Abstract
Yeast mitochondrial leucyl-tRNA synthetase (LeuRS) binds to the bI4 intron and collaborates with the bI4 maturase to aid excision of the group I intron. Deletion analysis isolated the inserted LeuRS CP1 domain as a critical factor in the protein’s splicing activity. Protein fragments comprised of just the LeuRS CP1 region rescued complementation of a yeast strain that expressed a splicing-defective LeuRS. Three-hybrid analysis determined that these CP1-containing LeuRS fragments, ranging from 214 to 375 amino acids, bound to the bI4 intron. In each case, interactions with only the LeuRS protein fragment specifically stimulated bI4 intron splicing activity. Substitution of a homologous CP1 domain from isoleucyl-tRNA synthetase or mutation within the LeuRS CP1 region of the smallest protein fragment abolished RNA binding and splicing activity. The CP1 domain is best known for its amino acid editing activity. However, these results suggest that elements within the LeuRS CP1 domain also play a novel role, independent of the full-length tRNA synthetase, in binding the bI4 group I intron and facilitating its self-splicing activity.
Keywords: amino acid editing/CP1/group I intron/NAM2/ribozyme
Introduction
The tRNA synthetases are a diverse enzyme family that share the responsibility of accurately translating the genetic code in the first step of protein synthesis (Carter, 1993; Martinis and Schimmel, 1996). This family is comprised of up to 20 essential proteins in every organism. Eukaryotes contain an additional set of tRNA synthetases for each organelle. Each tRNA synthetase is specific to just one amino acid, which it aminoacylates to a single or a set of tRNA isoacceptors for delivery of the correct amino acid during ribosomal translation.
The tRNA synthetases have also been recruited and adapted for alternate or secondary functions (Martinis et al., 1999a,b; Weiner and Maizels, 1999; Schimmel and Ribas de Pouplana, 2000). These protein synthesis enzymes may use pre-existing motifs and modules, e.g. that bind RNA, to aid novel functions, but can also acquire extra domains or insertions to confer or enhance these secondary activities. Two mitochondrial tRNA synthetases, leucine [LeuRS or NAM2p (Labouesse et al., 1985; Herbert et al., 1988; Labouesse, 1990)] and tyrosine [TyrRS or CYT-18p (Akins and Lambowitz, 1987; Kamper et al., 1992)], are required to facilitate group I intron splicing in Saccharomyces cerevisiae and Neurospora crassa, respectively (Lambowitz and Perlman, 1990; Dujardin and Herbert, 1997; Lambowitz et al., 1999). Both LeuRS (Rho and Martinis, 2000) and CYT-18p (Guo and Lambowitz, 1992) bind directly to the group I intron. CYT-18p utilizes its tRNA-binding domains for interactions with the group I intron (Kittle et al., 1991). It also relies on a unique N-terminal domain that can be important to CYT-18p-dependent splicing activity (Cherniack et al., 1990; Kamper et al., 1992; Mohr et al., 2001). This idiosyncratic CYT-18p domain is proposed to play a key structural role (Mohr et al., 2001) and is absent in other tyrosine enzymes and also LeuRS.
Yeast mitochondrial LeuRS promotes splicing of specific group I introns in collaboration with the bI4 maturase (Labouesse, 1990; Li et al., 1996). Suppressor mutations within LeuRS rescue splicing activity in the absence of a functional bI4 maturase (Labouesse et al., 1985; Herbert et al., 1988; Henke, 2000; R.M.Henke and P.S.Perlman, personal communication). Other mutations near one of these suppressor sites have been shown to impede LeuRS-dependent splicing, but minimally impact the enzyme’s aminoacylation function, suggesting that this localized region may be directly involved in interactions with the intron or aiding the group I intron splicing activity (Li et al., 1996). These suppressor and inhibitory mutations reside within a protein insertion called CP1 (connective polypeptide 1; Starzyk et al., 1987; Hou et al., 1991). X-ray crystal structures show that the majority of this primary sequence insert folds into a discrete tertiary domain (Nureki et al., 1998; Silvian et al., 1999; Cusack et al., 2000).
The CP1 domain is conserved amongst the leucine enzymes as well as certain other tRNA synthetases. The inserted module splits the ATP-binding fold that comprises the canonical catalytic core of the class I tRNA synthetase active site (Hou et al., 1991). It contains RNA-binding determinants (Rould et al., 1989; Burbaum and Schimmel, 1991; Wakasugi et al., 1998; Silvian et al., 1999) and is also responsible for amino acid editing in some tRNA synthetases (Lin et al., 1996; Hale et al., 1997; Chen et al., 2000; Hendrickson et al., 2000; Mursinna et al., 2001; Mursinna and Martinis, 2002). The CP1 domains of isoleucyl (IleRS)- and valyl (ValRS)-tRNA synthetases have been recombinantly isolated as stable proteins and shown to independently hydrolyze mis-aminoacylated tRNA in an RNA-specific manner (Lin et al., 1996).
LeuRS also exhibits a CP1-dependent editing activity that hydrolyzes amino acids that are charged to the tRNA (Chen et al., 2000; Mursinna et al., 2001; Larkin et al., 2002; Mursinna and Martinis, 2002), suggesting that it too has RNA-binding properties. Because splicing-specific mutations in mitochondrial LeuRS had mapped in close proximity to, or within, the folded CP1-based domain (Labouesse et al., 1985; 1987; Herbert et al., 1988; Li et al., 1996; Henke, 2000; R.M.Henke and P.S.Perlman, personal communication), we hypothesized that this inserted domain may interact directly with the group I intron RNA. We designed a series of LeuRS deletion mutants to isolate the CP1 region of the protein. Each of these protein fragments rescued a splicing-defective mutation of LeuRS in complementation assays. Further analysis determined that the fragments comprised of only the LeuRS CP1 domain interact directly with the bI4 group I intron and independently stimulate the group I intron’s splicing activity.
