T. and Jeang,K.T. (1997) Promoter activity of Tat at steps subsequent to TATA-binding protein recruitment. Mol. Cell. Biol., 17, 6898–6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Herrmann,C.H. and Rice,A.P. (1996) The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J. Virol., 70, 4576–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Gold,M.O., Tang,D.N., Lewis,D.E., Aguilar-Cordova,E., Rice,A.P. and Herrmann,C.H. (1997) TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc. Natl Acad. Sci. USA, 94, 12331–12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Pe’ery,T., Peng,J., Ramanathan,Y., Marshall,N., Marshall,T., Amendt,B., Mathews,M.B. and Price,D.H. (1997) Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev., 11, 2622–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]