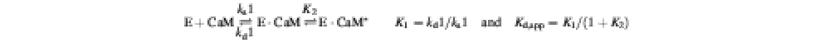Abstract
Edema factor (EF) and CyaA are calmodulin (CaM)-activated adenylyl cyclase exotoxins involved in the pathogenesis of anthrax and whooping cough, respectively. Using spectroscopic, enzyme kinetic and surface plasmon resonance spectroscopy analyses, we show that low Ca2+ concentrations increase the affinity of CaM for EF and CyaA causing their activation, but higher Ca2+ concentrations directly inhibit catalysis. Both events occur in a physiologically relevant range of Ca2+ concentrations. Despite the similarity in Ca2+ sensitivity, EF and CyaA have substantial differences in CaM binding and activation. CyaA has 100-fold higher affinity for CaM than EF. CaM has N- and C-terminal globular domains, each binding two Ca2+ ions. CyaA can be fully activated by CaM mutants with one defective C-terminal Ca2+-binding site or by either terminal domain of CaM while EF cannot. EF consists of a catalytic core and a helical domain, and both are required for CaM activation of EF. Mutations that decrease the interaction of the helical domain with the catalytic core create an enzyme with higher sensitivity to Ca2+–CaM activation. However, CyaA is fully activated by CaM without the domain corresponding to the helical domain of EF.
Keywords: adenylyl cyclase exotoxin/anthrax edema factor/Ca2+–calmodulin/CyaA/enzyme activation
Introduction
Calcium serves as a diffusible second messenger in response to extra- and intracellular signals, and calmodulin (CaM) is a key calcium sensor (Eldik and Watterson, 1998). CaM has two globular domains, each consisting of two helix–loop–helix calcium-binding motifs (Babu et al., 1985). The domains are linked by a flexible α-helix that is partially unfolded in solution (Barbato et al., 1992). Calcium induces a transition in both domains from a closed conformation with highly negatively charged surface to an open conformation with a large, exposed hydrophobic pocket (Finn et al., 1995). The flexibility of the central helix and the calcium-induced changes in surface properties enable CaM to bind and modulate a diverse array of physiologically important proteins, such as adenylyl cyclase, phosphodiesterase, nitric oxide synthase, protein kinase and phosphatase, receptor and ion channel (Eldik and Watterson, 1998; DeMaria et al., 2001). Consequently, CaM is involved in many intracellular processes including control of transcription, ion fluxes, signal transduction, vesicular transport and cytoskeleton functions (Deisseroth et al., 1998; Eldik and Watterson, 1998).
Structural and biochemical analyses have provided insights into CaM-dependent regulation of some target enzymes (Hoeflich and Ikura, 2002; Meador and Quiocho, 2002). The best-known activation mechanism is the release of autoinhibition exemplified by myosin light chain kinase (MLCK) and CaM kinase II (CaMKII). In this model, CaM is not associated with the target enzyme at the resting Ca2+ levels, and the enzyme’s catalytic activity is blocked by its own autoinhibitory domain (AID). Upon increase in the intracellular Ca2+ concentration, the Ca2+–CaM complex binds to an amphipathic α-helix that partially overlaps the AID. This presumably causes a conformational change that disrupts the interaction of the AID with the catalytic domain, resulting in kinase activation. A recent structure of the complex of CaM with the intracellular domain of the small conductance Ca2+-activated potassium channel, Ik(Ca), reveals a new activation mechanism (Schumacher et al., 2001). In this case, CaM is constitutively bound to Ik(Ca). Calcium loading allows CaM to act as a clamp to induce Ik(Ca) dimerization, which leads to an allosteric change and increased ion conductivity.
Several pathogenic bacteria, such as those that cause anthrax (Bacillus anthracis), whooping cough (Bordetella pertussis) and cholera (Vibrio cholerae), secrete toxins that increase the cAMP concentration in the host cells to a pathological level (Drum et al., 2002). Edema factor (EF) and CyaA are adenylyl cyclase toxins produced by B.anthracis and B.pertussis, respectively (Ladant and Ullmann, 1999; Mock and Fouet, 2001). Both EF and CyaA are activated in the host cell by CaM. EF is responsible for the massive edema seen in cutaneous anthrax and impairs the function of neutrophils and monocytes in systemic infection (Hoover et al., 1994). CyaA is important for the bacterial colonization of the respiratory tract, in part owing to its ability to induce apoptosis of macrophages (Khelef et al., 1993; Weingart and Weiss, 2000).
The CaM-activated adenylyl cyclase domain resides in the C-terminal 510 amino acid region of EF. The molecular structures of this domain with and without CaM were solved recently. These structures have provided a new model of CaM binding and activation of a target enzyme (Drum et al., 2002; Hoeflich and Ikura, 2002; Meador and Quiocho, 2002). Unlike the CaM complexes with MLCK and CaMKII, in which CaM takes a compact form, in the complex with EF, CaM has an extended conformation and makes extensive contacts (∼6000 Å2) with four discrete regions of EF. This interaction induces a 15 Å translation and 30° rotation of a 15 kDa helical domain of EF resulting in stabilization of a 12 amino acid-long loop. This loop contains several amino acid residues crucial for catalysis, and it is disordered in the structure of EF alone so that the catalytic site is open and incomplete. CaM-induced conformational changes stabilize this loop to enclose and complete the catalytic site, achieving over 1000-fold enhancement in the catalytic rate of EF.
There is considerable cross talk between the signaling pathways of calcium and cAMP, two key intracellular messengers. An increase in intracellular cAMP levels can elevate intracellular Ca2+ by the activation of calcium-permeable channels, such as L-type calcium channels, nicotinic acetylcholine receptors and cyclic nucleotide gated channels (Cooper et al., 1995). On the other hand, Ca2+–CaM can decrease the cAMP level by activation of phosphodiesterase, or increase it by activating specific isoforms of adenylyl cyclase (type I, III and VIII; Eldik and Watterson, 1998; Hanoune and Defer, 2001). Little is known regarding the effects of physiological Ca2+ concentrations on the adenylyl cyclase activity of EF and CyaA. This question is especially intriguing in view of the fact that Ca2+ ions were found only in the C-terminal domain of CaM in the structure of EF–CaM complex.
In this paper, we show that the physiological intracellular calcium concentration not only dramatically increases the CaM affinity of both EF and CyaA, enhancing the activation of adenylyl cyclase activity, but also directly interferes with binding of the catalytic metal, inhibiting catalysis. We also show that the interaction of the helical domain of EF with CaM plays a key role in EF activation.
Results
Physiological calcium and CaM concentrations are required for the optimal activation of EF
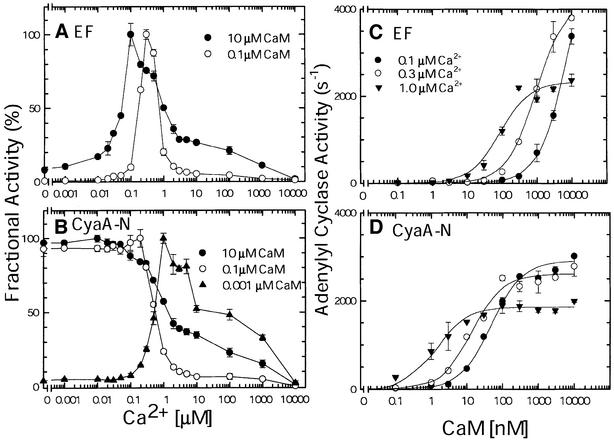
The activation of EF by CaM can be greatly reduced by the addition of EGTA, a calcium chelator (Leppla, 1984). However, it is not clear whether physiological concentrations of calcium could modulate the activation of EF by CaM. While the estimated total CaM concentration inside cells is ∼1–10 µM, free CaM concentration is significantly lower since it is associated with apo-CaM-binding proteins such as GAP-43 and RC3 (Gerendasy, 1999; Jurado et al., 1999). We therefore measured adenylyl cyclase activity of EF at a broad range of calcium concentrations (1 nM to 10 mM) in the presence of either 0.1 or 10 µM CaM, representing the low and high ends of the intracellular free CaM concentration range (Figure 1A). We found that, at both CaM concentrations, adenylyl cyclase activity exhibited a bell-shaped curve with optimal activity at the physiologically relevant calcium concentrations (0.1– 0.5 µM). While <5-fold change in the adenylyl cyclase activity of EF at the 0.03–10 µM calcium concentrations was observed in the presence of 10 µM CaM, the enzymatic activity of EF was sharply altered by the same calcium concentrations when 100-fold less CaM (0.1 µM) was used. This requirement of calcium and CaM for the optimal activation of EF was observed with both the physiological Mg2+ concentration (0.5 mM; Supplementary figure 1, available at The EMBO Journal Online) and the Mg2+ concentration optimal for EF activity (10 mM; Figure 1A). This data suggests that the optimal activation of EF requires the physiological calcium concentrations, particularly when CaM is limited.