Results
Design of CP1-containing LeuRS protein fragments
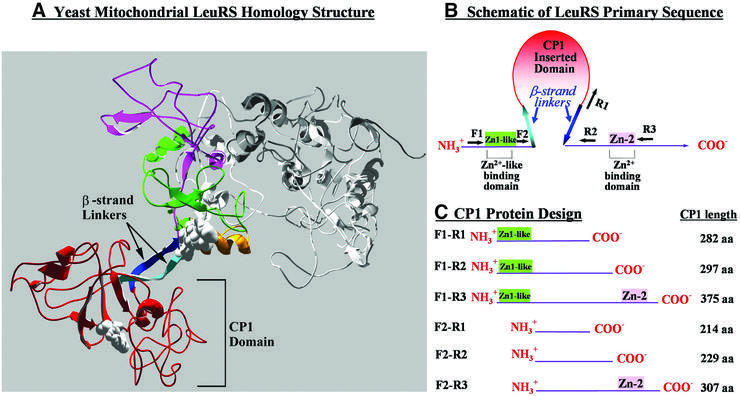
The CP1 domain was originally defined as a primary sequence insert that splits the sequence encoding the Rossmann fold-based aminoacylation core within certain tRNA synthetases (Starzyk et al., 1987; Hou et al., 1991). The X-ray crystal structure of Thermus thermophilus LeuRS shows that most of the CP1 insertion folds into a discrete domain that is linked to the conserved catalytic core of the class I tRNA synthetase via two β-strand tethers (Cusack et al., 2000). The β-strand tethers are flanked by two Zn2+-binding motifs, which are more closely associated with the main body of the class I tRNA synthetase. One Zn2+-binding motif is ∼20 amino acids upstream from the CP1 N-terminal β-strand, while the other is ∼5 amino acids downstream from the CP1 C-terminal β-strand.
The T.thermophilus and yeast mitochondrial LeuRSs share 36% identity. Both contain the Zn2+-binding motif called Zn-2, which resides on the C-terminal flank of the CP1 domain. The yeast mitochondrial LeuRS also contains a region on the N-terminal side of CP1 that is similar to the other T.thermophilus Zn2+-binding motif (called Zn-1). However, key histidine and cysteine residues that bind Zn2+ have been substituted in yeast mitochondrial LeuRS (denoted Zn1-like). Based on previous mutational analysis (Labouesse et al., 1985; Herbert et al., 1988; Henke, 2000; R.M.Henke and P.S.Perlman, personal communication), we hypothesized that the discrete CP1 domain as well as the C-terminal flanking Zn-2 site and/or the N-terminal Zn1-like region may be important to RNA–protein interactions between the yeast mitochondrial LeuRS and the group I intron.
We generated a homology model of yeast mitochondrial LeuRS that overlaps well with the T.thermophilus LeuRS crystal structure (Figure 1A). Previously identified splicing suppressor mutations were mapped to the core of the CP1 amino acid editing domain or proximal to its N-terminal β-strand linker on the main body of the LeuRS. These splicing-sensitive regions suggest that the CP1 domain of LeuRS may play an important role in the group I intron’s splicing activity.
Fig. 1. Deletion analysis of the yeast mitochondrial LeuRS splicing factor. (A) Homology model of the yeast mitochondrial LeuRS. The CP1 domain (red) is linked by two β-strand linkers, shown in light (N-terminal) and dark (C-terminal) blue. The C-terminal flanking region has a Zn2+-binding motif called Zn-2 (purple). An α-helical hairpin (yellow) links the N-terminal flanking region denoted as Zn1-like (green). Space-filling regions represent splicing-sensitive mutation sites. (B) Cartoon of the LeuRS CP1 primary sequence. Black arrows marked by ‘F’ and ‘R’, respectively, show forward and reverse primer sites used to amplify the gene regions encoding LeuRS fragments. Solid rectangles are Zn2+-like or Zn2+-binding motifs. (C) Schematic and sizes of LeuRS fragments that contained CP1.
In order to test for LeuRS CP1-specific effects on splicing activity or interactions with the group I intron, we isolated sub-genes encoding fragments of yeast mitochondrial LeuRS that contained the CP1 region (Figure 1B). Two forward PCR primers were utilized to include (primer F1) and exclude (primer F2) the Zn1-like region, which is linked to the N-terminal β-strand via an α-helical hairpin. In both designs, the β-strand tether as well as the splicing-sensitive regions were preserved as a linker to fuse the CP1 domain to other proteins or peptides for subsequent analysis via three-hybrid or complementation assays. Three reverse PCR primers were generated (Figure 1B). Primer R3 targeted the LeuRS gene sequence that is downstream of the encoded conserved C-terminal Zn-2 motif. The R2 primer omitted this sequence, but included that encoding the C-terminal β-strand linker of the CP1 domain. R1 eliminated DNA encoding the C-terminal β-strand.
Combinations of these forward and reverse primers were employed to amplify six sub-genes encoding LeuRS fragments of varying lengths that ranged from 214 (F2–R1) to 375 (F1–R3) amino acids (Figure 1C). All constructs preserved the portion of the CP1 insert that is folded into a discrete tertiary domain, as well as the known splicing-sensitive regions. The protein fragments also included one, both, or neither of the flanking Zn1-like and Zn-2 regions in yeast mitochondrial LeuRS.
LeuRS protein fragments rescue splicing-deficient mutants
We hypothesized that the isolated wild-type CP1 insert might compensate for a full-length LeuRS mutant that was deficient in its splicing role. Previously, we showed that Mycobacterium tuberculosis LeuRS complements a yeast null strain lacking the endogenous tRNA synthetase (Houman et al., 2000). A single-site mutation within the CP1 insert (W286C) of the M.tuberculosis LeuRS [or the homologous site (W238C) in yeast mitochondrial LeuRS (Li et al., 1996)] supports aminoacylation, but not spli cing activity (Houman et al., 2000). We used plasmids expressing six different protein fragments containing CP1 (Figure 1C), which were fused to a mitochondrial import sequence to transform null strains that contain the M.tuberculosis LeuRS CP1-based mutant. Figure 2A shows that each of the LeuRS protein fragments rescued the full-length LeuRS CP1-based splicing-defective mutant and complemented the yeast null strain on glycerol media. As expected, since the main catalytic core containing the aminoacylation active site was deleted in each protein fragment, these constructs alone did not complement the null strain that lacked a full-length LeuRS (data not shown).
