Introduction
When writing a scientific paper it makes sense to tell the story in a way that is clear to the reader rather than necessarily sticking to historical accuracy. A side effect of such revisionism is that the author can impress the reader with the incisive logic of his or her reasoning that led, with elegant inevitability, from the big question to the significant conclusion. But as we all know, the path of discovery actually meanders from one result to the next, with fresh insights appearing more rarely than we like to admit—it often resembles Brownian motion more than historical inevitability. I was hugely pleased to be awarded the 2001 EMBO gold medal and looking down the list of the 17 previous winners, I certainly felt the honour of the prize. But my pleasure was slightly tempered by the invitation to write a review of my work in an autobiographical style. I saw the danger of giving the impression that I considered myself the sole protagonist in a scientific odyssey where the only results were ‘key’, the only conclusions ‘significant’, and the only insights were ‘broad and general’. However, my predecessors had succeeded in avoiding the trap, and I enjoyed the personal element of their reviews; they present glimpses of the real choices and decisions they had faced. I therefore suppressed the urge to omit the autobiographical element of this review.
Pre-PhD
As a school science student trying to decide what to do at university, I was drawn to chemistry, mainly because I was better at it than physics and I wasn’t studying biology. But I had a hunch, based partly on an idealistic belief in the value of medical research, that biochemistry might be more exciting. This was despite not actually knowing what biochemistry really involved. At about the same time I had the opportunity to meet the Nobel Prize-winning immunologist Peter Medawar. In what was, I later discovered, a characteristically out rageous generalization (but like most of his opinions, insightful) he told me that chemistry was dead, physics was dying, and that biological science was where the future lay. As a seventeen year-old, this grand old man of science was a figure of some awe and a year later I found myself studying biochemistry at Oxford. The biochemistry course was in fact mostly chemistry in the first year and this knocked out of me any residual desire to be a chemist. In fact, no science figured very highly in my first 3 years at Oxford, which were occupied more by student journalism and politics. But by the fourth year I discovered the science and, with hindsight, a course on Drosophila genetics by David Roberts was a significant influence. I remember being struck by his introduction to a lecture one morning when he told us that he would be describing the ‘most important biological experiment of the last decade’: it was Gerry Rubin’s and Allan Spradling’s demonstration of P-element mediated transformation in Drosophila (Spradling and Rubin, 1982). Four years later I joined Gerry Rubin’s lab as a postdoc.
Imperial College
But before that, I went to David Glover’s group to help start a new project on the cell cycle. This was rather speculative and untested but immediately gripped me. Another attractive thing about Glover’s lab was that it formed one quarter of a Cancer Research Campaign-funded group at Imperial College in London, with the groups of Jean Beggs, David Lane and Peter Rigby all on the same floor. These four had been brought together as a gang of young group-leaders researching the nascent field of eukaryotic molecular genetics, and it was an exciting place to be. The first year and a half did not live up to my initial hopes and included a number of dead ends. I attempted to clone homologs of yeast cell cycle genes and, in collaboration with David Lane, a Drosophila p53. Apart from some early excitement about a homolog of a yeast gene provided by Paul Nurse (which evaporated after a couple of weeks when I realized that it was a contaminant from the yeast-based fly food that Drosophila live on), nothing worked. But then Christiane Nüsslein-Volhard sent us a number of maternal effect lethal mutations whose phenotypes suggested cell cycle defects. I jumped at the opportunity to study the most striking, which we named giant nuclei, or gnu (Freeman et al., 1986).
As its name implies, gnu embryos have huge nuclei: usually only two or three per embryo, each with the DNA content of hundreds of diploid nuclei (Figure 1). I discovered that, despite the complete absence of nuclear division, the centrosome division cycle was relatively normal (Freeman et al., 1986), and that the primary defect in gnu embryos was a failure of the normal block to DNA replication that occurs in unfertilized eggs (Freeman and Glover, 1987). Fifteen years later, the exact function of Gnu remains unclear but a long term effort by Terry Orr-Weaver’s lab has shown that in concert with two other proteins, Plutonium and Pan gu, it regulates the levels of cyclins during the specialized and rapid cell divisions that occur in the early embryo (Lee et al., 2001b).
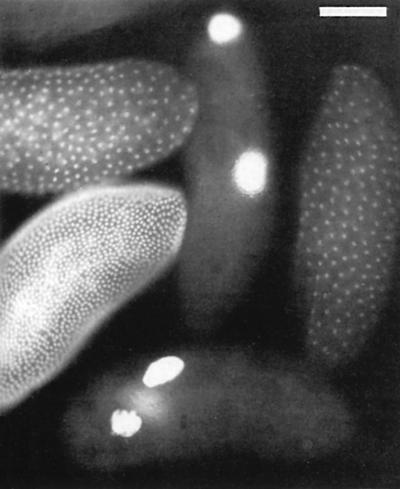
Fig. 1. Two gnu embryos alongside three wild-type embryos at different stages of early development. They are stained with a DNA dye to highlight the giant nuclei phenotype.
Berkeley
Working in David Glover’s group convinced me that Drosophila was a good organism in which to study complex questions, so I looked for a postdoc position in a lab where I could pursue this general approach. Gerry Rubin had just switched his group from working on P-elements to ‘neurobiology’ in a rather broad sense—on the grounds that there were interesting questions to be answered and it was then relatively under-populated. After meeting him while he was on sabbatical at the MRC Laboratory of Molecular Biology in Cambridge, where he had done his PhD, I was offered a place in his group. Gerry’s lab in Berkeley was a stimulating place to work. There were about 12 postdocs and four or five graduate students at the time, and all were working either on their own or in small groups on a wide range of different projects that had started out with the general theme of neurobiology but had rapidly diverged, although most did experimentally focus on the eye. Despite a depressingly familiar slow start featuring the disco gene, I loved working in Berkeley. The combination of strong science, the great outdoors, a lively city with excellent and cheap restaurants, good weather and views of the Golden Gate Bridge from my apartment added up to one of the best places I could imagine to be a postdoc. I also discovered, and enjoyed, the very strong American work ethic. Within Gerry’s lab there was a degree of commitment and hard work that I had not come across in England. Twelve-hour days and working through weekends were common and although this might not be sustainable forever, it taught me a lot about how much one can get done when necessary.