Fig. 1. The effect of calcium ions and CaM on the adenylyl cyclase activities of EF and CyaA-N. Adenylyl cyclase assays were performed in the presence of 1 nM EF (A) and 0.7 nM CyaA-N (B) under 10 µM CaM (filled circles), 0.1 µM CaM (open circles) and 1 nM CaM (filled triangles, CyaA-N only) at increasing [Ca2+]. They were also performed at 0.1 µM Ca2+ (filled circles), 0.3 µM Ca2+ (open circles) and 1.0 µM Ca2+ (filled triangles) at increasing [CaM] in the presence of 1 nM EF (C), and 0.7 nM CyaA-N (D). Maximal adenylyl cyclase activities (100%) for EF in the calcium titration are 1140 s–1 (10 µM CaM) and 228 s–1 (0.1 µM CaM) (A) and those for CyaA-N are 1465 s–1 (10 µM CaM), 713 s–1 (0.1 µM CaM) and 556 s–1 (1 nM CaM) (B). Means ± SE are representative of at least two experiments.
The 1706 amino acid CyaA is a bi-functional protein with both adenylyl cyclase and hemolytic activities. The N-terminal 45 kDa domain of CyaA has CaM-activated adenylyl cyclase activity and its C-terminal 140 kDa domain contains a transmembrane region, 42 glycine-rich motifs that bind calcium and a hemolytic domain. We expressed and purified the functional N-terminal adenylyl cyclase domain (aa 1–393, referred to here as CyaA-N) of CyaA without denaturant. We then addressed whether the activation of the adenylyl cyclase domain of CyaA also depends upon calcium and CaM concentrations. In contrast to EF, the activity of CyaA-N was optimally stimulated by CaM at 1 nM to 0.1 µM calcium with either 0.1 or 10 µM CaM, whereas its activity was significantly inhibited at calcium concentrations >0.1 µM (Figure 1B). With only 1 nM CaM, the adenylyl cyclase activity of CyaA-N exhibited a bell-shaped curve in the calcium concentrations range from 1 nM to 10 mM. This data suggests that, in contrast to EF, CyaA-N is optimally activated at resting calcium concentrations with free CaM ranging from 0.1 to 10 µM, and its activity is reduced with elevated intracellular calcium concentrations. A similar observation was found using the full-size CyaA (Gentile et al., 1990).
Physiological calcium concentrations modulate the affinity of CaM for EF and CyaA-N
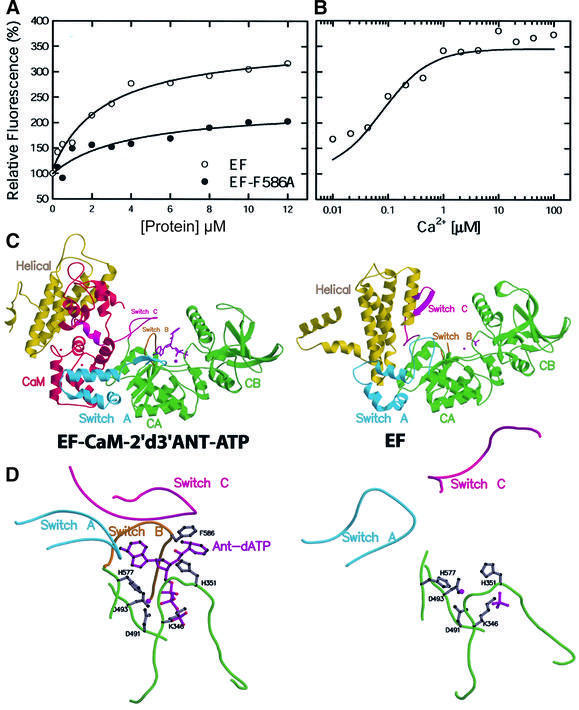
Calcium concentration may affect the interaction between CaM and EF. To address this question, we took advantage of the observation that the emission intensity of the fluorescent ATP analog 2′-deoxy,3′-anthraniloyl ATP (2′d3′ANT-ATP) increases ∼3-fold upon binding to the CaM–EF complex (Figure 2A; Sarfati et al., 1990). From the equilibrium titration monitored by fluorescence, we found that the Kd for the binding of 2′d3′ANT-ATP to CaM–EF is 1 µM, and the binding requires physiological calcium concentrations (0.05–2 µM; Figure 2A and B). In view of the observation that 2′d3′ANT-ATP is a potent inhibitor of CaM–EF (Sarfati et al., 1990), we then asked whether this compound binds to the enzyme in a manner similar to the normal substrate ATP. To answer this question and to explain the mechanism of the calcium/CaM-dependent enhancement of 2′d3′ANT-ATP fluorescence, we solved the structure of the EF–CaM– 2′d3′ANT-ATP complex (Table I; Figure 2C and D).
Fig. 2. The binding of 2′d3′ANT-ATP to EF. (A) Equilibrium titration of 2′d3′ANT-ATP–CaM with EF and EF-F586A. (B) Calcium titration of fluorescence enhancement by EF–CaM. 2′d3′ANT-ATP was added to a final concentration of 0.5 µM and the indicated free calcium concentrations were achieved by buffering with 10 mM EGTA. λexc = 320 nm and the optimal fluorescence emission of EF–CaM–2′d3′ANT-ATP (412 nm) was normalized to give the fold of enhancement. (C) Secondary structure of EF–CaM–2′d3′ANT-ATP in comparison with EF alone. (D) The active site of EF in the presence and absence of CaM and 2′d3′ANT-ATP.
Table I. Statistics of the EF–CaM–2′d3′ANT-ATP complex data set.
| Data collection |
|
|
|
| Beamline | APS, 14-BM-C | ||
| Space group | I222 | ||
| Unit cell (Å) | |||
| a | 116.92 | ||
| b | 167.92 | ||
| c | 341.74 | ||
| Resolution (Å) | 3.6 | ||
| Completeness (%) | 99.4 | ||
| Redundancya | 6.85 | ||
| Rsym (%)b | 8.3 | ||
|
I/σ |
11.3 |
|
|
| Refinement |
|
|
|
| Rcryst (%)c | Rfree (%)d | R.m.s.bond(Å) | R.m.s.angle(°) |
| 28.1 | 30.7 | 0.012 | 1.8 |
aNobs/Nunique.
bRsym = Σj|<I> – Ij|/Σ<I>, where Ij is the intensity of the jth reflection and <I> is the average intensity.
cRcryst = Σhkl|Fobs – Fcalc|/ΣhklFobs.
dRfree, calculated the same as for Rcryst but on the 5% data excluded from the refinement calculation.
There is a clear representation of 2′d3′ANT-ATP in the simulated annealing omit map of the EF–CaM–2′d3′ANT- ATP structure (Supplementary figure 2). The portions of the structure comprised of EF–CaM and the 3′d-ATP moiety of 2′d3′ANT-ATP are virtually identical to the structure of EF–CaM–3′d-ATP (Drum et al., 2002). The anthraniloyl group in 2′d3′ANT-ATP is covered by a catalytic loop called switch B (Figure 2C and D). Switch B is not visible in the EF alone structure, but it is stabilized by switch C and becomes ordered upon CaM binding. The anthraniloyl group in 2′d3′ANT-ATP is aligned with the phenyl group of F586, a residue in switch B. Consistent with the structural model, the mutation of F586 to alanine greatly reduced the ability of EF to enhance the fluorescence of 2′d3′ANT-ATP (Figure 2A). The F586A mutation had a relatively small effect on EC50 and Vmax values for CaM activation and Km for substrate binding (the EC50,CaM, Vmax and Km,ATP values are 2 nM, 718 s–1 and 0.7 mM for wild-type EF and 2.5 nM, 370 s–1 and 1.0 mM for EF-F586A).