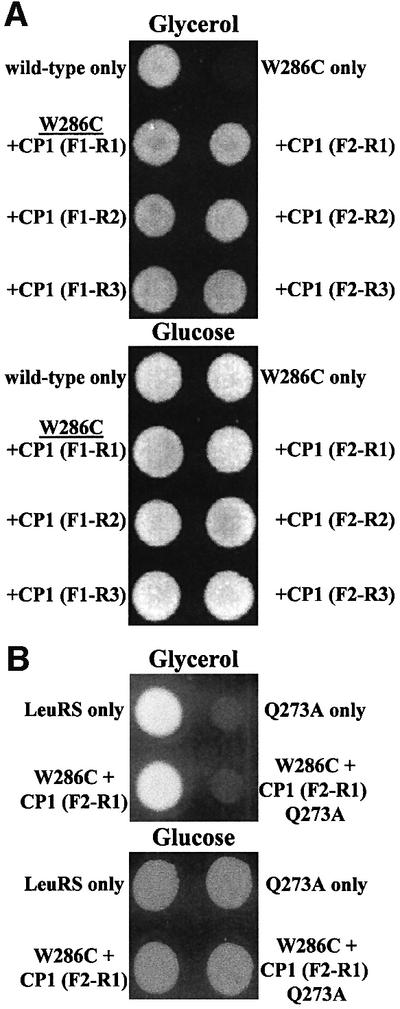
Fig. 2. Rescue of a splicing-deficient mutation by protein fragments that contained the LeuRS CP1 domain. (A) The yeast mitochondrial LeuRS deletion strain QBY320, which contains a splicing-deficient M.tuberculosis LeuRS mutant W286C [msl1Δ::HIS3 ρ–(pSBR-Mtb-W286C)] (Houman et al., 2000), was transformed with plasmids expressing different sub-genes for LeuRS fragments that contained CP1. Transformants were grown on glycerol (upper plate) and glucose (lower plate) medium (Houman et al., 2000). A positive control bearing the wild-type M. tuberculosis LeuRS (top left) grows on glycerol medium, while the W286C splicing mutant (top right) fails to complement the null strain, as described previously (Houman et al., 2000). Each of the six separate LeuRS CP1-containing protein fragments rescues the mutant W286C LeuRS. (B) Transformants included the wild-type (top left) and Q273A mutant (top right) yeast mitochondrial LeuRSs. The M.tuberculosis W286C mutant LeuRS is complemented by the wild-type LeuRS CP1-containing fragment (bottom left), but not the corresponding Q273A mutant (bottom right).
A suppressor site that restores splicing activity in the absence of an active bI4 maturase was identified within the LeuRS CP1 domain (Figure 1A; Henke, 2000; R.M.Henke and P.S.Perlman, personal communication). We changed this site to an alanine (Q273A) within the full-length enzyme as well as the smallest LeuRS fragment that contained CP1. Neither full-length nor the fragment of the yeast mitochondrial Q273A mutant LeuRS restored complementation of the yeast null strain that expresses the splicing-defective LeuRS (Figure 2B). These combined complementation results suggest that the isolated CP1 domain is either complementing the mutant LeuRS in trans or functioning independently in the splicing role.
Three-hybrid interactions between the CP1-containing LeuRS fragments and the bI4 group I intron
Similarly to the two-hybrid assay, the three-hybrid assay relies on the coalescence of transcriptional factor fusions or hybrids to identify specific RNA–protein interactions (Figure 3A; SenGupta et al., 1996; Zhang et al., 2000). The RNA-binding protein of interest is linked to a transcriptional activation domain (B42 AD), while the DNA-binding domain (DBD) is fused to the MS2 coat protein. The RNA of interest is linked to MS2 RNA, which binds to the MS2 coat protein and effectively anchors the MS2 RNA hybrid to the transcription complex. Interactions between the RNA hybrid bait and the B42 fusion prey activate transcription of reporter genes.
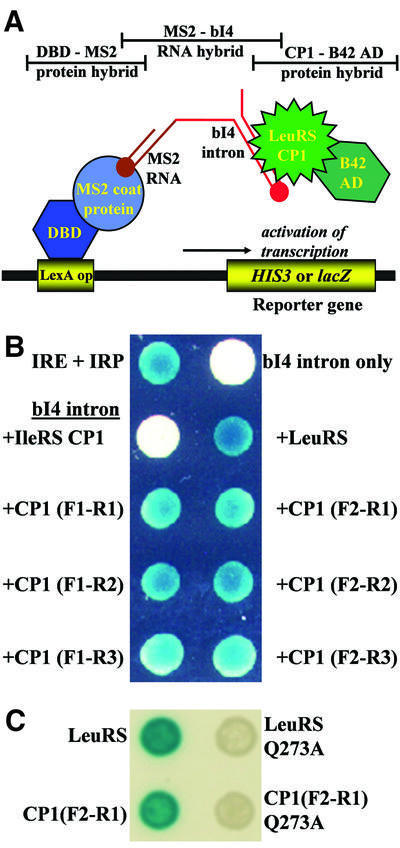
Fig. 3. Three-hybrid analysis detects binding interactions between the isolated LeuRS CP1 domains and the bI4 intron. (A) Cartoon of the three-hybrid model. The two protein hybrids are the DBD–MS2 coat protein fusion (shades of blue) and the LeuRS CP1 domain linked to B42 AD (shades of green). The bait RNA is comprised of MS2 RNA (brown) and the bI4 intron (red). Intracellular β-galactosidase reporter activity was detected by growing transformed yeast cells on solid agarose synthetic medium (Ura–, Trp–) which contained 2% galactose, 1% raffinose and 20 mg/ml X-gal (Rho and Martinis, 2000). (B) The bait bI4 intron RNA and prey proteins were expressed from the plasmids pRH3′ and pYESTrp2, respectively. Blue color developed for both the positive control (IRE and IRP) and each yeast colony that expressed bI4 intron as bait and either full-length or fragments of LeuRS that contained CP1 as prey. Expression of the bI4 intron alone (upper right corner) or with the IleRS CP1 domain (left, second from top) failed to yield blue colonies. (C) When the bait bI4 intron was tested the full-length (top right) or the smallest fragment (bottom right) of LeuRS that contained the Q273A mutation, a three-hybrid response was not detected.