My work began to take off when I collaborated with a small group of postdocs to carry out a large enhancer trap screen—then brand new technology—to look for genes with developmentally significant expression patterns in the eye. In about 6 months screening I found about 12 interesting-looking genes, from which I selected two to pursue. One I named argos (after the multiple-eyed shepherd from Greek mythology—argos mutants had extra photoreceptors), and the other turned out to be an insertion in the rhomboid gene, which had been identified in the Nüsslein-Volhard/Wieschaus screen for zygotic lethals (Jürgens et al., 1984). Although these projects were eventually to converge, at the time they appeared quite distinct and it was simply a matter of insurance against disappointment that I chose to work on both.
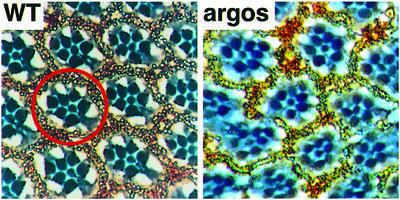
Before leaving Berkeley I had cloned argos and shown it to encode a secreted protein with an EGF motif that was involved in regulating the recruitment of cells in the developing eye (Freeman et al., 1992b). In the case of rhomboid, I had shown that its ectopic expression induced extra photoreceptors, and Bruce Kimmel and I had found it to be regulated by the rough homeobox gene (Freeman et al., 1992a). These two projects had an intriguing common phenotype: loss of function of argos and ectopic expression of rhomboid both led to excess recruitment of photoreceptor cells (Figure 2).
Fig. 2. Sections through wild-type and argos mutant adult eyes. A single ommatidium is circled. In the wild-type eye, each ommatidium has seven visible photoreceptors (highlighted by the dark circular rhabdomeres) which adopt a stereotypical trapezoid arrangement. argos mutants often have one or two extra photoreceptors per ommatidium. Overexpression of rhomboid causes a similar phenotype.
Cambridge and the LMB
Although in principle I wanted to return to Britain after my postdoc, science funding was not in good shape at the time and I was not certain that I would find a position that was attractive—I had been spoiled by the Bay Area. But when offered a place in the Cell Biology division of the Medical Research Council Laboratory of Molecular Biology (the LMB) in Cambridge, it was an easy decision to join a place with such a great reputation. With hindsight, this was an excellent move, and I have been strongly influenced and helped by the LMB’s approach to science, which includes small groups, being tough about what questions to address, encouraging interaction between groups with disparate experience, and not allowing any room for complacency. This last point often manifests itself as critical questioning of your experiments and the ideas behind them during lunch in the canteen—not great for the digestion but excellent for the science. Crucially, these attitudes are coupled with stable, long-term funding and scientific freedom.
For the first year or so of my time at the LMB, I worked on my own. The first person to join me was Richard Smith, an excellent technician with whom I worked from 1993 until his retirement in 2000. Without a group, I decided to focus on a single project and I opted for Argos. At this stage I knew only that it was a secreted protein with a function of preventing ‘mystery cells’ from becoming photoreceptors (Freeman et al., 1992b). To understand its function more clearly, I ectopically expressed it and found that it was able to inhibit the formation not only of photoreceptors but also of the other cell-types in the ommatidium—the non-neuronal cone and pigment cells (Freeman, 1994a). This hinted that there was some common element to the recruitment of these diverse cell types.
Despite trying to remain focused on one project, I continued to work on rhomboid as a sideline. Based on embryonic phenotypes, rhomboid belongs to the spitz group of genes (Mayer and Nüsslein-Volhard, 1988) and the striking similarity of phenotypes of this group suggested that they might act in a common pathway. spitz had just been cloned in Norbert Perrimon’s lab and encoded a protein similar to mammalian TGFα, a ligand for the EGF receptor, although it was not at all clear whether this similarity implied that Spitz was a ligand for the fly EGFR (Rutledge et al., 1992). Nevertheless, I tested whether loss of Spitz might cause a phenotype in the eye, and also looked to see whether other members of the spitz group and the EGFR itself might interact genetically with rhomboid. In fact, I was partially scooped on the genetic interactions between rhomboid and the EGFR by two groups working on different aspects of fly development—the oocyte and the wing (Ruohola-Baker et al., 1993; Sturtevant et al., 1993)—but we all reached the same general conclusion that Rhomboid mediated EGFR signalling by some undefined mechanism (Freeman, 1994b).
The work on Spitz suggested that the EGFR functioned in cell recruitment in the eye. In fact, the possible role of this receptor was first analysed by Nick Baker, when he was a postdoc in Gerry Rubin’s lab at the same time as me. He had shown that a dominant eye mutation, Ellipse, in which very few ommatidia form, was a gain-of-function allele of the EGFR (Baker and Rubin, 1989, 1992). This suggested that the normal function of the receptor was to inhibit ommatidial formation. A year or two later, Tian Xu, also a postdoc in Gerry Rubin’s lab, found that EGFR was necessary for photoreceptor recruitment, that is, its role was a positive one (Xu and Rubin, 1993). This appeared to contradict Baker’s conclusion, and the role of the EGFR in cell recruitment in the eye remained unclear (more recently, Nick Baker has explained the basis for the Ellipse phenotype; Lesokhin et al., 1999).