Based on fluorescence data on calcium-dependent CaM-binding to EF, we predicted that physiological calcium concentrations can alter the concentration of CaM required to achieve half maximal activation of EF (EC50). To test this prediction, we examined the ability of CaM to activate EF at 0.1, 0.3 and 1 µM calcium (Figure 1C). Our results showed that with increasing calcium concentrations, the EC50 values for CaM activation of EF decreased from 2 µM to 50 nM. Calcium concentrations >1 µM did not cause further significant decrease of the EC50 values (Supplementary figure 3). These results agree with the notion that physiological calcium concentrations can affect the affinity of EF to CaM and highlights the free energy coupling between Ca2+ and EF binding to CaM.
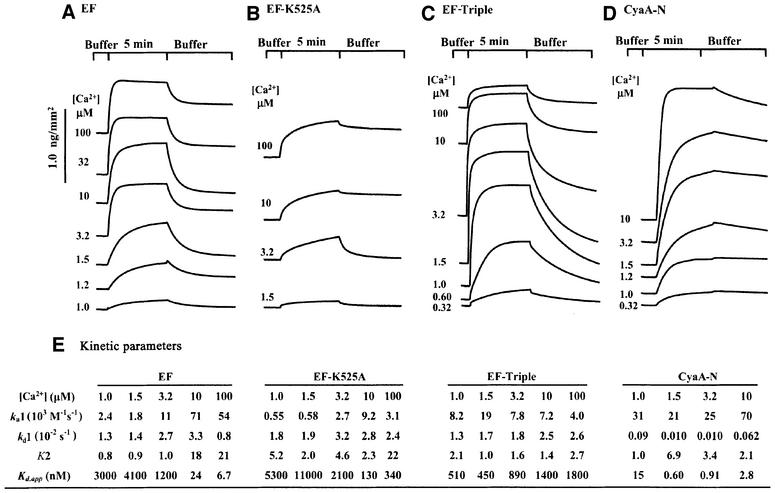
We then studied the interaction of EF with immobilized CaM by surface plasmon resonance (SPR) spectroscopy (Figure 3). We immobilized CaM to a self-assembled monolayer using a recently reported active site-directed immobilization method (Hodneland et al., 2002). CaM was fused to the C-terminal end of cutinase from the fungus Fusarium salani and the cutinase–CaM fusion protein (cut-CaM) was covalently bound to a gold-coated glass surface previously coated with 4-nitrophenyl- phosphonate ligand, a transition state substrate analog of cutinase. Cut-CaM activated EF with 5-fold reduced affinity (Supplementary figure 4). The SPR analysis showed that the fraction of immobilized CaM bound to EF increases at elevated calcium concentrations (Figure 3A), providing further evidence that calcium ions enhance the binding of these two proteins. These experiments reveal that CaM binds EF with a Kd of 20 nM at 10 µM calcium, in agreement with the reported affinity (Figure 3E; Drum et al., 2002). The SPR analysis also showed that calcium ions predominantly increased the rate of association (ka1) and that of the conformational transition (K2) rather than decreasing the dissociation rate.
Fig. 3. The effect of calcium ions on the interaction of EF and CyaA-N with immobilized CaM by SPR sensorgram analysis. Sensorgrams were recorded in the presence of 1.4 µM wild-type EF (A), 1.3 µM EF-K525A (B), 0.37 µM CyaA-N (D) and 8.0 µM EF-triple mutant (C) at the indicated free calcium concentrations. (E) Kinetic analysis of SPR sensorgrams based on a ‘two-state conformational change’ model.
At the lowest calcium concentration, signal from the bound EF could be reduced to the base line with the wash using the same calcium concentration. However, a fraction of the signal could not be reduced at the higher calcium concentrations until the buffer with the lowest calcium concentration was used in the wash step. The signal from the bound CyaA-N could not be reduced to the base line without a prolonged wash.
To ensure that the SPR experiments reflected the binding of CaM to EF, we examined EF-K525A, an EF mutant with a point mutation changing lysine 525 to alanine. The structure of the EF–CaM complex reveals that lysine 525 of EF forms a salt bridge with glutamate 114 of CaM (Drum et al., 2002). Our SPR analysis showed that at the highest calcium concentration assayed, EF-K525A achieved a signal comparable with wild-type EF at low levels of calcium (Figure 3B). Consistent with our biochemical analysis, the affinity of EF-K525A for CaM was two orders of magnitude lower than that of the wild-type EF at 100 µM calcium (Figure 3E; Drum et al., 2002).
We also examined whether calcium concentrations can alter the EC50 value for the activation of CyaA-N by CaM (Figure 1D). We found that calcium concentrations from 0.1 to 1 µM reduced the EC50 value for CaM activation of CyaA-N from 100 to 2 nM, and calcium concentrations beyond 1 µM did not further reduce EC50 values (Supplementary figure 3). To evaluate the binding of CaM to CyaA-N directly, we then examined the interaction of CyaA-N with cut-CaM using SPR (Figure 3D). Cut-CaM activated CyaA-N with an EC50 similar to that of wild-type CaM, and with 2-fold higher Vmax (Supplementary figure 4). The SPR analysis showed that CyaA-N reached levels of binding to cut-CaM comparable with those observed for EF, but at much lower calcium concentrations (Figure 3). Although the rates of association (Ka1) and of the conformational transition (K2) for the cut-CaM binding were similar between EF and CyaA-N, the dissociation rate from cut-CaM (Kd1) was significantly reduced for CyaA-N as compared with EF (Figure 3E).
Calcium ions directly inhibit adenylyl cyclase activity of EF and CyaA-N
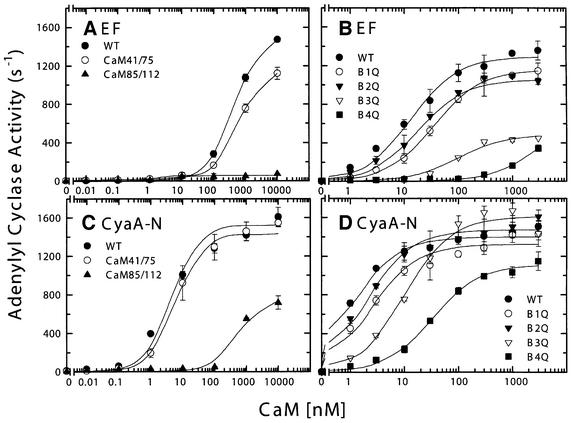
In the EF–CaM structure, only the calcium-binding sites in the C-terminal domain of CaM are loaded with calcium, whereas the N-terminal domain does not contain Ca2+ and adopts the conformation of apo-CaM (Drum et al., 2002). As described above, we found that calcium concentrations >1 µM significantly reduce the adenylyl cyclase activities of both EF and CyaA-N. We hypothesized that the inhibitory effect of calcium might be caused by calcium binding to the N-terminal domain of CaM and a subsequent change in the interaction between this domain and EF. To test this hypothesis, we examined the ability of a mutated form of CaM, CaM41/75, to activate EF and CyaA-N (Figure 4A and C). CaM41/75 has cysteines replacing residues 41 and 75, capable of forming a disulfide bond and locking the N-terminal domain of CaM in the closed conformation, irrespective of calcium (Tan et al., 1996). If calcium binding in the N-terminal domain of CaM and the resulting conformational change cause the reduced activation of EF and CyaA-N by CaM, then the activation by CaM41/75 should not decrease at high Ca2+ concentrations. Our result showed that CaM41/75 activates EF and CyaA-N in a manner indistinguishable from the wild-type CaM (Figure 4A and C; data not shown for Ca2+ titration). Thus, the inhibition for the activities of EF and CyaA-N by Ca2+ is unlikely to be mediated through the Ca2+ binding to the N-terminal domain of CaM.