Previously, we demonstrated via three-hybrid analysis that LeuRS binds specifically to the bI4 group I intron (Rho and Martinis, 2000). Here, we employed three-hybrid analysis to determine whether the bI4 intron bound directly to the LeuRS fragments that contained CP1. Three-hybrid yeast host cells (L40uraMS2) that contained the plasmid p3MbI4R encoding the bI4 intron–MS2 hybrid bait RNA were transformed with plasmids expressing the LeuRS fragments fused to B42 AD. Transformed cells were grown on histidine-deficient medium to select for expression of the HIS3 reporter gene. Three-hybrid analysis indicated that each of the LeuRS protein fragments could interact sufficiently with the bI4 intron to stimulate growth reporter activity (data not shown).
A second screen that monitors β-galactosidase activities determined that co-expression of the bI4 intron and the LeuRS fragment fusions that contained CP1 developed varying intensities of blue (Figure 3B). A fragment containing the homologous CP1 domain from IleRS that overlaps completely with the smallest LeuRS fragment did not stimulate significant reporter activity. We also determined that introducing the Q273A mutation into full-length or the smallest fragment of LeuRS abolished the three-hybrid response, supporting the view that this suppressor site is important to interactions with the group I intron (Figure 3C). These results suggest that the LeuRS CP1 domain contains determinants that specifically interact with the bI4 group I intron.
Three-hybrid-based β-galactosidase activity was also quantitated via solution-based ONPG assays (data not shown; Rho et al., 1996). The strength of the full-length LeuRS interaction with the bI4 intron was similar to the well-characterized positive IRP (iron regulatory protein) and IRE (iron response element) control. In comparison, three-hybrid reporter activities that were dependent on binding of the bI4 intron to the CP1-containing LeuRS fragments were decreased to ∼30–40%. The presence or absence of the zinc or zinc-like binding domains had minimal, if any, effects on the three-hybrid interactions between the CP1-containing LeuRS fragment and the bI4 intron. Increases in the size of the LeuRS protein fragments only modestly enhanced reporter activity.
Decreased three-hybrid binding interactions relative to the full-length LeuRS may result from one or more factors. It is possible that the truncated LeuRS protein fragments are unstable and/or not readily imported into the nucleus, which would lead to lower levels of the protein for three-hybrid interactions. The nucleus-based CP1 (F2–R1) fusion, which produced one of the weakest three-hybrid signals, was quantitated compared with the full-length LeuRS via western blot analysis. Figure 4 suggests that these prey proteins were present at similar concentrations in the yeast nucleus for three-hybrid interactions. It is likely, then, that other regions of the LeuRS that are not present in these CP1-containing fragments enhance binding of the protein splicing factor to the bI4 intron, even if these regions are not necessarily critical to the protein’s splicing activity. As one example, the C-terminal end of LeuRS has been shown to influence the protein’s splicing role (Li et al., 1996; Houman et al., 2000), but is missing in each of the LeuRS fragments.
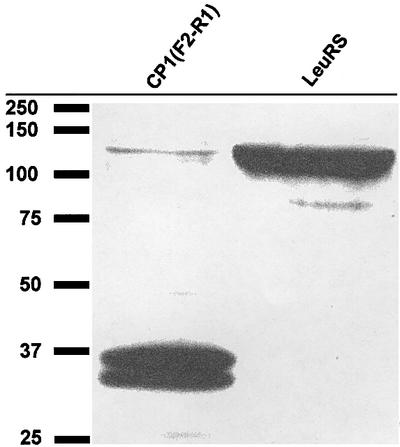
Fig. 4. Western blot of B42 AD-fused prey proteins that are imported into the nucleus of the yeast three-hybrid cells. Nuclear proteins were extracted from L40uraMS2 cells that contained the bI4 intron–MS2 hybrid bait RNA and either the full-length LeuRS (right) or the smallest CP1 (F2–R1)-containing LeuRS fragment (left), which were fused to B42 AD as prey. These prey proteins also contain a 14 amino acid V5 epitope that could be detected using an anti-V5 antibody. Protein markers (data not shown) were used to identify the prey proteins based on molecular weight. The CP1 (F2–R1)–B42 AD fusion is shown in the left lane at ∼36 kDa, while the full-length LeuRS–B42 AD fusion is in the right lane at ∼112 kDa.
The LeuRS protein fragments support group I intron splicing activity
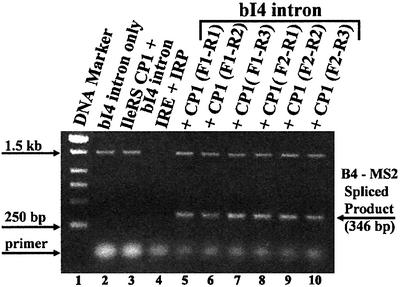
Our complementation experiments suggest that LeuRS fragments that contain CP1 might directly stimulate splicing activity. Previously, we had shown that the bI4 intron expressed in the nucleus of the three-hybrid cells yielded spliced product when LeuRS was present (Rho and Martinis, 2000). We isolated RNA from three-hybrid cells that expressed each of the LeuRS protein fragments and the bI4 intron, and tested for spliced product by RT–PCR. The bI4 intron used in this investigation contains fused 3′ B5–B6 exons and is synthesized for three-hybrid analysis with a 3′ link to two copies of MS2 RNA (SenGupta et al., 1996; Rho and Martinis, 2000). Splicing of this artificial RNA in the nucleus would yield the natural spliced B4–B5–B6 exons, but should also be fused at the 3′ end to the tandem MS2 RNA motif. We utilized this artificial MS2 fusion as a marker by designing a reverse primer (P-MS2) that specifically targeted the junction of the two linked MS2 RNAs.