My results with Spitz in the eye suggested that the EGFR probably did have a positive role in cell determination and this led me to wonder whether Argos, the secreted inhibitor of determination with an EGF-like motif, could be an inhibitory ligand of the EGFR. At this stage, this was wishful thinking—there were no other inhibitory ligands known for any receptor tyrosine kinase in any organism, and the mere presence of an EGF domain is not diagnostic of an EGFR ligand. Nevertheless, my first PhD student, Rob Howes, obtained convincing genetic evidence for the idea. But we wanted to demonstrate it more directly, leading to a fruitful collaboration with Benny Shilo at the Weizmann Institute. His lab had just developed (but not yet published) a tissue culture assay for measuring Drosophila EGFR activation by Spitz (Schweitzer et al., 1995b). He offered to try Argos in their assay and, as I had already purified Argos protein, I sent some immediately. The experiment worked first time: Argos was clearly able to inhibit EGFR activation by Spitz in a competitive and reversible fashion, thereby confirming the genetic evidence (Schweitzer et al., 1995a). To date, Argos is the only known inhibitory ligand of the EGF receptor in any organism; it acts by directly binding the receptor, and appears to block dimerization rather than simply competing for the Spitz binding site (Howes et al., 1998; Jin et al., 2000; Vinós and Freeman, 2000). Importantly for its function, Argos participates in a negative feedback loop, in which activation of the EGFR pathway induces its own inhibitor (Golembo et al., 1996b). This has helped to highlight the significance of feedback as a regulator of many different kinds of signalling (Wasserman and Freeman, 1998; Perrimon and McMahon, 1999; Freeman, 2000).
EGFR in the eye
At that time (1995), most of what was known about cell recruitment in the developing eye was based on the example of the receptor tyrosine kinase Sevenless, which is necessary for the recruitment of the R7 photoreceptor (Campos-Ortega et al., 1979; Harris et al., 1976; Tomlinson and Ready, 1986; Banerjee et al., 1987; Hafen et al., 1987). This example of a receptor dedicated to a single cell type had led to a model of combinatorial induction in which a number of such receptors specified the recruitment of the different photoreceptor cell types by a combinatorial code of signals (Tomlinson et al., 1987; Tomlinson and Ready, 1987). Our results with Argos, Spitz and Rhomboid suggested an alternative hypothesis—that the EGFR was responsible for recruiting all cells in the developing ommatidium. This idea was also supported by the EGFR mutant clones made by Tian Xu (Xu and Rubin, 1993). The problem with this experiment was that the EGFR mutant cells were sick, so it was impossible to know whether their failure to be recruited was a secondary effect. I tried a different approach, by expressing a dominant-negative (DN) version of the receptor that had been made by the Shilo lab. By expressing the DN receptor after the period when the EGFR was required for cell survival, it became clear that there was a primary requirement for EGFR signalling in recruiting all cells apart from the R8 founder cell of each ommatidium (Freeman, 1996).
EGFR activation was sufficient, as well as necessary, to recruit the different cell types: overexpression of either Spitz or a constitutively active form of the receptor triggered the recruitment of all the different photoreceptors, the cone cells and the pigment cells (Freeman, 1996). The decision about which fate a cell adopted once it was recruited appeared to be a function of the stage in development at which the signal was received, rather than being dependent on the nature of the signal: the signals themselves are information-poor. On the basis of these results, I proposed an alternative model to explain the recruitment and specification of the cells within the ommatidium (Freeman, 1996, 1997; see Box 1).
Box 1. EGFR in ommatidial recruitment—a model.
| I proposed that the founder R8 cell of each ommatidium is specified independently of EGFR signalling but that all other cells are recruited by consecutive waves of EGFR activation, triggered by Spitz, Rhomboid and Star (see main text). A negative feedback loop mediated by Argos, the inhibitor, limits the range of each of these waves of signalling. Although this model accounts for current data quite well, it has the obvious disadvantage of not explaining in any detail how the same receptor can trigger so many different cell types: presumably it reflects the ‘state’ of the cell that receives the triggering signal from the EGFR, but what specifies this? The existence of a number of transcription factors which affected the identity of recruited cells hinted that the answer would be in the repertoire of transcription factors that cells express at any stage in their developmental history (Freeman, 1997). Subsequent work from the labs of Utpal Banerjee and Richard Carthew solidified this general but vague notion and extended it importantly by determining the precise complement of transcription factors that encode the different cell fates in the eye (Flores et al., 2000; Xu et al., 2000). |
| More recently, we have extended our analysis of the role of the EGFR in Drosophila eye development, showing that it has several functions beyond recruiting cells (e.g. regulation of proliferation, cell survival and spacing the ommatidia) (Domínguez et al., 1998; Baonza et al., 2001). These multiple functions emphasize the point that EGFR signalling is not dedicated to a particular function; instead the response is dependent on the context and developmental history of the cell that receives the signal. |
Patterning the egg
By 1997 my group had grown to four people and my focus was the EGF receptor—the molecular details of how it is regulated and the logic of how it controls developmental decisions and pattern. Not only does this receptor have multiple important functions in Drosophila—from cell fate determination to regulation of cell division and death (reviewed in Schweitzer and Shilo, 1997)—but it acts as a model for human EGFR signalling. Since loss of control of the human receptor is implicated in many diseases, the hope is that the regulatory mechanisms we identify in flies will ultimately have clinical relevance (albeit indirectly).
We turned back to Rhomboid and a question that had been nagging us and others for several years: how does Rhomboid function and, as a preliminary issue, does it mediate EGFR signalling by working in the signal-sending or signal-receiving cell? Since the Rhomboid protein gave no hints about its function (Bier et al., 1990), this was far from obvious. Several models had been proposed over the years but the predominant one (which finally proved to be correct) came most strongly from Benny Shilo’s lab. Using ectopic expression of Rhomboid in the embryo, they had shown that Rhomboid could induce EGFR signalling at a distance from the Rhomboid-expressing cells, suggesting that it functioned in signal emission (Golembo et al., 1996a). In the light of earlier evidence that Spitz required proteolytic cleavage (Freeman, 1994b; Schweitzer et al., 1995b), they postulated that it might be involved in cleaving the membrane-tethered Spitz. The experiments were not absolutely conclusive but they felt right. Yet there was a contradictory experiment that had been done in Yuh-Nung Jan’s lab. They had used genetic mosaics to show that in the developing egg, Rhomboid was required in the somatic follicle cells, which are the cells that express the EGFR and receive the signal, and is definitely not required in the oocyte, the source of the ligand (in this case not Spitz but another TGFα-like ligand, Gurken) (Ruohola-Baker et al., 1993). These two results both appeared solid but were mutually incompatible and therefore needed explaining.