Fig. 4. The activation of EF and CyaA-N by wild-type CaM and two series of CaM mutants. EF (1 nM) and CyaA-N (0.7 nM) were used for an adenylyl cyclase activity assay in the presence of 0.1 µM free Ca2+ with CaM mutants CaM 41/75 and 85/112. Each mutant has two cysteine mutations to lock either the N- or the C-terminal domain of CaM in the closed conformation (A and B). The same concentrations of EF and CyaA-N were used for an adenylyl cyclase activity assay in the presence of 1 µM free Ca2+ with CaM mutants B1Q, B2Q, B3Q and B4Q. Each mutant has a mutation inactivating one of four calcium-binding sites (C and D). Means ± SE are representative of at least two experiments.
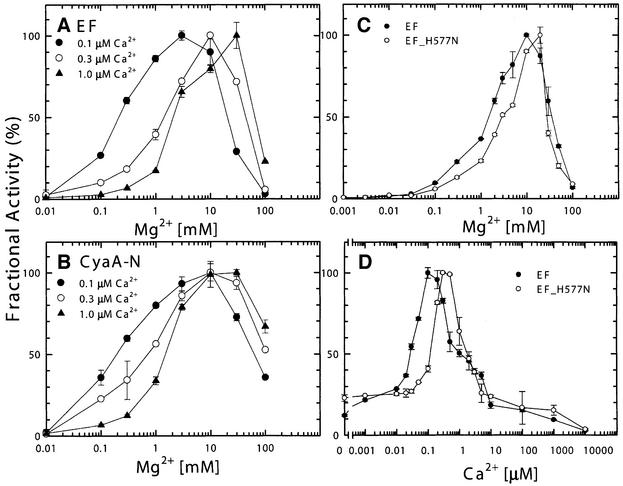
We then examined whether calcium bound to the substrate in place of Mg2+ could cause inhibition of the catalytic activity. We found <0.2% of adenylyl cyclase activity for both EF and CyaA-N when Ca2+–ATP was used as the substrate as compared with the magnesium– ATP (data not shown; Labruyure et al., 1990). Thus, Ca2+–ATP is a poor substrate for both EF and CyaA. To understand the molecular mechanism behind calcium-mediated inhibition, we analyzed the dependence of adenylyl cyclase activity of EF and CyaA-N on magnesium at 0.1, 0.3 and 1 µM calcium (Figure 5A and B). We observed a bell-shape curve for the magnesium requirement with the optimum between 1 and 20 mM for both EF and CyaA-N. In the magnesium concentrations ranging from 0.1 to 10 mM, we found that the higher the calcium concentration, the greater the magnesium concentration required for maximal activity. Assuming that the calcium titration curves in Figure 1A and B were composed of a calcium-dependent activation, fit by a simple rectangular hyperbola, and a calcium-dependent inhibition using
Fig. 5. Effect of calcium and magnesium ions on adenylyl cyclase activity of EF and CyaA-N (A and B) and an EF mutant, EF-H577N (C and D). Adenylyl cyclase activity assays were performed with 10 µM CaM at 0.1 µM Ca2+ (filled circles), 0.3 µM Ca2+ (open circles) and 1.0 µM Ca2+ (filled triangles) in the presence of 1 nM EF (A), and 0.7 nM CyaA-N (B). To analyze the mutant form of EF, adenylyl cyclase activities were measured with 10 µM CaM and 1 nM EF or 66 nM EF-H577N with either 0.3 µM Ca2+ (C) or 10 mM Mg2+ (D). Both were buffered by 10 mM EGTA. Maximal activities for EF in the magnesium titration are 1616 s–1 (0.1 µM Ca2+), 1074 s–1 (0.3 µM Ca2+), and 1009 s–1 (1.0 µM Ca2+) (A) and those for CyaA-N are 2106 s–1 (0.1 µM Ca2+), 1674 s–1 (0.3 µM Ca2+) and 1385 s–1 (1.0 µM) (B). Maximal activities for EF and EF-H577N were 1208 s–1 and 4 s–1 (C) and those for EF and EF-H577N were 2726 s–1 and 6 s–1 (D), respectively. Means ± SE are representative of at least two experiments.
v = Vmax × ([Ca2+]/(EC50 + [Ca2+])) × (1/(1 + ([Ca2+]/IC50)))
we estimated the half maximal concentration for calcium inhibition (IC50) of EF and CyaA-N to be ∼0.3–1 µM, suggesting that the calcium inhibition of EF and CyaA is mediated by the high nanomolar affinity calcium-binding site(s).
In our EF–CaM–3′dATP structure, the catalytic site of EF has one catalytic metal ion, which is coordinated by two aspartates, D491 and D493 (Drum et al., 2002). Histidine 577 of EF forms hydrogen bonds with these aspartates in addition to contacting the catalytic metal (Figure 2D). We found that EF-H577N, an EF mutant with asparagine substituted for histidine 577, had adenylyl cyclase activity reduced by two orders of magnitude even though this mutant had normal ATP-binding activity and CaM activation (Drum et al., 2002). We used the metal sensitivity of EF-H577N to determine whether H577 plays a role in the binding of calcium and magnesium ions. We found that, with Mg2+ ranging from 0.1 to 10 mM, the sensitivity of EF-H577N to magnesium was reduced 5- to 10-fold compared with that of the wild-type EF (Figure 5C). Similar reduction in sensitivity to calcium inhibition was also observed with Ca2+ concentrations ranging from 0.1 to 1 µM (Figure 5D). Together our data suggest that a magnesium ion binds to the catalytic metal site of EF and that calcium interferes with the binding of this magnesium ion.
EF and CyaA-N have different requirements for their activation by CaM
Our SPR and kinetic analyses demonstrate that CaM binds CyaA-N with 20- to 100-fold higher affinity than it does EF. In order to identify the roles of the different structural elements of CaM in activating these two enzymes, we examined their activation by three sets of CaM mutants. CaM 85/112 has cysteine substituted for residues 85 and 112, resulting in the C-terminal domain of CaM being locked in the closed conformation by a disulfide bond (Tan et al., 1996). We found that CaM85/112 activated EF poorly, while it could activate CyaA-N with an increased EC50 value and 50% decrease in Vmax (Figure 4A and C)
We also used a set of CaM mutants (Q-series), each of which had a mutation in one of four calcium-binding sites (Maune et al., 1992), to probe the contribution of individual calcium-binding sites to the activation of EF and CyaA-N (Figure 4B and D). In this series of CaM mutants, the conserved glutamate at position 12 of each calcium-binding site was mutated to glutamine, a change that effectively eliminates calcium binding. As observed previously, the mutation at either site 3 or 4 severely reduced the Vmax and EC50 values for CaM activation of EF, whereas those at either site 1 or 2 had a minimal effect (Figure 4B; Drum et al., 2000). In contrast, only the mutation at site 4 of CaM had a significant effect on the EC50 value for CaM activation of CyaA-N with little change in Vmax value. Mutations at the other three sites caused little change (Figure 4D).