The P-B4 forward and P-MS2 reverse primers were combined in an RT–PCR reaction using RNA template isolated from the three-hybrid cells. For each sample that expressed the LeuRS protein fragments, RT–PCR amplification yielded a clear band at the expected size of 346 bp, which indicates product formation (Figure 5). An upper band at ∼1.7 kb represents unspliced bI4 intron with its flanking exons. Lane 4 is a control using cells that expressed the IRE RNA and IRP protein, which interact and elicit a three-hybrid response. Lack of any bands supports the view that background or contaminating mitochondrial RNA was not amplified via RT–PCR. In the absence of plasmid expressing the CP1-containing fragment (lane 2) or, alternatively, when the IleRS CP1 domain (lane 3) was substituted, the 346 bp band was missing; this finding indicates that the nuclear splicing reaction is dependent specifically on the LeuRS CP1 domain.
Fig. 5. RT–PCR detects spliced product of the nuclear-expressed bI4 intron in the presence of CP1-containing LeuRS fragments. The RT–PCR products were produced and analyzed on a 1% agarose gel as described previously (Rho and Martinis, 2000). DNA markers are in lane 1. A diffuse band near the gel bottom is excess primer. RT–PCR primers target the B4 exon and MS2–RNA 3′ fusion. A band at ∼1.7 kb represents unspliced RNA that includes flanking exons and the intron. A band at 346 bp indicates ligated B4–B5 exons. Lane 2 shows amplified DNA from three-hybrid cells that only express the bI4 intron. A control in lane 3 shows RT–PCR-amplified RNA from three-hybrid cells that contained the bI4 intron and IleRS CP1 domain. Lane 4 represents an IRE and IRP control. Lanes 5–10 contain samples isolated from three-hybrid cells that expressed bI4 intron from p3MbI4R in the presence of the LeuRS fragments that contained CP1.
Discussion
We have demonstrated that a small protein fragment, comprised of the LeuRS CP1 region, is sufficient to stimulate the LeuRS-mediated bI4 intron splicing reaction. Deletion analysis showed that the canonical main body of the tRNA synthetase, as well as the flanking zinc and zinc-like binding domains of the CP1 insert, are dispensable for its splicing activity. This suggests that the full-length LeuRS has two distinct RNA-binding sites. In contrast, the only other known tRNA synthetase that aids splicing, TyrRS, requires multiple, non-contiguous regions of the protein that include the tRNA aminoacylation binding sites (Cherniack et al., 1990; Kittle et al., 1991; Kamper et al., 1992; Mohr et al., 2001). These differences suggest that LeuRS and TyrRS use different mechanisms to aid splicing.
Our finding that the CP1 region of LeuRS is essential to the bI4 splicing reaction is somewhat unexpected because the CP1 domain has become best known for its role in amino acid editing (Chen et al., 2000; Mursinna et al., 2001; Mursinna and Martinis, 2002). Homologous CP1 domains of LeuRS, ValRS and IleRS bind mischarged tRNA and hydrolytically edit the non-cognate amino acid (Lin et al., 1996; Chen et al., 2000; Hendrickson et al., 2000; Mursinna and Martinis, 2002). Only the LeuRS CP1 domain is known to play a role in RNA splicing. Our results show that the homologous IleRS CP1 domain can not substitute for the LeuRS CP1 to enable splicing of the bI4 intron. We hypothesize that the LeuRS CP1 domain binds cognate tRNALeu for editing in a similar manner as the bI4 intron RNA for splicing. Because LeuRSs from bacteria and other mitochondrial origins can substitute for the yeast mitochondrial LeuRS in splicing (Houman et al., 2000), it is possible that ancient adaptations for interactions of the CP1 domain with tRNALeu, which have been conserved through evolution, are also important to binding the group I intron.
The only other example of a tRNA synthetase involved in splicing is CYT-18p from N.crassa, which amino acylates tyrosine (Akins and Lambowitz, 1987). CYT-18p lacks a region that is analogous to the LeuRS CP1 insert. Rather, CYT-18p-mediated splicing activity involves a unique N-terminal extension, as well as the canonical class I aminoacylation core and the TyrRS C-terminal domain, both of which also bind to tRNATyr (Cherniack et al., 1990; Kittle et al., 1991; Kamper et al., 1992; Mohr et al., 2001). The idiosyncratic N-terminus that is critical to splicing activity does not actually bind to the group I intron or the tRNA, but appears to stabilize another portion of the protein that interacts directly with the RNA (Mohr et al., 2001). Thus, CYT-18p relies on different domains of the protein to facilitate ribozyme activity, while only the CP1 portion of LeuRS is required to aid splicing of the bI4 intron.
Extensive investigations of the molecular binding interface between CYT-18p and the group I intron suggest that the protein interacts with a tRNA-like motif on the group I intron (Caprara et al., 1996a,b; Myers et al., 2002). Interestingly, this motif possesses a structural element that mimics the unique extra long variable loop of a type II tRNA (Caprara et al., 1996a; Myers et al., 2002). LeuRS and TyrRS (from eubacteria and organelles) are two of the three tRNA synthetases that interact with type II tRNAs. (Seryl-tRNA synthetase also interacts with type II tRNAs, but as yet has not been shown to be a splicing factor.) It is quite possible that LeuRS and TyrRS were recruited because they were both predisposed to interact with the rarer type II tRNA. Since other diverse LeuRSs can function in place of the yeast mitochondrial LeuRS to enable group I intron splicing (Houman et al., 2000), it appears then that minimal, if any, additional adaptations would be required to initially confer its secondary splicing role. Interestingly, previous work supports the view that the CP1 domain was probably present in the last common ancestor of LeuRS (Schimmel and Ribas de Pouplana, 2001). Thus, it is possible that the LeuRS CP1 domain could also have been responsible for an early splicing role.
The tRNA synthetases have been suggested to be amongst the oldest family of proteins (Burbaum and Schimmel, 1991). Their evolutionary age and large diversity have facilitated their recruitment to perform many varied alternate activities (Martinis et al., 1999a,b). The canonical aminoacylation module is attached to appendages and also interrupted by insertions that range from short peptide sequences to large polypeptide domains (Starzyk et al., 1987; Burbaum and Schimmel, 1991). In some cases, unique or adapted modules confer secondary functions to the tRNA synthetase (Cherniack et al., 1990; Kamper et al., 1992; Lin et al., 1996; Lechler and Kreutzer, 1998; Schimmel and Wang, 1999; Wakasugi and Schimmel, 1999). Others have been shown to enhance aminoacylation efficiency and specificity, for example, by interacting with the tRNA (Gale and Schimmel, 1995; Wang and Schimmel, 1999; Wang et al., 2000). Although the roles of many of these protein additions remain undefined, it is clear that either just the isolated regions or combinations of the tRNA synthetase catalytic core with other modules provide a rich RNA-binding environment that would be attractive for the cell to exploit for other purposes.
