Abstract
The exoribonuclease polynucleotide phosphorylase (PNPase) has been implicated in mRNA processing and degradation in bacteria as well as in chloroplasts of higher plants. Here, we report the first comprehensive in vivo study of chloroplast PNPase function. Modulation of PNPase activity in Arabidopsis chloroplasts by a reverse genetic approach revealed that, although this enzyme is essential for efficient 3′-end processing of mRNAs, it is insufficient to mediate transcript degradation. Surprisingly, we identified PNPase as also being indispensable for 3′-end maturation of 23S rRNA transcripts. Analysis of tRNA amounts in transgenic Arabidopsis plants suggests a direct correlation of PNPase activity and tRNA levels, indicating an additional function of this exoribo nuclease in tRNA decay. Moreover, the extent of polyadenylated mRNAs in chloroplasts is negatively correlated with PNPase activity. Together, our results attribute novel functions to PNPase in the metabolism of all major classes of plastid RNAs and suggest an unexpectedly complex role for PNPase in RNA processing and decay.
Keywords: chloroplast/PNPase/polyadenylation/ribonuclease/RNA processing
Introduction
RNA stability is one of the parameters determining gene expression in the cell. A universal feature of mRNAs is their transcription as 3′ extended precursor molecules that subsequently undergo processing events resulting in the formation of mature 3′ ends. Studies in a wide range of organisms have established that efficient 3′ processing is required for RNA stabilization and that unprocessed or incorrectly processed RNA is subject to rapid degradation by ribonucleases (Regnier and Arraiano, 2000; Hilleren et al., 2001). Since these enzymes not only are usually involved in mediating processing reactions but also convey the degradation of RNAs, their strict regulation appears to be of particular importance for cellular RNA metabolism (Garcia-Mena et al., 1999).
All eukaryotic exoribonucleases known to be essential for viability appear to form multiprotein complexes designated as exosomes (Decker, 1998; van Hoof and Parker, 1999; Mitchell and Tollervey, 2000). In contrast, most of the eight identified 3′→5′ exoribonucleases from Escherichia coli function independently of each other and without detectable complex formation. The only noted exception is the polynucleotide phosphorylase (PNPase), a phosphorolytically processive 3′→5′ exoribonuclease constituting a component of a high-molecular-weight complex, the degradosome. In addition to PNPase, this complex consists of the endoribonuclease RNase E, the helicase RhlB, enolase and sub-stoichiometric amounts of polyphosphate kinase (Carpousis et al., 1999; Regnier and Arraiano, 2000).
A PNPase was also purified from spinach chloroplasts, and extensive biochemical analyses suggested its function in plastid mRNA processing (Hayes et al., 1996). As chloroplasts represent endosymbiotic descendants of formerly free-living bacteria, their RNA processing and degradation mechanisms exhibit numerous prokaryotic characteristics, but also involve novel processing components and unique features (Hayes et al., 1999). Most plastid RNA precursors undergo a series of complex maturation events, including splicing, editing and 5′ and 3′ processing. Crucial for efficient 3′ processing is an inverted repeat structure in the 3′ untranslated region (UTR) of mRNAs that folds into a stem–loop structure (Hayes et al., 1999). These stem–loops do not function as effective transcription termination signals. Instead, they represent structures acting as barriers to prevent 3′→5′ degradation, possibly by supporting efficient 3′-end processing and subsequent protection. A 28 kDa RNA-binding protein with no functional or structural counterpart in bacteria has been shown to interact with these RNA structures. This protein is essential for efficient plastid 3′ processing, but its exact mode of action remains unclear (Hayes et al., 1999).
Biochemical analyses revealed the existence of several endoribonucleases in plastids. The p54 endoribonuclease from mustard has been reported to mediate 3′-end formation of several precursor RNAs in vitro (Liere and Link, 1997). In contrast, the p67 endonuclease purified from spinach plastids appears to cut mRNAs upstream of the stem–loop structure or shortly downstream of the coding region. Therefore, it is unlikely to be involved in RNA 3′-end formation and rather appears to initiate degradation of plastid RNAs, although its exact function in vivo remains to be established (Kudla et al., 1996).