The answer came by asking a simple question (and ignoring some already published work). If the function of Rhomboid in other places is to activate Spitz signalling, could it be that Spitz as well as Gurken functions during oogenesis? Jonathan Wasserman, the first of a string of excellent Canadian PhD students in my group, tested this possibility by making loss-of-function clones of spitz, either in the follicle cells or in the oocyte: the result was clear. Eggs developed normally when Spitz was removed from the oocyte but showed phenotypes characteristic of a reduction of EGFR signalling when it was removed from the follicle cells (Wasserman and Freeman, 1998). This result was the key to a two-stage model. The initial EGFR signalling event is, as shown earlier by Trudi Schupbach, Gurken signalling from the oocyte to the follicle cells (Schüpbach, 1987). This causes Rhomboid to be expressed in the follicle cells (Ruohola-Baker et al., 1993) leading to signal amplification, since Spitz in the presence of Rhomboid now becomes an effective autocrine signal (Wasserman and Freeman, 1998). Although this did not prove that Rhomboid functions to trigger active Spitz production, it did remove the glaring contradiction about the site of Rhomboid function, since the same follicle cells both send and receive EGFR signalling (see Box 2).
Box 2.
| Our work on egg patterning led us to propose how the EGFR actually patterns one aspect of the egg. This derived from the striking phenotype of removing Spitz from the follicle cells. The two large appendages that emerge from either side of the dorsal midline of the egg became fused into a single central appendage (Figure 3). Rather strangely, loss of Argos caused a very similar phenotype, despite Argos being an inhibitor not an activator of EGFR signalling (Wasserman and Freeman, 1998). We realized that this must be telling us something about how the dorsolateral position of the wild-type appendages is specified, and eventually we were able to explain how the receptor controls this simple example of pattern ing (Wasserman and Freeman, 1998). The model relies on interlocking and sequential feedback loops: initially positive feedback—the amplification by Rhomboid and Spitz described above—which then triggers negative feedback, mediated by Argos. These two cycles of regulation produce an initial peak of EGFR activity at the dorsal midline, at which stage the dorsal/ventral axis is specified (Schüpbach, 1987), followed by a splitting into twin peaks of signalling, laterally displaced, which specify the position of the appendages (Wasserman and Freeman, 1998). This mechanism is interesting because it illustrates the logic of how a single signalling pathway, despite being quite information-poor (all that happens is a cell knows that it has been nudged by a neighbour), can control pattern formation, albeit of a simple kind. |
Screens and Sprouty
As well as analysing the logic of how already-identified components of the EGFR pathway regulate development, we have used fly genetics to identify new regulators of signalling. We turned to the approach that had been largely pioneered by Mike Simon when we were both postdocs in Gerry Rubin’s lab—the dominant modifier screen (Simon et al., 1991). This is a very simple concept. You perturb your favourite pathway, in our case by overexpressing argos in the eye and thereby partially blocking EGFR signalling. This leads to a visible phenotype (rough eyes) and, importantly, the severity of the phenotype is sensitive to small changes in overall signalling levels. Randomly mutagenized chromosomes are crossed into this background and selected if they dominantly suppress or enhance the original phenotype. If they do, it implies that halving the dose of the mutated gene causes a change in EGFR signalling intensity. The real key to these screens is to have secondary and, if possible, even tertiary, counter-screens with which to re-test the primary candidates.
My student Tanita Casci performed the first of what has turned out to be many such screens we have done, and identified an important new inhibitor of EGFR signalling, Sprouty, which is conserved in mammals (Casci et al., 1999). Our task of identifying the mutated gene was made much easier by the fact that Mark Krasnow cloned Sprouty just as we started to work on it (Hacohen et al., 1998). Intriguingly, like Argos (and another inhibitor, Kekkon, identified in Norbert Perrimon’s group; Ghiglione et al., 1999), Sprouty participates in negative feedback control of the EGFR. But unlike Argos and Kekkon, Tanita Casci and Javier Vinós showed that Sprouty is an intracellular protein that acts downstream of the receptor, blocking Ras activation, implying that it inhibits signalling by all receptor tyrosine kinases that use the Ras signal transduction pathway (Casci et al., 1999) (Figure 4). The plethora of inhibitors of EGFR signalling, and their role on feedback control, is one of the themes that has emerged over the last few years and their discovery illustrates the power of using fly genetics to identify physiologically important signal regulators (Freeman, 2000).
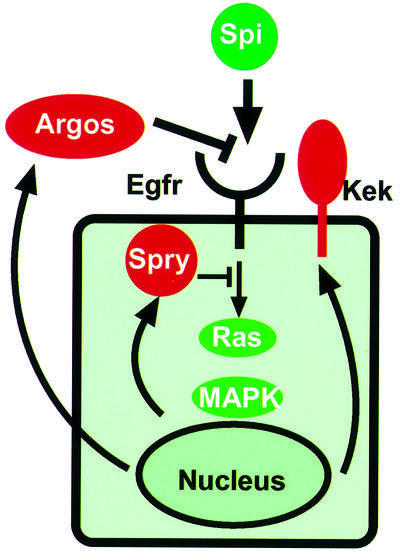
Fig. 4. A simplified view of EGFR regulation by its ligand Spitz and three well-defined inhibitors, Argos, Sprouty and Kekkon-1. Note that the expression of all three depends on EGFR signalling, implying that each forms a negative feedback loop.