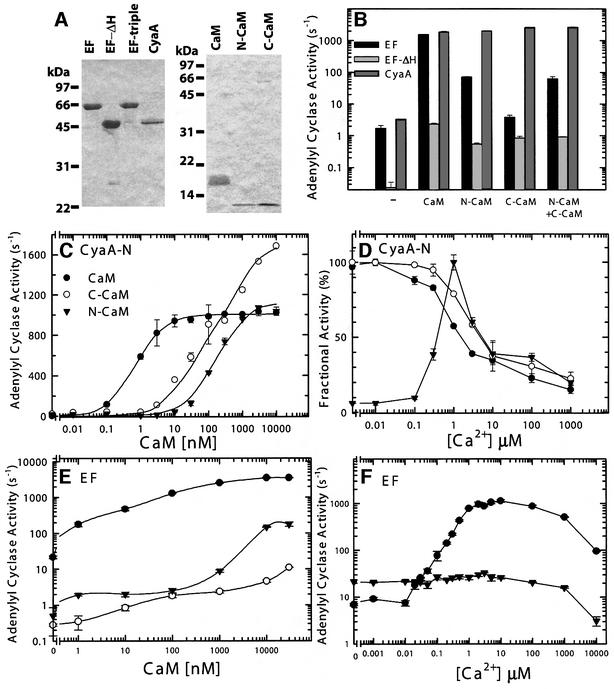
We purified N- and C-terminal domains of CaM (N-CaM and C-CaM) and examined their abilities to activate EF and CyaA-N (Figure 6A and B). We found that either N-CaM or C-CaM alone could fully activate CyaA-N, although the EC50 values were increased (Figure 6C and D). Similar to wild-type CaM, C-CaM can optimally activate CyaA-N in resting physiological calcium concentrations (0.03–0.1 µM; Figure 6D). However, the activation of CyaA-N by N-CaM required the elevated calcium concentrations (0.1–1 µM; Figure 6D). We also found that N-CaM could partially activate EF (5% of wild-type CaM) and this activation was independent of the physiological concentrations of calcium (Figure 6B, E and F). We observed similar partial activation when CaM85/112 was used (Figure 4A). C-CaM had significantly reduced ability to activate EF and the addition of C-CaM did not further enhance the activation of EF by N-CaM (Figure 6B and E).
Fig. 6. The activation of EF and CyaA-N by wild-type CaM, N- and C-terminal domain of CaM (N-CaM and C-CaM). (A) Coomassie Blue staining of purified EF, EF mutants, CyaA-N, CaM and CaM mutants (2 µg each) on SDS–PAGE. (B) EF (1 nM), EF-ΔH (80 nM) and CyaA-N (0.7 nM) were assayed for adenylyl cyclase activity at 1.0 µM free Ca2+ and 10 µM CaM in the absence or the presence of full-length CaM, N-CaM, C-CaM or both N-CaM and C-CaM (N+C CaM). (C and E) Activation of CyaA-N and EF by wild-type and mutant CaM. Adenylyl cyclase toxin (0.7 nM CyaA-N or 1 nM EF) and 1.0 µM free Ca2+ were used in an adenylyl cyclase activity assay in the presence of wild-type CaM (filled circles), C-CaM (open circles) and N-CaM (filled triangles). (D and F) Calcium titration assay in CaM activation of CyaA-N and EF. Adenylyl cyclase activity of CyaA-N (0.7 nM) was measured with 10 µM wild-type CaM (filled circles), C-CaM (open circles) and N-CaM (filled triangles) and that of EF (1 nM) was examined with 1 µM wild-type CaM (filled circles) and 10 µM N-CaM (filled triangles). Maximal activity for wild-type CaM, N-CaM, and C-CaM is 1465, 230 and 1219 s–1, respectively. Means ± SE are representative of at least two experiments.
The role of the EF helical domain in EF activity and CaM activation
The adenylyl cyclase domain of EF consists of a catalytic core (CA and CB) and a helical domain that are linked by switch C (Figure 1C). Based on sequence comparison, the adenylyl cyclase domain of CyaA-N (aa 1–393) does not contain sequences corresponding to the helical domain of EF, nor does ExoY, a related adenylyl cyclase toxin from Pseudomonas aeruginosa (Yahr et al., 1998). However, an EF mutant, EF-N (aa 291–640) that contains only the catalytic core and part of switch C had 10-fold reduction in basal activity and 20-fold reduction in CaM activation, highlighting the importance of the helical domain in the activity of EF and its activation by CaM (Drum et al., 2000). This interpretation was compromised by the subsequent finding that the portion of switch C that was deleted in EF-N (aa 641–655) is involved in CaM binding (Drum et al., 2002). Thus, we made EF-ΔH (aa 291–655) that had both the catalytic core and the entire switch C and examined its ability to be activated by CaM (Figure 6A and B). EF-ΔH was tagged with His6 at the C-terminal end and was purified using a Ni-NTA column to ensure that switch C was present in the protein. We found that, like EF-N, EF-ΔH had ∼50-fold reduction in basal activity and ∼10-fold reduction in its activation by wild-type CaM.
The helical domain of EF makes substantial contact with the CA domain and switch C in the absence of CaM (combined contact surface = 3600 Å2) and most of this contact is disrupted upon CaM binding (Figure 2C and D; Drum et al., 2002). Thus, we hypothesize that one of the roles for the helical domain is to lock EF in an inactive state. The insertion of CaM between CA and the helical domain leads to the extensive interaction between C-CaM with switch A and C. Such interaction triggers conformational changes in switch C which then stabilize the catalytic loop (switch B) to achieve over 1000-fold catalytic activation (Figure 2C). This hypothesis predicts that wild-type EF will be activated by C-CaM poorly since C-CaM cannot insert itself between CA and the helical domain, while EF without the helical domain can interact with C-CaM thus be better activated by C-CaM. Consistent with this prediction, our result showed that C-CaM could activate EF-ΔH ∼50-fold, but could only activate wild-type EF 3-fold (Figure 6B).
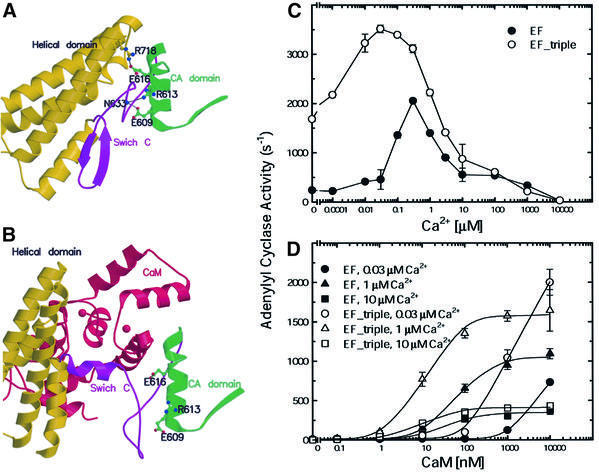
Our model also predicts that an EF mutant with a mutation that disrupts the interaction of the helical domain with CA and switch C but does not alter CaM binding should be more sensitive to CaM activation than wild-type EF, since less energy is needed for CaM insertion. Only a few residues, including E609, R613 and E616, fit this criterion since most residues that make contact between the CA and the helical domain are also involved in CaM interaction (Figure 7A and B). We thus made an EF mutant that had a single point mutation at E609, R613 or E616 to alanine (EF-E609A, EF-R613A or EF-R616A) and a mutant that had all three mutations (EF-triple). We found that the adenylyl cyclase activity of Escherichia coli lysates that contained EF-E609A, EF-R613A, EF-E616A and EF-triple mutants had 0.5-, 2-, 4- and 7-fold higher adenylyl cyclase activity, respectively, than one containing wild-type EF (data not shown). We purified EF-triple and found that EF-triple had 2- to 8-fold higher adenylyl cyclase activity than wild-type EF at 0.001–0.1 µM calcium (Figures 6A and 7C). At relatively low calcium concentrations, the EF-triple mutant had increased affinities for CaM based on both SPR and kinetic analyses (Figures 3D and 77). This result is consistent with our hypothesis that the interaction between the CA and the helical domain locks EF in the inactive state.
Fig. 7. Calcium–CaM activation of EF and EF-triple mutant. The locations of E609, R613 and E616 and their interacting residues, R718 and N633, are shown in the ribbon representation of EF structure in the absence (A) and presence (B) of CaM. CA–CB domain, switch C, helical domain and CaM are colored in green, purple, yellow and red, respectively. Ca2+ titration assay in the presence of 10 µM CaM (C) and CaM titration assay at 0.03, 1 and 10 µM calcium (D) were performed with 1 nM EF and 0.5 nM of EF-triple mutant.