Despite the observed complexity of exoribonucleases in E.coli, study of chloroplast RNA metabolism has so far identified only a single exoribonuclease (PNPase) as potentially executing transcript processing and degradation. This protein exhibits extensive similarity to bacterial PNPases and was originally purified as a 100 kDa protein that appeared to be part of a 550 kDa protein complex (Hayes et al., 1996). Recent analysis of this complex has revealed that it is formed by the assembly of three PNPase monomers (Baginsky et al., 2001). Biochemical analyses have suggested that PNPase is required for mRNA 3′-end processing in vitro (Hayes et al., 1996) and, moreover, forms the major activity triggering the degradation of RNA molecules following endonucleolytic cleavage and polyadenylation (Hayes et al., 1999). Polyadenylation enhances the decay of RNA molecules by tagging them for degradation and is a common feature of bacteria and plant organelles (Carpousis et al., 1999; Hayes et al., 1999). Studies of RNA polyadenylation in E.coli have benefited greatly from the use of mutant strains deficient in one or more exoribonuclease activities (Sarkar, 1997; Coburn and Mackie, 1999). This approach has allowed us to efficiently detect poly(A) tails, quantitate their occurrence and measure their length. Moreover, analysis of the half-life times and processing patterns of specific mRNAs in such strains has provided important insights into in vivo functions of polyadenylation (Sarkar, 1997; Coburn and Mackie, 1999).
In the green alga Chlamydomonas reinhardtii, numerous mutants affected in plastid RNA processing and stability have been isolated (Rochaix, 1996). Their molecular analysis revealed important insights into processing components but also disclosed that most of the characterized loci function in the maturation of individual messages and do not represent factors with a general role in RNA metabolism. In addition, recent studies suggest that plastid RNA processing and/or degradation in Chlamydomonas could be different from processing in bacteria and higher plant plastids, since in Chlamydomonas a 5′→3′ exonuclease appears to play an important role in RNA metabolism (Drager et al., 1999). A similar activity has so far not been found in bacteria or higher plant organelles.
In higher plants, genetic approaches based on the identification of hcf (high chlorophyll fluorescence) mutants from maize and Arabidopsis have provided important insights into the molecular identity and function of components mediating processing events in plastids (Barkan, 1993; Meurer et al., 1996). However, a common feature of all higher plant processing mutants characterized so far is their functional specificity for distinct messages. Thus far, general processing or degradation factors have been investigated exclusively in vitro by biochemical approaches (Hayes et al., 1999). Although this work has contributed significantly to our current understanding of RNA metabolism in plastids, their validity for the situation in planta remains to be established.
Here, we have applied a reverse genetic approach to determine the function of the plastid exoribonuclease PNPase in vivo and report the effects of modulating PNPase activity on plastid RNA metabolism. Our findings suggest that the function of PNPase in vivo and its contributions to RNA metabolism in plastids are much more complex than previously recognized.
Results
Isolation and characterization of a plastid PNPase from Arabidopsis thaliana
Previous studies identified a 100 kDa RNA-binding protein (100RNP) from spinach chloroplasts as a PNPase involved in mRNA 3′-end processing (Hayes et al., 1996). However, since spinach is hardly accessible for genetic manipulations, the exact function of this enzyme in vivo could not be addressed. Therefore, we sought to establish the in vivo function of PNPase in higher plant chloroplast RNA metabolism, taking a transgenic approach in the model plant Arabidopsis.