Mechanism of Rhomboid and Star
The work of a number of labs had highlighted the significance of Rhomboid in controlling EGFR signalling. The completion of the Drosophila genome sequence revealed that there are in fact seven Rhomboid-like genes in flies (Guichard et al., 2000; Wasserman et al., 2000) and we showed that one of the earliest eye mutations to be discovered, roughoid, was a mutation in rhomboid-3 (Wasserman et al., 2000). Rhomboid-3 replaces the role of Rhomboid-1 in the eye and we now believe that all EGFR signalling by membrane-tethered ligands requires the action of a Rhomboid (Urban et al., 2002a). Moreover, in those places where Rhomboid-1 functions, it can be genetically defined as being the paramount regulator of EGFR activity (Bier et al., 1990; Freeman et al., 1992a; Ruohola-Baker et al., 1993; Sturtevant et al., 1993; Golembo et al., 1996a; zür Lage et al., 1997; Guichard et al., 1999; Wasserman et al., 2000). This increasing spotlight on Rhomboid made it intolerable that we had no idea about its molecular function (and it was a continuing embarrassment at seminars and conference talks, where the function of Rhomboid was an inevitable question). We therefore decided to focus hard on the mechanism of Rhomboid.
At the same time we thought about Star, another gene identified as a member of the spitz group and which, although genetically implicated in Rhomboid function, was equally mysterious (Mayer and Nüsslein-Volhard, 1988; Heberlein and Rubin, 1991; Heberlein et al., 1993; Kolodkin et al., 1994; Pickup and Banerjee, 1999; Ruden et al., 1999). Actually, I was not sure that we should commit to focusing on two difficult and uncertain projects at the same time. But Star was clearly a factor to be considered and the significance of the link between it and Rhomboid were confirmed by Ethan Bier’s lab, which showed that ectopic Rhomboid required Star in order to activate EGFR signalling (Guichard et al., 1999). The significance of Star and Rhomboid in Spitz activation was later confirmed mechanistically, although their molecular functions remained unclear (Bang and Kintner, 2000).
Our approach to tackling Rhomboid was simple—we directly tested the model that Rhomboid somehow triggered the cleavage of the membrane-tethered form of Spitz, which was thought to be a necessary activation step in EGFR signalling. But this proved to be quite difficult, largely due to the low concentration of the cleaved form of Spitz. Marc Sohrmann started the project by engineering a tagged form of Spitz; another postdoc, Clare Garvey, continued this approach after Marc left the lab. Eventually, 2 years after starting, it was a graduate student, Sin Urban, who performed the decisive experiment. In order to get enough eggs, several thousand virgin flies with the tagged Spitz were mated to males which led to their offspring expressing tagged Spitz with inducible Star and/or Rhomboid. Even with several grams of embryos as the starting material, the bands on the resulting western blot were weak. But they unambiguously showed that Spitz was cleaved in response to Rhomboid, but only in the presence of Star (Lee et al., 2001a). The second important development was made only a few days later by another graduate student, Jeff Lee, who showed that Rhomboid and Star-dependent cleavage of Spitz could be recapitulated by coexpressing the proteins in COS cells—a mammalian cell line that is easy to work with in culture. This freed us from the constraints of working with thousands of hand-picked flies and allowed us to do many experiments in a few months, where the equivalent in flies would have taken years.
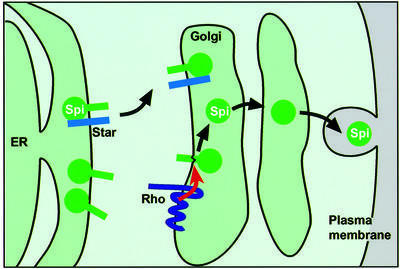
The work with COS cells (later confirmed in flies) showed that, unexpectedly, full-length, membrane- tethered Spitz is retained in the endoplasmic reticulum rather than passing through the secretory pathway to the plasma membrane (Lee et al., 2001a). Equally unexpectedly, neither is Rhomboid at the plasma membrane, but is instead confined to the Golgi apparatus. The key result was the observation that coexpression of Star leads to the relocalization of Spitz from the ER to the Golgi apparatus, where Rhomboid induces its cleavage. This led to the development of our current model, in which the primary regulation of EGFR signalling activity occurs at the level of the ER to Golgi transport of its ligand Spitz, thereby allowing Spitz to be proteolytically activated by Rhomboid in the Golgi, from where it is freely exported through the secretory pathway (Lee et al., 2001a; Figure 5).
Fig. 5. Regulation of Spitz activation by Star and Rhomboid. Regulated translocation and cleavage of its activating ligand Spitz is the primary control of EGFR activation in Drosophila.
The most obvious question that this model posed was how Rhomboid induced Spitz cleavage—it has no obvious homologies to proteases known previously. The answer has emerged from a number of mutational and biochemical analyses done by Sin Urban, all of which pointed to the conclusion that Rhomboid is a new kind of serine protease that cleaves Spitz within its transmembrane domain (Urban et al., 2001). It is unlike all other members of this well defined class of enzymes in that the putative active site is located within the lipid bilayer of the membrane, yet it resembles them strikingly by apparently having the same catalytic triad of residues that comprise the heart of the serine protease mechanism. On the basis of this work we have proposed that the Rhomboid proteins, which are conserved in all the major branches of evolution (Pascall and Brown, 1998; Wasserman et al., 2000; Urban et al., 2002b), are all members of this new family of intramembrane proteases. This proposal has an historical resonance within the LMB, as it was here that much of the ground-breaking work on serine protease structure and function was carried out in the 1960s by David Blow, Brian Hartley and others, including Richard Henderson, the LMB’s current director.
Presumably, this is a case of convergent evolution of serine proteases: as with bacterial subtilisins, it appears that the nucleophilic serine, activated by charge relay in a catalytic triad, has evolved independently. The age of the Rhomboids, predating the divergence of bacteria, archaea, fungi, plants and animals, further supports the idea that they are not derived from the soluble serine proteases. This ancient origin, coupled with their role in intercellular signalling—at least in flies—raises the question of what the primordial function of the Rhomboids might have been. One way of addressing this is to discover their function in modern unicellular organisms and we are now pursuing this in yeast and bacteria. Intriguingly, the Rhomboid homologue in the bacterium Providencia stuartii has been implicated previously in signal emission during quorum sensing, the generic term for intercellular signalling in bacteria (Rather et al., 1999; Gallio and Kylsten, 2000). It has only recently been appreciated how widespread a phenomenon this is (Miller and Bassler, 2001). It is therefore a tantalizing possibility that Rhomboid molecules have a role in intercellular signalling from bacteria to multicellular animals (Gallio et al., 2002; Urban et al., 2002b).