Discussion
In response to the elevation of intracellular Ca2+, CaM can bind to and modulate the activity of numerous cellular proteins (Eldik and Watterson, 1998; Jurado et al., 1999). Its ubiquitous expression and relative abundance in eukaryotic cells make CaM ideally suited to be the activator of anthrax and pertussis adenylyl cyclase toxins, EF and CyaA, ensuring that these toxins are only active inside the host cells. Proteins that interact with CaM can be divided into two broad classes; those that have higher affinity for Ca2+-loaded CaM than for apo-CaM and those that prefer to bind apo-CaM. The first class is exemplified by CaM kinase, calcineurin, cAMP phosphodiesterase and type I adenylyl cyclase, while the second class is exemplified by brush-border myosin I and neuromodulin (GAP-43). Although CyaA was previously categorized as a member of the second class based on its ability to bind CaM in the presence of millimolar concentrations of EGTA, our results using the combination of SPR, kinetic and mutational analyses clearly show that CyaA, like EF, requires physiological concentrations of calcium ions to bind CaM at low CaM concentration (1 nM) and the calcium-loaded CaM has higher affinity for CyaA and EF than apo-CaM (Jurado et al., 1999).
CyaA has 100-fold higher affinity to CaM than EF does. Our study and the previous characterization of these two toxins reveal significant differences in binding and activation of EF and CyaA by CaM. CyaA can be fully activated by CaM mutants with one defective C-terminal Ca2+-binding site or by the N- or C-terminal domain of CaM while EF cannot. A small region of CyaA (aa 225–267) contributes over 80% of binding free energy (Bouhss et al., 1993) and the region corresponding to the helical domain of EF is not required for its activation by CaM. However, a large binding surface from four discrete regions of EF (∼3000 Å2) makes contact to CaM and the helical domain is a major part of such interaction (Drum et al., 2002).
Our biochemical and structural analysis in conjunction with a recent NMR study using EF in complex with 15N- and 13C-labeled CaM reveals a new insight into the dynamic interaction among EF, CaM and calcium ions (Drum et al., 2002; T.Ulmer, S.Soelaiman, W.-J.Tang and A.Bax, submitted). Our structural models of EF and EF–CaM reveal that the N-terminal domain of CaM interacts with the helical domain of EF, which is solvent exposed in the EF alone structure, while the C-terminal domain of CaM interacts with several regions of EF that are mostly buried in the EF alone structure. This suggests that CaM most likely makes the initial contact with EF via its N-terminal domain. Consistent with this notion, our mutational analysis shows that N-CaM, not C-CaM, can partially activate EF and that this activation is independent of calcium concentration. The NMR study further supports this notion by showing that the N-terminal, but not C-terminal domain, of CaM makes contact with EF in the absence of calcium (T.Ulmer, S.Soelaiman, W.-J.Tang and A.Bax, submitted).
Based on our structural models, we hypothesize that the initial contact between N-CaM and the helical domain of EF leads to the insertion of C-CaM between CA and the helical domain. Such insertion triggers the conformational changes of switch C, which stabilizes the catalytic loop, leading to catalytic activation. Consistent with this notion, the mutations that weaken the interaction between CA and the helical domain results in an EF mutant, EF-triple, that has higher sensitivity to Ca2+–CaM. The interaction of C-CaM with the helical domain and CA of EF requires calcium binding to the C-terminal domain of CaM as seen in the X-ray crystallographic structure of EF–CaM (Drum et al., 2002) and NMR analysis (T.Ulmer, S.Soelaiman, W.-J.Tang and A.Bax, submitted). Interestingly, the N-terminal domain of CaM in the EF–CaM complex can dynamically interact with calcium ions. The recent NMR analysis reveals that the addition of two more moles of calcium allows calcium to bind the N-terminal domain of CaM to promote further interaction between the N-terminal domain of CaM and EF (T.Ulmer, S.Soelaiman, W.-J.Tang and A.Bax, submitted) even though no calcium ion is bound to the N-terminal domain of CaM in the X-ray crystallographic structure of the EF–CaM complex. However, the calcium loading to the N-terminal domain of CaM does not affect the ability of CaM to activate EF based on the fact that neither the CaM mutant with a mutated calcium-binding site at the N-terminal domain nor that with mutation to lock N-terminal domain in the calcium-free conformation has reduced ability to activate EF.
Both EF and CyaA have about three orders of magnitude higher adenylyl cyclase activity than those of host cells (Drum et al., 2002), thus the entry of EF and CyaA should raise the cAMP concentration of host cells to the supra-physiological level. In this paper, we show that the optimal activation of both EF and CyaA by CaM are highly active at the resting intracellular calcium concentration when CaM is not limited (i.e. 10 µM). However, there is a major difference between these two toxins at relatively low free CaM concentrations (i.e. 0.1 µM). Our data show that the apparent affinity of EF for CaM is low (Kd > 10 µM) at the resting calcium concentration (20–50 nM) so that EF may be minimally activated. EF becomes tightly associated with CaM (apparent Kd = 5– 20 nM) and is fully active only when the intracellular Ca2+ is elevated to 1 µM. In contrast, CyaA has 100-fold higher affinity to CaM than EF so that CyaA is still optimally activated at the resting calcium concentration. Physio logical calcium concentrations only affect the activation of CyaA-N by CaM when the free CaM is reduced to the concentration that is unlikely to occur in the host cells (i.e. 1 nM CaM; Gerendasy, 1999; Jurado et al., 1999).
This raises the possibility that EF activity can be regulated by the intracellular Ca2+ concentration while CyaA is fully active regardless of the intracellular calcium concentrations of its host cells. Consistent with this notion, a recent study using CHO cells, neutrophils, macrophages and lymphocytes shows that the extracellular calcium will enter into the host cells upon the intoxication of edema toxin, and such calcium entry is required for the optimal cAMP production by EF (Kumar et al., 2002). This study also shows that intoxication with edema toxin induces a transient calcium spike in human lymphocytes, consistent with the notion that increases in intracellular cAMP can elevate intracellular calcium concentration (Cooper et al., 1995). This cAMP-induced rise of Ca2+ might then further enhance the activity of EF in its host cells.
cAMP synthesis by adenylyl cyclase and degradation by phosphodiesterase are tightly controlled in mammalian cells (Hanoune and Defer, 2001). Similar to the observation that the elevated intracellular Ca2+ can inhibit the enzymatic activity of adenylyl cyclases (type V and VI; Cooper et al., 1995; Hu et al., 2002), we observe inhibition of EF and CyaA by elevated calcium concentrations (IC50 = 0.3–1.0 µM). Although the role of Ca2+ inhibition of EF and CyaA remains elusive, one might speculate that intracellular Ca2+ can control the rate of cAMP production of adenylyl cyclase toxins. This control may reduce ATP deprivation by adenylyl cyclase, which could lead to cell apoptosis and necrosis (McClintock et al., 2002). This could also set up the cAMP wave that is predicted to optimize cAMP-mediated signaling (Cooper et al., 1995).
EF is secreted by B.anthracis, and spores of this organism were used in the 2001 anthrax attacks in the US (Inglesby et al., 2002). This incident has heightened the need for anti-anthrax pharmaceuticals. Immobilizing CaM–EF complex on a glass surface and determining the structural basis for the fluorescence enhancement of 2′d3′ANT–ATP on binding to EF–CaM provides a viable means by which to screen compounds that can block either CaM activation of, or nucleotide binding to, EF. Adenylyl cyclase toxins are known to be secreted by two other human pathogens, B.pertussis and P.aeruginosa, which cause whooping cough and 10–20% of hospital-acquired infections, respectively (Yahr et al., 1998; Ladant and Ullmann, 1999). Biochemical studies have shown that Yersinia pestis, the bacterium that causes plague also secretes an adenylyl cyclase toxin and genome sequencing identified a gene in Y.pestis for such a toxin (Shevchenko and Mishankin, 1987; Michankin et al., 1992; Parkhill et al., 2001). Thus, adenylyl cyclase toxins are shared among several potent human pathogens and the development of therapeutic agents that block the activity of adenylyl cyclase toxins could have a broader usage against infections with pathogenic bacteria.
Materials and methods
Plasmid construction of EF mutants and CyaA-N mutants
The desired mutations were introduced using the Stratagene QuikChange Kit and the resulting mutations were confirmed by DNA sequencing. To construct the plasmid for the expression of EF-triple, pProExH6-EF was used as a template with primers that introduced three point mutations from E609, R613 and E616 to alanine. The C-terminal His6-tagged EF-ΔH (aa 291–658) was made using pProEx-EF-CH6 as a template, and a NotI site was introduced from residue 659 to 661 to remove the coding region of the helical domain (aa 659–800). The plasmid for the expression of CyaA-N(aa 1–393) was generated by introducing a termination codon at residue 394 using pEX-CyaA-1–412 as a template (Bejerano et al., 1999).