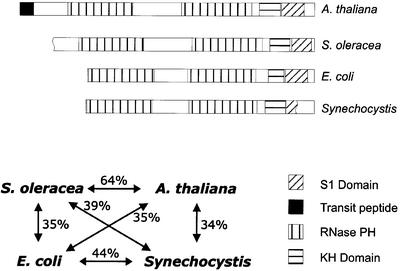
Degenerate primers were derived from the nucleotide sequences of spinach PNPase and the corresponding E.coli enzyme and were used to amplify an Arabidopsis PNPase cDNA fragment. Subsequent screening of a cDNA library and 5′ RACE yielded a full-length cDNA potentially encoding a protein of 922 amino acids with striking similarity to known PNPases from other organisms. Sequence comparison revealed 64% identical amino acids with the spinach 100RNP and 34 and 35% identity with the PNPase enzymes from Synechocystis and E.coli, respectively. Moreover, conserved domains typical of functional PNPases, including two RNase PH domains and two RNA-binding domains, were identified in the Arabidopsis protein (Figure 1). In addition, it carries a putative N-terminal chloroplast import sequence. The protein was therefore designated Arabidopsis thaliana chloroplast PNPase (AtcpPNPase). Analysis of the Arabidopsis genome database revealed no other related sequence predicted to be imported into chloroplasts. However, an additional putatively mitochondrial localized PNPase is also found in the Arabidopsis genome (Yehudai-Resheff et al., 2001).
Fig. 1. Structural comparison of different plant and bacterial PNPases. The domain structures of plastid PNPases from Arabidopsis and spinach are compared with PNPase enzymes from E.coli and Synechocystis. The conserved S1, RNase PH and KH domains and the transit peptide are marked as indicated in the bottom right of the figure. The degree of amino acid identity is depicted in the bottom left.
Since we intended to use a FLAG-tagged version of PNPase to modulate chloroplast PNPase activity in vivo, several controls were performed to ensure the functionality of the protein and the validity of our approach. To this end, a FLAG tag sequence was C-terminally fused to the PNPase cDNA sequence lacking the putative transit peptide. After over-expression in E.coli and subsequent purification, the recombinant protein possessed 3′→5′ exonuclease activity and released nucleoside diphos phates, thus excluding an interference of the FLAG tag with the innate exonuclease activity of the protein in vitro (data not shown).
For ectopic over-expression in transgenic plants, a FLAG epitope sequence was C-terminally fused to the full-length AtcpPNPase cDNA and cloned into a binary vector harboring a 35S promoter and a BASTA resistance selection cassette (Figure 2A). After transformation and selection of herbicide-resistant Arabidopsis plants, the integration of the T-DNA was verified by Southern blotting and PCR (data not shown) and the expression of the transgene was monitored by western blotting with a monoclonal antibody against the FLAG epitope (Figure 2B). To verify chloroplast localization of AtcpPNPase, plastids from transgenic plants over-expressing the full-length FLAG-tagged PNPase were isolated and co-purification of the FLAG-tagged AtcpPNPase with chloroplasts was verified by western blotting (Figure 2C, lane 4). Incubation of isolated chloroplasts with the protease thermolysin did not affect detection of the FLAG-tagged PNPase, indicating that the protein was efficiently imported into chloroplasts (Figure 2C, lane 1). When the plastids were disrupted by sonication prior to protease addition, the signal for the transgenic form of the PNPase disappeared (Figure 2C, lane 2).