Materials and methods
Materials
The plasmid pBN2 encoding the CP1 domain of Escherichia coli IleRS was kindly provided by Dr P.Schimmel (The Scripps Research Institute, La Jolla, CA). The RNA–Protein Hybrid Hunter Kit, zeocin, anti-V5 antibody and plasmid pHybLex/Zeo were purchased from Invitrogen (Carlsbad, CA).
Homology modeling of yeast mitochondrial LeuRS
The percent identity and conservation between LeuRS primary sequences were calculated using BLAST2 (Karlin and Altschul, 1993). Local sequence alignments were performed with ClustalW Multiple Sequence Alignment (Ver. 1.8; Thompson et al., 1994) and then used to manually align the primary sequences of yeast mitochondrial LeuRS with T.thermophilus LeuRS within the Homology module in Insight II (Molecular Simulations, Inc.). Conserved regions of yeast mitochondrial LeuRS were assigned coordinates based on the X-ray crystal structure coordinates of T.thermophilus LeuRS (Cusack et al., 2000). The Homology module was used to predict secondary structures of unconserved regions that link adjacent conserved regions.
Plasmid constructions
Sub-genes encoding fragments of yeast mitochondrial LeuRS were amplified by PCR using plasmid pYM3-17 (Rho and Martinis, 2000) as template and a series of 5′ and 3′ primers that contained HindIII and EcoRI restriction sites, respectively. The purified PCR fragment was cleaved with HindIII and EcoRI, and then cloned into these same restriction-digested sites of plasmid pYESTrp2 (Invitrogen). Six constructs were made that would express CP1-containing LeuRS fragments in varied sizes (Figure 1). Similarly, a control encoding the E.coli IleRS CP1 domain was amplified via PCR from pBN2, which introduced 5′ HindIII and 3′ EcoRI sites, and cloned into pYESTrp2.
Likewise, a second parallel set of CP1-encoding genes was amplified using 5′ and 3′ primers that contained XbaI and KpnI restriction sites, respectively. The plasmid pQB153 (Houman et al., 2000) and each of the PCR fragments were digested with XbaI and KpnI, and then ligated. These resultant plasmids were also digested with ScaI and NsiI to remove the LEU2 marker, which was replaced by the ScaI–NsiI fragment from plasmid YEplac112 (Sugino and Gietz, 1988) to introduce the Trp1 marker, yielding plasmids pSBR/F1-R1, pSBR/F1-R2, pSBR/F1-R3, pSBR/F2-R1, pSBR/F2-R2 and pSBR/F2-R3.
Three-hybrid and RT–PCR analysis
Standard yeast molecular biology methods and media preparation were carried out according to established procedures (Lundblad, 1995). Three-hybrid reporter activity, β-galactosidase and ONPG assays, and RT–PCR analysis have been described previously (Rho and Martinis, 2000). Upon cell lysis, yeast nuclei and mitochondria were separated from each other and other cellular components using differential buffer and centrifugation conditions (Ausubel et al., 1994). Western blot analysis utilized anti-V5 antibodies (Invitrogen) and was carried out according to the commercial protocol. The RT–PCR MS2-RNA targeted primer is 5′-ATGTTTT CTAGAGTCGACCTGCAG-3′.
Complementation of yeast mitochondrial LeuRS deletion strain
The yeast mitochondrial LeuRS deletion strain QBY320 [MATa ade2-1 ura3-1 his3-11,15 trp1Δ63 leu2-3,112 can1-100 msl1Δ::HIS3 ρ+(pQB223)], containing a maintenance plasmid pQB223 (URA3 marker) that expresses mitochondrial LeuRS, has been described previously (Houman et al., 2000) and was used for complementation studies. QBY320 was transformed with plasmids (containing a LEU2 selectable marker) that expressed wild-type yeast mitochondrial (pQB184) and M.tuberculosis LeuRS (pC3-Mtb) (Houman et al., 2000) as well as the yeast mitochondrial mutant Q273A LeuRS (pQB184-273A). QBY320 was also transformed with a plasmid that encodes the W286C mutant of M.tuberculosis LeuRS (Houman et al., 2000), which abolishes LeuRS-dependent mitochondrial splicing activity. Selected transformants were grown on 5-FOA medium to induce loss of the maintenance plasmid pQB223. Subsequently, the cells were tested for growth on glycerol medium and, as described previously, both wild-type genes complemented the null strain, while the M.tuberculosis (Houman et al., 2000) and yeast mitochondrial gene mutants failed to rescue the strain.
Competent cells harboring plasmids expressing the M.tuberculosis LeuRS W286C mutants were transformed with a second series of plasmids (pSBR/F1-R1, pSBR/F1-R2, pSBR/F1-R3, pSBR/F2-R1, pSBR/F2-R2 and pSBR/F2-R3), which encoded varied sizes of LeuRS fragments that contained CP1. These cells were also transformed with a plasmid encoding the site-specific mutant (Q273A) form of the F2–R1 LeuRS fragment. This same series of plasmids was also used to transform QBY320, followed by incubation on 5-FOA to select for loss of the maintenance plasmid. Each of these isolated colonies was tested for complementation activity on glycerol media (Houman et al., 2000).
Acknowledgments
Acknowledgements
We thank Dr P.S.Perlman for release of unpublished data. We are grateful to Dr J.Briggs for aid in our computational work and also to Dr P.Schimmel for helpful advice and providing the clone for the IleRS CP1 domain. This work was supported in part by The National Institutes of Health (GM63107), The Robert A.Welch Foundation (E-1404), the Texas Advanced Research Program (003652-0017-1999) and the University of Houston Grants to Enhance and Advance Research (GEAR-116346).