Another implication of the mechanism of Star and Rhomboid is the extension of the function of regulated intramembrane proteolysis (RIP). RIP has been revealed over the last few years to be a mechanism for regulating transcriptional activation by membrane-tethered transcription factors (Brown et al., 2000; Kopan and Goate, 2000). For example, cholesterol biosynthesis in mammalian cells is controlled by the intramembrane proteolysis of the membrane-tethered factor SREBP, in response to a sterol-sensing mechanism. Spitz cleavage is the first example of RIP being used to control extracellular signalling (Urban and Freeman, 2002).
Conclusions and thanks
Even 2 or 3 years ago, I would not have predicted that our route would lead to the enzymology of intramembrane proteases, let alone cell biology of mammals, yeast and bacteria, so there’s little point in trying to predict where our current work will take us over the next 5 years. It is one of the pleasures of working at the LMB that it is easy to change fields as necessary, and this has been a major component of the stimulating environment it has provided. Our current work is focused on the roles of Rhomboids in mammals, yeast and bacteria, and their enzymology (Jeff Lee, Olli Lohi, Angus McQuibban, Sin Urban); the genetic identification of new regulatory components of EGFR signalling, coupled to a long-term interest in their role in eye development (Antonio Baonza, Katherine Brown, Clare Garvey, Saroj Saurya); and extracellular aspects of signalling (Birgitta Olofsson).
Finally, the most important single influence on the science my lab has done since arriving in Cambridge in 1992 has been my colleagues. To be surrounded by such people has been a tremendous privilege and without their continuing help, support and friendship I would not have won the EMBO medal. This includes all those who have worked in my group as well as many others at the LMB. Outside my own group, I am particularly indebted to Mariann Bienz and Sean Munro, but I also thank the many other colleagues who have contributed in so many ways to the work we have done.
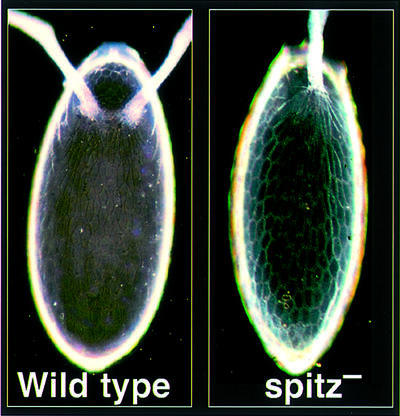
Fig. 3. See Box 2.
References
- Baker N.E. and Rubin,G.M. (1989) Effect on eye development of dominant mutations in Drosophila homologue of the EGF receptor. Nature, 340, 150–153. [DOI] [PubMed] [Google Scholar]
- Baker N.E. and Rubin,G.M. (1992) Ellipse mutations in the Drosophila homologue of the EGF receptor affect pattern formation, cell division, and cell death in eye imaginal discs. Dev. Biol., 150, 381–396. [DOI] [PubMed] [Google Scholar]
- Banerjee U., Renfranz,P.J., Hinton,D.R., Rabin,B.A. and Benzer,S. (1987) The sevenless+ protein is expressed apically in cell membranes of developing Drosophila retina; it is not restricted to cell R7. Cell, 51, 151–158. [DOI] [PubMed] [Google Scholar]
- Bang A.G. and Kintner,C. (2000) Rhomboid and Star facilitate presentation and processing of the Drosophila TGF-α homolog Spitz. Genes Dev., 14, 177–186. [PMC free article] [PubMed] [Google Scholar]
- Baonza A., Casci,T. and Freeman,M. (2001) A primary role for the EGF receptor in ommatidial spacing in the Drosophila eye. Curr. Biol., 11, 396–404. [DOI] [PubMed] [Google Scholar]
- Bier E., Jan,L.Y. and Jan,Y.N. (1990) rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes Dev., 4, 190–203. [DOI] [PubMed] [Google Scholar]
- Brown M.S., Ye,J., Rawson,R.B. and Goldstein,J.L. (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell, 100, 391–398. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega J.A., Jurgens,G. and Hofbauer,A. (1979) Cell clones and pattern formation: studies on sevenless, a mutant of Drosophila melanogaster. Roux’s Arch. Ent. Org., 186, 27–50. [DOI] [PubMed] [Google Scholar]
- Casci T., Vinós,J. and Freeman,M. (1999) Sprouty, an intracellular inhibitor of Ras signalling. Cell, 96, 655–665. [DOI] [PubMed] [Google Scholar]
- Domínguez M., Wasserman,J.D. and Freeman,M. (1998) Multiple functions of the EGF receptor in Drosophila eye development. Curr. Biol., 8, 1039–1048. [DOI] [PubMed] [Google Scholar]
- Flores G.V., Duan,H., Yan,H., Nagaraj,R., Fu,W., Zou,Y., Noll,M. and Banerjee,U. (2000) Combinatorial signaling in the specification of unique cell fates. Cell, 103, 75–85. [DOI] [PubMed] [Google Scholar]
- Freeman M. (1994a) Misexpression of the Drosophila argos gene, a secreted regulator of cell determination. Development, 120, 2297–2304. [DOI] [PubMed] [Google Scholar]
- Freeman M. (1994b) The spitz gene is required for photoreceptor determination in the Drosophila eye where it interacts with the EGF receptor. Mech. Dev., 48, 25–33. [DOI] [PubMed] [Google Scholar]
- Freeman M. (1996) Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell, 87, 651–660. [DOI] [PubMed] [Google Scholar]
- Freeman M. (1997) Cell determination strategies in the Drosophila eye. Development, 124, 261–270. [DOI] [PubMed] [Google Scholar]
- Freeman M. (2000) Feedback control of intercellular signalling in development. Nature, 408, 313–319. [DOI] [PubMed] [Google Scholar]
- Freeman M. and Glover,D.M. (1987) The gnu mutation of Drosophila causes inappropriate DNA synthesis in unfertilized and fertilized eggs. Genes Dev., 1, 924–930. [DOI] [PubMed] [Google Scholar]