Fig. 2. Modulation of chloroplast PNPase activity in transgenic plants. (A) Schematic diagram of the transformed T-DNA. The PNPase cDNA sequence was fused to the FLAG tag sequence (FT) and expressed under control of the CaMV 35S promoter (35S) and the TMV translation enhancing omega element (Ω). Transformed plants were selected for BASTA resistance (BAR). uidA, β-glucuronidase; NosT, termination signal. (B–E) PNPase expression in transgenic plants. (B) Expres sion of the FLAG-tagged and the endogenous PNPases was surveyed by western analyses after separation by SDS–PAGE and transfer to PVDF membranes. For the analysis of transgene expression, the monoclonal FLAG-M2 antibody (Sigma) was used (Anti-FLAG). The transgenic and endogenous form of PNPase was detected with a polyclonal antiserum against spinach PNPase (Anti-PNP; Hayes et al., 1996). The arrowhead marks the signal for the endogenous PNPase in the wild-type (WT) lane. The asterisk marks unspecific signals due to long exposure. The figure exemplarily depicts the analyses of a wild-type line and two representative transgenic lines either over-expressing the transgene (PNP+, +) or exhibiting reduction of PNPase expression caused by co-suppression (PNP–, –). (C) Chloroplasts from PNPase over-expressing plants were isolated and, following the indicated treatments, 8 µg of soluble proteins were analyzed by SDS–PAGE and western blotting. The FLAG-tagged PNPase co-purified together with isolated chloroplasts (lane 4). Resistance to thermolysin treatment verified the integrity of the isolated chloroplasts and the efficient import of the recombinant protein (lane 1). Disruption of the plastids by sonication prior to protease addition led to digestion of the protein (lane 2), whereas sonication treatment alone did not affect PNPase content of the sample (lane 3). (D) UV cross-linking analysis of PNPase expression. Leaf total soluble proteins (10 µg) were cross-linked to a radioactive probe corresponding to the spinach petD 3′ UTR (Hayes et al., 1996). PNPase protein was detected by autoradiography after separation by SDS–PAGE. Total RNAs from wild-type plants and from three independent transgenic lines showing moderate over-expression (+), strong over-expression (++) or no expression (–) of the FLAG-tagged PNPase were used. C, no protein added. (E) Quantification of PNPase protein amounts in wild-type and transgenic Arabidopsis lines. A dilution series of soluble proteins from wild-type and different amounts of protein from transgenic plants were analyzed by UV cross-linking as described in (D). Signals were quantified by phosphoimaging.
Since PNPases exist as part of high-molecular-weight protein complexes (Baginsky et al., 2001; Symmons et al., 2002), we next investigated whether the affinity tag interferes with the assembly of the protein into larger protein complexes. Soluble chloroplast protein extracts from wild-type and transgenic plants were separated by gel filtration and analyzed by UV cross-linking. The elution profiles of the native and the affinity-tagged AtcpPNPases were identical (data not shown), indicating that the subunit composition of the recombinant protein is similar to that of the native protein.
PNPase activity mediates efficient 3′-end processing of chloroplast mRNAs
Having established that the FLAG-tagged PNPase is correctly targeted and fully functional in planta, we analyzed transgene expression in a larger number of transgenic lines using the FLAG antibody. Most of these lines exhibited moderate to strong (3- to >20-fold) over-expression of the transgene. However, we also obtained transgenic plants, which showed no detectable expression of the transgene. In these plants, the expression of the endogenous PNPase was also strongly reduced (Figure 2B), most likely caused by co-suppression. Since the faint signal observed in western analyses of PNP– plants could result from cross-reaction of polyclonal PNPase antisera with structurally related proteins, we also performed comparative UV cross-linking experiments. In these analyses, we did not detect any PNPase protein in PNP– plants (Figure 2D and E). In addition, comparative UV cross-linking assays revealed that even 3% remaining PNPase protein would be detectable by this sensitive assay (Figure 2E). Therefore, if there were any remaining PNPase protein in PNP– plants, the amount would have been reduced at least to levels clearly below 3% of the protein present in wild-type plants (Figure 2E). When we analyzed the T2 and T3 generations, we observed significant epigenetic variability of PNPase expression in the progeny from each of these lines. Therefore, in all subsequent experiments, individual transgenic plants were tested for expression of the FLAG-tagged PNPase before further analysis. Plants without detectable PNPase expression were designated as co-suppressed plants.