References
- Akins R.A. and Lambowitz,A.M. (1987) A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell, 50, 331–345. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidmen,J.G., Smith,J.A. and Struhl,K. (1994) Current Protocols in Molecular Biology. Wiley, New York, NY, pp. 13.13.1–13.13.9.
- Burbaum J.J. and Schimmel,P. (1991) Structural relationships and the classification of aminoacyl-tRNA synthetases. J. Biol. Chem., 266, 16965–16968. [PubMed] [Google Scholar]
- Caprara M.G., Lehnert,V., Lambowitz,A.M. and Westhof,E. (1996a) A tyrosyl-tRNA synthetase recognizes a conserved tRNA-like structural motif in the group I intron catalytic core. Cell, 87, 1135–1145. [DOI] [PubMed] [Google Scholar]
- Caprara M.G., Mohr,G. and Lambowitz,A.M. (1996b) A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J. Mol. Biol., 257, 512–531. [DOI] [PubMed] [Google Scholar]
- Carter C.W. Jr (1993) Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem., 62, 715–748. [DOI] [PubMed] [Google Scholar]
- Chen J.F., Guo,N.N., Li,T., Wang,E.D. and Wang,Y.L. (2000) CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry, 39, 6726–6731. [DOI] [PubMed] [Google Scholar]
- Cherniack A.D., Garriga,G., Kittle,J.D.,Jr, Akins,R.A. and Lambowitz,A.M. (1990) Function of Neurospora mitochondrial tyrosyl-tRNA synthetase in RNA splicing requires an idiosyncratic domain not found in other synthetases. Cell, 62, 745–755. [DOI] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk,A. and Tukalo,M. (2000) The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J., 19, 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G. and Herbert,C.J. (1997) Aminoacyl tRNA synthetases involved in group I intron splicing. In Green,R. and Schroeder,R. (eds), Ribosomal RNA and Group I Introns. Landes Bioscience, Austin, TX, pp. 179–198.
- Gale A.J. and Schimmel,P. (1995) Isolated RNA binding domain of a class I tRNA synthetase. Biochemistry, 34, 8896–8903. [DOI] [PubMed] [Google Scholar]
- Guo Q. and Lambowitz,A.M. (1992) A tyrosyl-tRNA synthetase binds specifically to the group I intron catalytic core. Genes Dev., 6, 1357–1372. [DOI] [PubMed] [Google Scholar]
- Hale S.P., Auld,D.S., Schmidt,E. and Schimmel,P. (1997) Discrete determinants in transfer RNA for editing and aminoacylation. Science, 276, 1250–1252. [DOI] [PubMed] [Google Scholar]
- Hendrickson T.L., Nomanbhoy,T.K. and Schimmel,P. (2000) Errors from selective disruption of the editing center in a tRNA synthetase. Biochemistry, 39, 8180–8186. [DOI] [PubMed] [Google Scholar]
- Henke R.M. (2000) Molecular and biochemical studies on a bifunctional group I intron encoded protein of yeast mitochondria. PhD dissertation, The University of Texas Southwestern Medical Center at Dallas, Dallas, TX.
- Herbert C.J., Labouesse,M., Dujardin,G. and Slonimski,P.P. (1988) The NAM2 proteins from S.cerevisiae and S.douglasii are mitochondrial leucyl-tRNA synthetases and are involved in mRNA splicing. EMBO J., 7, 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.-M., Shiba,K., Mottes,C. and Schimmel,P. (1991) Sequence determination and modeling of structural motifs for the smallest monomeric aminoacyl-tRNA synthetase. Proc. Natl Acad. Sci. USA, 88, 976–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houman F., Rho,S.B., Zhang,J., Shen,X., Wang,C.-C., Schimmel,P. and Martinis,S.A. (2000) A prokaryote and human tRNA synthetase provide an essential RNA splicing function in yeast mitochondria. Proc. Natl Acad. Sci. USA, 97, 13743–13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamper U., Kuck,U., Cherniack,A.D. and Lambowitz,A.M. (1992) The mitochondrial tyrosyl-tRNA synthetase of Podospora anserina is a bifunctional enzyme active in protein synthesis and RNA splicing. Mol. Cell. Biol., 12, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S. and Altschul,S.F. (1993) Applications and statistics for multiple high-scoring segments in molecular sequences. Proc. Natl Acad. Sci. USA, 90, 5873–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittle J.D. Jr, Mohr,G., Gianelos,J.A., Wang,H. and Lambowitz,A.M. (1991) The Neurospora mitochondrial tyrosyl-tRNA synthetase is sufficient for group I intron splicing in vitro and uses the carboxy-terminal tRNA-binding domain along with other regions. Genes Dev., 5, 1009–1021. [DOI] [PubMed] [Google Scholar]
- Labouesse M. (1990) The yeast mitochondrial leucyl-tRNA synthetase is a splicing factor for the excision of several group I introns. Mol. Gen. Genet., 224, 209–221. [DOI] [PubMed] [Google Scholar]
- Labouesse M., Dujardin,G. and Slonimski,P.P. (1985) The yeast nuclear gene NAM2 is essential for mitochondrial DNA integrity and can cure a mitochondrial RNA-maturase deficiency. Cell, 41, 133–143. [DOI] [PubMed] [Google Scholar]
- Labouesse M., Herbert,C.J., Dujardin,G. and Slonimski,P.P. (1987) Three suppressor mutations which cure a mitochondrial RNA maturase deficiency occur at the same codon in the open reading frame of the nuclear NAM2 gene. EMBO J., 6, 713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz A.M. and Perlman,P.S. (1990) Involvement of aminoacyl-tRNA synthetases and other proteins in group I and group II intron splicing. Trends Biochem. Sci., 15, 440–444. [DOI] [PubMed] [Google Scholar]
- Lambowitz A.M., Caprara,M.G., Zimmerly,S. and Perlman,P.S. (1999) Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–485.