- Freeman M., Nüsslein-Volhard,C. and Glover,D.M. (1986) The dissociation of nuclear and centrosomal division in gnu, a mutation causing giant nuclei in Drosophila. Cell, 46, 457–468. [DOI] [PubMed] [Google Scholar]
- Freeman M., Kimmel,B.E. and Rubin,G.M. (1992a) Identifying targets of the rough homeobox gene of Drosophila: Evidence that rhomboid functions in eye development. Development, 116, 335–346. [DOI] [PubMed] [Google Scholar]
- Freeman M., Klämbt,C., Goodman,C.S. and Rubin,G.M. (1992b) The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell, 69, 963–975. [DOI] [PubMed] [Google Scholar]
- Gallio M. and Kylsten,P. (2000) Providencia may help find a function for a novel, widespread protein family. Curr. Biol., 10, R693–R694. [DOI] [PubMed] [Google Scholar]
- Gallio M., Sturgill,G., Rather,P. and Kylsten,P. (2002) A conserved mechanism for extracellular signaling in eukaryotes and prokaryotes. Proc. Natl Acad. Sci. USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione C., Carraway,K.L.,III, Amundadottir,L.T., Boswell,R.E., Perrimon,N. and Duffy,J.B. (1999) The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis. Cell, 96, 847–856. [DOI] [PubMed] [Google Scholar]
- Golembo M., Raz,E. and Shilo,B.Z. (1996a) The Drosophila embryonic midline is the site of Spitz processing, and induces activation of the EGF receptor in the ventral ectoderm. Development, 122, 3363–3370. [DOI] [PubMed] [Google Scholar]
- Golembo M., Schweitzer,R., Freeman,M. and Shilo,B.-Z. (1996b) argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development, 122, 223–230. [DOI] [PubMed] [Google Scholar]
- Guichard A., Biehs,B., Sturtevant,M.A., Wickline,L., Chacko,J., Howard,K. and Bier,E. (1999) rhomboid and Star interact synergistically to promote EGFR/MAPK signaling during Drosophila wing vein development. Development, 126, 2663–2676. [DOI] [PubMed] [Google Scholar]
- Guichard A., Roark,M., Ronshaugen,M. and Bier,E. (2000) brother of rhomboid, a rhomboid-related gene expressed during early Drosophila oogenesis, promotes EGF-R/MAPK signaling. Dev. Biol., 226, 255–266. [DOI] [PubMed] [Google Scholar]
- Hacohen N., Kramer,S., Sutherland,D., Hiromi,Y. and Krasnow,M.A. (1998) sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell, 92, 253–263. [DOI] [PubMed] [Google Scholar]
- Hafen E., Basler,K., Edstroem,J.E. and Rubin,G.M. (1987) Sevenless, a cell-specific homeotic gene of Drosophila, encodes a putative transmembrane receptor with a tyrosine kinase domain. Science, 236, 55–63. [DOI] [PubMed] [Google Scholar]
- Harris W.A., Stark,W.S. and Walker,J.A. (1976) Genetic dissection of the photoreceptor system in the compound eye of Drosophila melanogaster. J. Physiol., 256, 415–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein U. and Rubin,G.M. (1991) Star is required in a subset of photoreceptor cells in the developing Drosophila retina and displays dosage sensitive interactions with rough. Dev. Biol., 144, 353–361. [DOI] [PubMed] [Google Scholar]
- Heberlein U., Hariharan,I.K. and Rubin,G.M. (1993) Star is required for neuronal differentiation in the Drosophila retina and displays dosage-sensitive interactions with Ras1. Dev. Biol., 160, 51–63. [DOI] [PubMed] [Google Scholar]
- Howes R., Wasserman,J.D. and Freeman,M. (1998) In vivo analysis of Argos structure-function: sequence requirement for inhibition of the Drosophila epidermal growth factor receptor. J. Biol. Chem., 273, 4275–4281. [DOI] [PubMed] [Google Scholar]
- Jin M.H., Sawamoto,K., Ito,M. and Okano,H. (2000) The interaction between the Drosophila secreted protein argos and the epidermal growth factor receptor inhibits dimerization of the receptor and binding of secreted spitz to the receptor. Mol. Cell. Biol., 20, 2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G., Wieschaus,E., Nüsslein-Volhard,C. and Kluding,H. (1984) Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II. Zygotic loci on the third chromosome. Wilhelm Roux’s Arch. Dev. Biol., 193, 283–295. [DOI] [PubMed] [Google Scholar]
- Kolodkin A.L., Pickup,A.T., Lin,D.M., Goodman,C.S. and Banerjee,U. (1994) Characterization of Star and its interactions with sevenless and EGF receptor during photoreceptor cell development in Drosophila. Development, 120, 1731–1745. [DOI] [PubMed] [Google Scholar]
- Kopan R. and Goate,A. (2000) A common enzyme connects Notch signaling and Alzheimer’s disease. Genes Dev., 14, 2799–2806. [DOI] [PubMed] [Google Scholar]
- Lee J.R., Urban,S., Garvey,C.F. and Freeman,M. (2001a) Regulated intracellular ligand transport and proteolysis controls EGF signal activation in Drosophila. Cell, 107, 161–171. [DOI] [PubMed] [Google Scholar]
- Lee L.A., Elfring,L.K., Bosco,G. and Orr-Weaver,T.L. (2001b) A genetic screen for suppressors and enhancers of the Drosophila PAN GU cell cycle kinase identifies cyclin B as a target. Genetics, 158, 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesokhin A.M., Yu,S.Y., Katz,J. and Baker,N.E. (1999) Several levels of EGF receptor signaling during photoreceptor specification in wild-type, Ellipse, and null mutant Drosophila. Dev. Biol., 205, 129–144. [DOI] [PubMed] [Google Scholar]
- Mayer U. and Nüsslein-Volhard,C. (1988) A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes Dev., 2, 1496–1511. [DOI] [PubMed] [Google Scholar]
- Miller M.B. and Bassler,B.L. (2001) Quorum sensing in bacteria. Annu. Rev. Microbiol., 55, 165–199. [DOI] [PubMed] [Google Scholar]