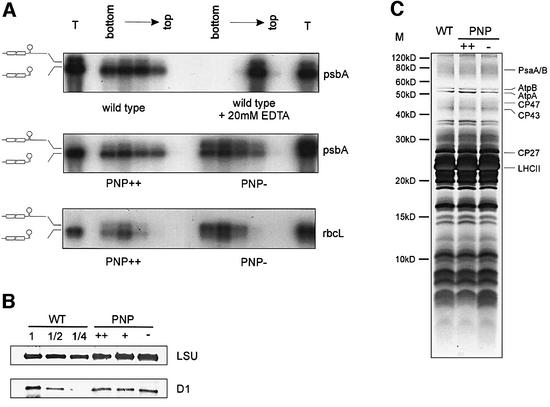
To analyze the function of AtcpPNPase in vivo, we first investigated the influence of PNPase expression level on mRNA accumulation and processing in a series of northern hybridizations. We included RNAs from plants with moderate (PNP+; Figure 3B–F, lane +) and high (PNP++; Figure 3B–F, lane ++) over-expression of the AtcpPNPase as well as from wild-type (Figure 3B–F, lane WT) and co-suppressed (PNP–; Figure 3B–F, lane –) plants. All hybridizations were reproduced with RNA samples from several independent transgenic lines and gave identical results (data not shown). Northern analyses with probes specific for the coding region of the rbcL and psbA mRNAs revealed that over-expression of AtcpPNPase had no significant effect on either RNA processing or accumulation levels of the respective mature mRNAs (Figure 3B and D). In contrast, reduced PNPase expression led to a strong decrease in correctly processed rbcL and psbA mRNAs. Interestingly, an increased accumulation of mRNA molecules that were ∼200–300 nucleotides longer than the mature mRNA was observed (Figure 3B and D, lane –). Despite the weaker appearance of the rather heterogeneous rbcL specific signals in PNP– plants in northern analyses, additional quantification of total rbcL and psbA mRNA amounts (mature plus unprocessed mRNA) by phosphoimaging and by slot blot hybridizations with radioactively labeled chloroplast cDNAs from PNP–, PNP++ and wild-type plants revealed no significant differences in total transcript accumulation between the different lines investigated (data not shown). It seems reasonable to assume that the longer mRNA molecules represent unprocessed mRNAs with prolonged 3′ ends, since additional northern analyses with probes simultaneously covering the 3′ UTR and regions 3′ of the stem–loop that are normally not part of the mature RNA gave rise to strong signals in PNP– plants (Figure 3C and E). Interestingly, the same 3′ unprocessed RNA molecules were also detectable in wild-type RNA preparations, albeit at much lower concentrations. Intriguingly, in PNPase over-expressing plants, the amount of 3′ unprocessed RNA species decreases further and is negatively correlated with increasing amounts of PNPase. This clearly indicates that the efficiency of mRNA 3′-end processing is strictly dependent on the PNPase activity in the chloroplast. In contrast, the stability of mature psbA and rbcL mRNAs appears to remain unaffected by PNPase over-expression or co-suppression. Taken together, these data suggest that the PNPase activity is required for efficient 3′-end processing of mRNAs in vivo but is not sufficient to mediate their degradation.
Fig. 3. Northern analysis of plants transformed with FLAG-tagged PNPase. (A) Schematic diagram of the location of the probes used for northern hybridizations within the rbcL and psbA mRNA (black bars). (B and D) Northern hybridizations with rbcL- and psbA-specific probes corresponding to the respective coding regions (CDS). The labeling of the investigated lines corresponds to Figure 2D. (C and E) Identical membranes were analyzed with probes specific for the unprocessed 3′ trailer (3′ UTR) of rbcL and psbA, respectively. (F) Control hybridization with a probe for the nuclear-encoded 18S rRNA. The numbers indicate the sizes of the detected RNA species (in kb).