Purification of recombinant proteins
For all the EF mutants, plasmids carrying the desired mutation were transformed into RNaseE-deficient E.coli BL21 Star (DE3) cells harboring pUBS520. The mutant proteins were purified using DEAE, SP-Sepharose and Ni-NTA columns as described (Drum et al., 2000). The yield for EF mutants was ∼30 mg/l E.coli culture. To express CyaA-N, pEX-CyaA-N was transformed into E.coli B834 cells containing pUBS520. The resulting cells were grown in a modified T7 medium with 50 µg/ml ampicillin and 25 µg/ml kanamycin at 24°C to A600 = 0.4, induced by adding isopropyl-1-thiogalactopyranoside to a final concentration of 100 µM and harvested overnight post-induction. E.coli cells were lyzed in T20β5N500P0.1 buffer [20 mM Tris–HCl pH 8.0, 5 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 500 mM NaCl] by lysozyme (0.1 mg/ml) and sonication. After the lysate was spun at 200 000 g for 30 min, the supernatant of E.coli lysate was loaded directly onto a Ni2+-NTA column that was equilibrated with T20β5N100P0.1. The Ni2+-NTA column was then washed with three column volumes of T20β5N100P0.1 and with T20β5N100P0.1 containing 20 mM imidazole (pH 7). The His6-tagged CyaA-N was then eluted with buffer T20β5N100P0.1 containing 150 mM imidazole. The peak fractions were loaded onto a Q-Sepharose column after 5-fold dilution with T20D1P0.1 (20 mM Tris–HCl pH 8.0, 1 mM dithiothreitol, 0.1 mM PMSF) and the proteins were eluted by NaCl gradient. The purified CyaA-N was then concentrated to >10 mg/ml and stored at –80°C. The yield for CyaA-N was ∼100 mg/l culture. Human and fruit fly full-length CaM, human CaM mutants (N-CaM and C-CaM), human cross-linked CaM and fruit fly CaM mutants (B1Q, B2Q, B3Q and B4Q) were expressed in bacteria and purified as described previously (Maune et al., 1992; Huber et al., 1996). The molecular weight of N-CaM and C-CaM was confirmed by LC/MS.
Structure determination and fluorescence measurement of EF–CaM–2′d3′ANT-ATP complex
2′d3′ANT-ATP was synthesized by reacting 2′deoxy-ATP with isatoic anhydride as described previously (Hiratsuka, 1983). To determine the structure of EF–CaM–2′d3′ANT-ATP, crystals of EF3-CH6–CaM complex were grown using vapor diffusion, soaked with 2 mM 2′d3′ANT-ATP during cryoprotection, and frozen in liquid nitrogen as described (Drum et al., 2001). Data were collected at 100 K at APS Biocars 14-BM-C and processed with the program Denzo and Scalepack (Otwinowski and Minor, 1997). The initial phase was obtained by difference Fourier method using program CNS and the model of EF–CaM complex. The 3.6 Å model was refined using the programs Turbo-Frodo, O and CNS. The coordinates for EF–CaM–2′d3′ANT-ATP have been deposited with the Protein Data Bank (accession code 1LVC). Steady-state fluorescence emission spectra were recorded on a Jobin Yvon-Spex Fluoromax-2 photon counting spectrofluorometer (Edison, NJ). The indicated free calcium concentrations in the fluorescence assays as well as the subsequent SPR and adenylyl cyclase assays were obtained by buffering with EGTA based on the MAXC program website, http://www.stanford.edu/∼cpatton/maxc.html (Bers et al., 1994) and the conditions for each experiment are listed in the tables in the supplementary data.
SPR spectroscopy
SPR measurements were performed with BIAcore 1000 instrument at 25°C. Cut-CaM fusion protein was immobilized on a self-assembled monolayer as described (Hodneland et al., 2002). Following incubation in buffer X (10 mM Tris–HCl pH 7.0, 150 mM KCl, 1.0 mM EGTA, 10 mM MgCl2 and variable CaCl2 concentrations to reach the desired free Ca2+ concentrations), buffer X containing adenylyl cyclase proteins was passed over the immobilized calmodulin with a flow rate of 1 µl/min for 5 min to allow association. The monolayer was then washed with buffer X for >10 min to allow dissociation. Following each use, EGTA solution (100 mM) was washed over the surface to completely dissociate the complex. Protein–protein interactions were measured at three different adenylyl cyclase concentrations ranging from 10 nM to 8 µM, and the data were fit into the conformational change model provided by the manufacturer of the instrument.
Adenylyl cyclase assay
Adenylyl cyclase activities were measured after 10 min at 30°C in the presence of 20 mM HEPES pH 7.2, 5 mM ATP with a trace amount of [32P]ATP, 10 mM EGTA, 0.1 M KCl, 10 mM MgCl2 (unless indicated) and the indicated free calcium concentrations buffered by 10 mM EGTA. ATP and cAMP were separated by Dowex and alumina columns as described previously (Drum et al., 2000).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to P.Gardner (HHMI, University of Chicago) for help with oligonucleotide synthesis and DNA sequencing, to Dr Emanuel Hanski at Hebrew University–Hadassah Medical School for the gene encoding CyaA, and to Drs Keith Brister, Gary Navrotski, Bill Desmarais and Robert Henning at APS BioCars 14-BMC for their help in data collection. This research was supported by National Institute of Health GM53459, GM62548 and American Heart Association Established Investigator Award to W.-J.T., Defense Advanced Research Projects Agency N00173–01-G010 and National Science Foundation DMR-9808595 to M.M., National Institute of Health AR41637 to Z.G. and Welch Foundation C-1119 to K.B. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under contract No. W-31-109-ENG-38.
References
- Babu Y.S., Sack,J.S., Greenhough,T.J., Bugg,C.E., Means,A.R. and Cook,W.J. (1985) Three-dimensional structure of calmodulin. Nature, 315, 37–40. [DOI] [PubMed] [Google Scholar]
- Barbato G., Ikura,M., Kay,L.E., Pastor,R.W. and Bax,A. (1992) Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry, 31, 5269–5278. [DOI] [PubMed] [Google Scholar]
- Bejerano M., Nisan,I., Ludwig,A., Goebel,W. and Hanski,E. (1999) Characterization of the C-terminal domain essential for toxic activity of adenylate cyclase toxin. Mol. Microbiol., 31, 381–392. [DOI] [PubMed] [Google Scholar]
- Bers D., Patton,C. and Nuccitelli,R. (1994) A practical guide to the preparation of Ca buffers. In Nuccitelli,R. (ed.), Methods Cell Biology–A Practical Guide to the Study of Ca2+in Living Cells. Vol. 40. Academic Press, San Diego, CA, pp. 3–29. [DOI] [PubMed]
- Bouhss A., Krin,E., Munier,H., Gilles,A.M., Danchin,A., Glaser,P. and Barzu,O. (1993) Cooperative phenomena in binding and activation of Bordetella pertussis adenylate cyclase by calmodulin. J. Biol. Chem., 268, 1690–1694. [PubMed] [Google Scholar]
- Cooper D.M., Mons,N. and Karpen,J.W. (1995) Adenylyl cyclases and the interaction between calcium and cAMP signalling. Nature, 374, 421–424. [DOI] [PubMed] [Google Scholar]
- Deisseroth K., Heist,E.K. and Tsien,R.W. (1998) Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature, 392, 198–202. [DOI] [PubMed] [Google Scholar]
- DeMaria C.D., Soong,T.W., Alseikhan,B.A., Alvania,R.S. and Yue,D.T. (2001) Calmodulin bifurcates the local Ca2+ signal that modulates P/Q-type Ca2+ channels. Nature, 411, 484–489. [DOI] [PubMed] [Google Scholar]
- Drum C.L., Yan,S.Z., Sarac,R., Mabuchi,Y., Beckingham,K., Bohm,A., Grabarek,Z. and Tang,W.J. (2000) An extended conformation of calmodulin induces interactions between the structural domains of adenylyl cyclase from Bacillus anthracis to promote catalysis. J. Biol. Chem., 275, 36334–36340. [DOI] [PubMed] [Google Scholar]
- Drum C.L., Shen,Y., Rice,P.A., Bohm,A. and Tang,W.J. (2001) Crystallization and preliminary X-ray study of the edema factor exotoxin adenylyl cyclase domain from Bacillus anthracis in the presence of its activator, calmodulin. Acta Crystallogr. D Biol. Crystallogr., 57, 1881–1884. [DOI] [PubMed] [Google Scholar]
- Drum C.L., Yan,S.Z., Bard,J., Shen,Y., Lu,D., Soelaiman,S., Grabarek,Z., Bohm,A. and Tang,W.-J. (2002) Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature, 415, 396–402. [DOI] [PubMed] [Google Scholar]
- Eldik L.V. and Watterson,D.M. (1998) Calmodulin and Signal Transduction. Academic Press, San Diego, CA.