- Larkin D.C., Williams,A.M., Martinis,S.A. and Fox,G.E. (2002) Identification of essential domains for Escherichia coli tRNAleu aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res., 30, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler A. and Kreutzer,R. (1998) The phenylalanyl-tRNA synthetase specifically binds DNA. J. Mol. Biol., 278, 897–901. [DOI] [PubMed] [Google Scholar]
- Li G.-Y., Becam,A.-M., Slonimski,P.P. and Herbert,C.J. (1996) In vitro mutagenesis of the mitochondrial leucyl-tRNA synthetase of Saccharomyces cerevisiae shows that the suppressor activity of the mutant proteins is related to the splicing function of the wild type protein. Mol. Gen. Genet., 252, 667–675. [DOI] [PubMed] [Google Scholar]
- Lin L., Hale,S.P. and Schimmel,P. (1996) Aminoacylation error correction. Nature, 384, 33–34. [DOI] [PubMed] [Google Scholar]
- Lundblad V. (1995) Saccharomyces cerevisiae. In Ausubel,F., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds), Short Protocols in Molecular Biology, 3rd edn. Wiley, New York, NY, pp. 13–51.
- Martinis S.A. and Schimmel,P. (1996) Aminoacyl tRNA synthetases: general structures and relationships. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella Cellular and Molecular Biology, 2nd edn. ASM Press, Washington, DC, pp. 887–901.
- Martinis S.A., Plateau,P., Cavarelli,J. and Florentz,C. (1999a) Aminoacyl-tRNA synthetases: a family of expanding functions. EMBO J., 18, 4591–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinis S.A., Plateau,P., Cavarelli,J. and Florentz,C. (1999b) Aminoacyl-tRNA synthetases: a new image for a classical family. Biochimie, 81, 683–700. [DOI] [PubMed] [Google Scholar]
- Mohr G., Rennard,R., Cherniack,A.D., Stryker,J. and Lambowitz,A.M. (2001) Function of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase in RNA splicing. Role of the idiosyncratic N-terminal extension and different modes of interaction with different group I introns. J. Mol. Biol., 307, 75–92. [DOI] [PubMed] [Google Scholar]
- Mursinna R.S. and Martinis,S.A. (2002) Rational design to block amino acid editing of a tRNA synthetase. J. Am. Chem. Soc., 124, 7286–7287. [DOI] [PubMed] [Google Scholar]
- Mursinna R.S., Lincecum,T.L.,Jr and Martinis,S.A. (2001) A conserved threonine within Escherichia coli leucyl-tRNA synthetase prevents hydrolytic editing of leucyl-tRNALeu. Biochemistry, 40, 5376–5381. [DOI] [PubMed] [Google Scholar]
- Myers C.A., Kuhla,B., Cusack,S. and Lambowitz,A.M. (2002) tRNA-like recognition of group I introns by a tyrosyl-tRNA synthetase. Proc. Natl Acad. Sci. USA, 99, 2630–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nureki O. et al. (1998) Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science, 280, 578–582. [DOI] [PubMed] [Google Scholar]
- Rho S.B. and Martinis,S.A. (2000) The bI4 group I intron binds directly to both its protein splicing partners, a tRNA synthetase and maturase, to facilitate RNA splicing activity. RNA, 6, 1882–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho S.B., Lee,K.H., Kim,J.W., Shiba,K., Jo,Y.J. and Kim,S. (1996) Interaction between human tRNA synthetases involves repeated sequence elements. Proc. Natl Acad. Sci. USA, 93, 10128–10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rould M.A., Perona,J.J., Söll,D. and Steitz,T.A. (1989) Structure of E.coli glutaminyl-tRNA synthetase complexed with tRNAGln and ATP at 2.8 Å resolution. Science, 246, 1135–1142. [DOI] [PubMed] [Google Scholar]
- Schimmel P. and Ribas de Pouplana,L. (2000) Footprints of aminoacyl-tRNA synthetases are everywhere. Trends Biochem. Sci., 25, 207–209. [DOI] [PubMed] [Google Scholar]
- Schimmel P. and Ribas de Pouplana,L. (2001) Formation of two classes of tRNA synthetases in relation to editing functions and genetic code. Cold Spring Harb. Symp. Quant. Biol., LXVI, 161–166. [DOI] [PubMed] [Google Scholar]
- Schimmel P. and Wang,C.-C. (1999) Getting tRNA synthetases into the nucleus. Trends Biochem. Sci., 24, 127–128. [DOI] [PubMed] [Google Scholar]
- SenGupta D.J., Zhang,B., Kraemer,B., Pochart,P., Fields,S. and Wickens,M. (1996) A three-hybrid system to detect RNA–protein interactions in vivo. Proc. Natl Acad. Sci. USA, 93, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvian L.F., Wang,J. and Steitz,T.A. (1999) Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science, 285, 1074–1077. [PubMed] [Google Scholar]
- Starzyk R.M., Webster,T.A. and Schimmel,P. (1987) Evidence for dispensable sequences inserted into a nucleotide fold. Science, 237, 1614–1618. [DOI] [PubMed] [Google Scholar]
- Sugino A. and Gietz,R.D. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL 2: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi K., Quinn,C.L., Tao,N. and Schimmel,P. (1998) Genetic code in evolution: switching species-specific aminoacylation with a peptide transplant. EMBO J., 17, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi K. and Schimmel,P. (1999) Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science, 284, 147–151. [DOI] [PubMed] [Google Scholar]
- Wang C.C. and Schimmel,P. (1999) Species barrier to RNA recognition overcome with nonspecific RNA binding domains. J. Biol. Chem., 274, 16508–16512. [DOI] [PubMed] [Google Scholar]
- Wang C.C., Morales,A.J. and Schimmel,P. (2000) Functional redundancy in the nonspecific RNA binding domain of a class I tRNA synthetase. J. Biol. Chem., 275, 17180–17186. [DOI] [PubMed] [Google Scholar]
- Weiner A.M. and Maizels,N. (1999) A deadly double life. Science, 284, 63–64. [DOI] [PubMed] [Google Scholar]
- Zhang B., Kraemer,B., SenGupta,D., Fields,S. and Wickens,M. (2000) Yeast three-hybrid system to detect and analyze RNA–protein interactions. Methods Enzymol., 318, 399–419. [DOI] [PubMed] [Google Scholar]