- Pascall J.C. and Brown,K.D. (1998) Characterization of a mammalian cDNA encoding a protein with high sequence similarity to the Drosophila regulatory protein Rhomboid. FEBS Lett., 429, 337–340. [DOI] [PubMed] [Google Scholar]
- Perrimon N. and McMahon,A. (1999) Negative feedback mechanisms and their roles during pattern formation. Cell, 97, 13–16. [DOI] [PubMed] [Google Scholar]
- Pickup A.T. and Banerjee,U. (1999) The role of Star in the production of an activated ligand for the EGF receptor signaling pathway. Dev. Biol., 205, 254–259. [DOI] [PubMed] [Google Scholar]
- Rather P.N., Ding,X., Baca-DeLancey,R.R. and Siddiqui,S. (1999) Providencia stuartii genes activated by cell-to-cell signaling and identification of a gene required for production or activity of an extracellular factor. J. Bacteriol., 181, 7185–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruden D.M., Wang,X., Cui,W., Mori,D. and Alterman,M. (1999) A novel follicle-cell-dependent dominant female sterile allele, StarKojak, alters receptor tyrosine kinase signaling in Drosophila. Dev. Biol., 207, 393–407. [DOI] [PubMed] [Google Scholar]
- Ruohola-Baker H., Grell,E., Chou,T.B., Baker,D., Jan,L.Y. and Jan,Y.N. (1993) Spatially localized rhomboid is required for establishment of the dorsal–ventral axis in Drosophila oogenesis. Cell, 73, 953–965. [DOI] [PubMed] [Google Scholar]
- Rutledge B.J., Zhang,K., Bier,E., Jan,Y.N. and Perrimon,N. (1992) The Drosophila spitz gene encodes a putative EGF-like growth factor involved in dorsal–ventral axis formation and neurogenesis. Genes Dev., 6, 1503–1517. [DOI] [PubMed] [Google Scholar]
- Schüpbach T. (1987) Germline and soma cooperate during oogenesis to establish the dorsoventral pattern of egg shell and embryo in Drosophila melanogaster. Cell, 49, 699–707. [DOI] [PubMed] [Google Scholar]
- Schweitzer R. and Shilo,B.-Z. (1997) A thousand and one roles for the Drosophila EGF receptor. Trends Genet., 13, 191–196. [DOI] [PubMed] [Google Scholar]
- Schweitzer R., Howes,R., Smith,R., Shilo,B.-Z. and Freeman,M. (1995a) Inhibition of Drosophila EGF receptor activation by the secreted protein Argos. Nature, 376, 699–702. [DOI] [PubMed] [Google Scholar]
- Schweitzer R., Shaharabany,M., Seger,R. and Shilo,B.-Z. (1995b) Secreted Spitz triggers the DER signalling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes Dev., 9, 1518–1529. [DOI] [PubMed] [Google Scholar]
- Simon M.A., Bowtell,D.D., Dodson,G.S., Laverty,T.R. and Rubin,G.M. (1991) Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell, 67, 701–716. [DOI] [PubMed] [Google Scholar]
- Spradling A.C. and Rubin,G.M. (1982) Transposition of cloned P elements into Drosophila germ line chromosomes. Science, 218, 341–347. [DOI] [PubMed] [Google Scholar]
- Sturtevant M.A., Roark,M. and Bier,E. (1993) The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev., 7, 961–973. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. and Ready,D.F. (1986) Sevenless a cell-specific homeotic mutation of the Drosophila eye. Science, 231, 400–402. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. and Ready,D.F. (1987) Neuronal differentiation in the Drosophila ommatidium. Dev. Biol., 120, 366–376. [DOI] [PubMed] [Google Scholar]
- Tomlinson A., Bowtell,D.D., Hafen,E. and Rubin,G.M. (1987) Localization of the sevenless protein, a putative receptor for positional information, in the eye imaginal disc of Drosophila. Cell, 51, 143–150. [DOI] [PubMed] [Google Scholar]
- Urban S. and Freeman,M. (2002) Intramembrane proteolysis controls diverse signaling pathways throughout evolution. Curr. Opin. Genet. Dev., 12, 512–518. [DOI] [PubMed] [Google Scholar]
- Urban S., Lee,J.R. and Freeman,M. (2001) Drosophila Rhomboid-1 defines a family of putative intramembrane serine proteases. Cell, 107, 173–182. [DOI] [PubMed] [Google Scholar]
- Urban S., Lee,J.R. and Freeman,M. (2002a) A family of Rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. EMBO J., 21, 4277–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S., Schlieper,D. and Freeman,M. (2002b) Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic Rhomboids. Curr. Biol., 12, 1507–1512. [DOI] [PubMed] [Google Scholar]
- Vinós J. and Freeman,M. (2000) Evidence that Argos is an antagonistic ligand of the EGF receptor. Oncogene, 19, 3560–3562. [DOI] [PubMed] [Google Scholar]
- Wasserman J.D. and Freeman,M. (1998) An autoregulatory cascade of EGF receptor signalling patterns the Drosophila egg. Cell, 95, 355–364. [DOI] [PubMed] [Google Scholar]
- Wasserman J.D., Urban,S. and Freeman,M. (2000) A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signalling. Genes Dev., 14, 1651–1663. [PMC free article] [PubMed] [Google Scholar]
- Xu C., Kauffmann,R.C., Zhang,J., Kladny,S. and Carthew,R.W. (2000) Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell, 103, 87–97. [DOI] [PubMed] [Google Scholar]
- Xu T. and Rubin,G.M. (1993) Analysis of genetic mosaics in developing and adult Drosophila tissues. Development, 117, 1223–1237. [DOI] [PubMed] [Google Scholar]
- zürLage P., Jan,Y.N. and Jarman,A.P. (1997) Requirement for EGF receptor signalling in neural recruitment during formation of Drosophila chordotonal sense organ clusters. Curr. Biol., 7, 166–175. [DOI] [PubMed] [Google Scholar]