- Finn B.E., Evenas,J., Drakenberg,T., Waltho,J.P., Thulin,E. and Forsen,S. (1995) Calcium-induced structural changes and domain autonomy in calmodulin. Nat. Struct. Biol., 2, 777–783. [DOI] [PubMed] [Google Scholar]
- Gentile F., Knipling,L.G., Sackett,D.L. and Wolff,J. (1990) Invasive adenylyl cyclase of Bordetella pertussis. Physical, catalytic and toxic properties. J. Biol. Chem., 265, 10686–10692. [PubMed] [Google Scholar]
- Gerendasy D. (1999) Homeostatic tuning of Ca2+ signal transduction by members of the calpacitin protein family. J. Neurosci. Res., 58, 107–119. [PubMed] [Google Scholar]
- Hanoune J. and Defer,N. (2001) Regulation and role of adenylyl cyclase isoforms. Ann. Rev. Pharm. Toxicol., 41, 145–174. [DOI] [PubMed] [Google Scholar]
- Hiratsuka T. (1983) New ribose-modified fluorescent analogs of adenine and guanine nucleotides available as substrates for various enzymes. Biochim. Biophys. Acta, 742, 496–508. [DOI] [PubMed] [Google Scholar]
- Hodneland C.D., Lee,Y.-S., Min,D.-H. and Mrksich,M. (2002) Selective immobilization of protein to self-assembled monolayers presenting active site directed capture ligands. Proc. Natl Acad. Sci. USA, 99, 5048–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich K.P. and Ikura,M. (2002) Calmodulin in action: diversity in target recognition and activation mechanisms. Cell, 108, 739–742. [DOI] [PubMed] [Google Scholar]
- Hoover D.L., Friedlander,A.M., Rogers,L.C., Yoon,I.K., Warren,R.L. and Cross,A.S. (1994) Anthrax edema toxin differentially regulates lipopolysaccharide-induced monocyte production of tumor necrosis factor α and interleukin-6 by increasing intracellular cyclic AMP. Infect. Immun., 62, 4432–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Nakata,H., Gu,C., de Beer,T. and Cooper,D.M. (2002) A critical interplay between Ca2+-inhibition and activation by Mg2+ of AC5 revealed by mutants and chimeric constructs. J. Biol. Chem., 277, 33139–33147. [DOI] [PubMed] [Google Scholar]
- Huber P.A., El-Mezgueldi,M., Grabarek,Z., Slatter,D.A., Levine,B.A. and Marston,S.B. (1996) Multiple-sited interaction of caldesmon with Ca2+–calmodulin. Biochem. J., 316, 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglesby T.V. et al. (2002) Anthrax as a biological weapon, 2002: updated recommendations for management. JAMA, 287, 2236–2252. [DOI] [PubMed] [Google Scholar]
- Jurado L.A., Chockalingam,P.S. and Jarrett,H.W. (1999) Apocal modulin. Physiol. Rev., 79, 661–682. [DOI] [PubMed] [Google Scholar]
- Khelef N., Zychlinsky,A. and Guiso,N. (1993) Bordetella pertussis induces apoptosis in macrophages: role of adenylate cyclase-hemolysin. Infect. Immun., 61, 4064–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Ahuja,N. and Bhatnagar,R. (2002) Anthrax edema factor requires influx of calcium for inducing cyclic AMP toxicity in target cells. Infect. Immun., 70, 4997–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labruyure E., Mock,M., Ladant,D., Michelson,S., Gilles,A.M., Laoide,B. and Barzu,O. (1990) Characterization of ATP and calmodulin-binding properties of a truncated form of Bacillus anthracis adenylate cyclase. Biochemistry, 29, 4922–4928. [DOI] [PubMed] [Google Scholar]
- Ladant D. and Ullmann,A. (1999) Bordetella pertussis adenylate cyclase: a toxin with multiple talents. Trends Microbiol., 7, 172–176. [DOI] [PubMed] [Google Scholar]
- Leppla S.H. (1984) Bacillus anthracis calmodulin-dependent adenylate cyclase: chemical and enzymatic properties and interactions with eucaryotic cells. Adv. Cyclic Nucleotide Protein Phosphorylation Res., 17, 189–198. [PubMed] [Google Scholar]
- Maune J.F., Klee,C.B. and Beckingham,K. (1992) Ca2+ binding and conformational change in two series of point mutations to the individual Ca2+-binding sites of calmodulin. J. Biol. Chem., 267, 5286–5295. [PubMed] [Google Scholar]
- McClintock D.S., Santore,M.T., Lee,V.Y., Brunelle,J., Budinger,G.R., Zong,W.X., Thompson,C.B., Hay,N. and Chandel,N.S. (2002) Bcl-2 family members and functional electron transport chain regulate oxygen deprivation-induced cell death. Mol. Cell. Biol., 22, 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador W.E. and Quiocho,F.A. (2002) Man bites dog. Nat. Struct. Biol., 9, 156–158. [DOI] [PubMed] [Google Scholar]
- Michankin B.N., Chevchenko,L.A. and Asseeva,L.E. (1992) Adenylate-cyclase. Un eventuel facteur de la pathogenicite de Yersinia pestis. Bull. Soc. Pathol. Exot., 85, 17–21. [PubMed] [Google Scholar]
- Mock M. and Fouet,A. (2001) Anthrax. Annu. Rev. Microbiol., 55, 647–671. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Parkhill J. et al. (2001) Genome sequence of Yersinia pestis, the causative agent of plague. Nature, 413, 523–527. [DOI] [PubMed] [Google Scholar]
- Sarfati R.S., Kansal,V.K., Munier,H., Glaser,P., Gilles,A.M., Labruyere,E., Mock,M., Danchin,A. and Barzu,O. (1990) Binding of 3′-anthraniloyl-2′-deoxy-ATP to calmodulin-activated adenylate cyclase from Bordetella pertussis and Bacillus anthracis. J. Biol. Chem., 265, 18902–18906. [PubMed] [Google Scholar]
- Schumacher M.A., Rivard,A.F., Bachinger,H.P. and Adelman,J.P. (2001) Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature, 410, 1120–1124. [DOI] [PubMed] [Google Scholar]
- Shevchenko L.A. and Mishankin,B.N. (1987) Adenylyl cyclase of the causative agent of plague: its purification and properties. Zh Mikrobiol. Epidemiol. Immunobiol., 7, 59–63. [PubMed] [Google Scholar]
- Tan R.Y., Mabuchi,Y. and Grabarek,Z. (1996) Blocking the Ca2+-induced conformational transitions in calmodulin with disulfide bonds. J. Biol. Chem., 271, 7479–7483. [DOI] [PubMed] [Google Scholar]
- Weingart C.L. and Weiss,A.A. (2000) Bordetella pertussis virulence factors affect phagocytosis by human neutrophils. Infect. Immun., 68, 1735–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahr T.L., Vallis,A.J., Hancock,M.K., Barbieri,J.T. and Frank,D.W. (1998) ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl Acad. Sci. USA, 95, 13899–13904. [DOI] [PMC free article] [PubMed] [Google Scholar]