Impaired 3′-end processing does not affect mRNA translation
Despite the reduced amount of mature rbcL and psbA mRNAs, the PNP– plants did not exhibit any obvious phenotype under various growth conditions (data not shown). It has recently been reported that, in chloroplasts from Chlamydomonas, correct processing of mRNA 3′ ends represents a prerequisite for efficient translation initiation (Rott et al., 1998). As chloroplasts are highly flexible in regulating their gene expression, it appeared possible that, in the PNP– plants, only correctly processed mRNAs are associated with ribosomes but are translated more efficiently. In order to test this hypothesis, polysomal RNA from PNP++, PNP– and wild-type plants was isol ated and separated by sucrose gradient centrifugation. Subsequently, the RNA from individual fractions was isolated and analyzed by northern hybridizations using probes with enhanced specificity for 3′ unprocessed RNA molecules (Figure 3). For both rbcL and psbA, mature and unprocessed RNA molecules were found to be associated equally efficiently with polysomes in all plant lines investigated (Figure 4A). Accordingly, western analyses with polyclonal antisera revealed that the amount of large subunit of RubisCO (encoded by rbcL) and PSII D1 proteins (encoded by psbA) were also not affected in plants with either an increase or decrease in PNPase activity (Figure 4B). To test whether possible changes in processing patterns of other transcripts have influences on protein expression, thylakoid membrane proteins were analyzed by high-resolution SDS–PAGE. This approach enabled us to comprehensively monitor and compare the quantity of chloroplast proteins in the different Arabidopsis lines. As shown in Figure 4C, these analyses did not reveal any significant differences in the protein accumulation patterns of wild-type, PNP++ and PNP– plants. These findings imply that, in higher plant chloroplasts, correct mRNA 3′-end maturation is not necessarily required for efficient translation.
Fig. 4. Plastid protein biosynthesis in plants with modified PNPase expression. (A) Analysis of polysome association of rbcL and psbA mRNAs in wild-type, PNPase over-expressing (PNP++) and PNPase co-suppressed (PNP–) plants. Polysomal RNA was separated in a continuous sucrose gradient. As a control, a sample of wild-type polysomes was separated in the presence of 20 mM EDTA to dissociate the ribosomal subunits. RNAs from different fractions were separated by denaturing agarose gel electrophoresis, transferred to nylon membranes and hybridized with probes containing the 3′ trailer and sequences downstream (described in Figure 3A) for psbA (upper and middle) and rbcL (lower). The structure of the mRNA molecules is depicted schematically on the left. (B) Western analysis of chloroplast proteins. After electrophoretic separation, leaf soluble protein extracts and membrane-enriched fractions (upper and lower panel, respectively) were transferred onto PVDF membranes. Immunodetections were performed with antisera against the large subunit of RubisCO (LSU) and the D1 subunit of PSII (D1), respectively. Proteins (10 µg) from wild-type (WT1), PNPase over-expressing (PNP+/++) and PNPase co-suppressed (PNP–) plants as well as dilutions of wild-type protein extracts (WT1/2 and WT1/4) were analyzed. (C) Analysis of protein composition of thylakoid membranes in wild-type, PNPase over-expressing (++) and PNPase co-suppressed (–) Arabidopsis plants. Thylakoid membrane proteins were isolated, separated on a 16% SDS–Tris–Tricine polyacrylamide gel and stained with silver. Proteins identified according to their known separation characteristics are indicated on the right. M, molecular weight marker.
AtcpPNPase activity is indispensable for proper rRNA processing
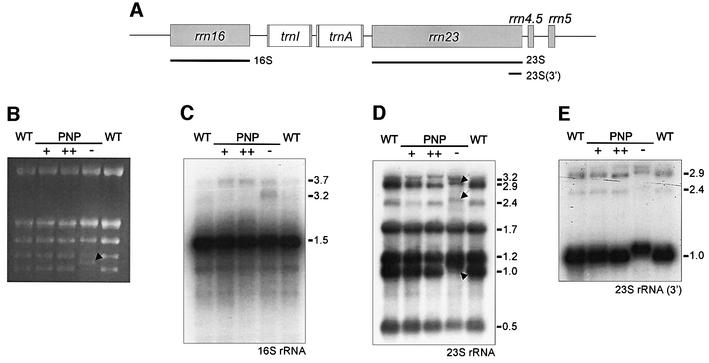
Most studies on RNA processing in bacteria and plant organelles have focused on mRNAs. Accordingly, the relationship of mRNA processing and maturation of rRNAs in terms of common or diverse enzymatic and mechanistic characteristics has not been elucidated. In addition, a function of bacterial or organellar PNPases in rRNA processing has not been ascertained so far. Interestingly, the most obvious molecular phenotype displayed by PNP– plants is a shift of an rRNA species, which can easily be detected when total RNA is separated on agarose gels (Figure 5B, arrow). The 23S rRNA is processed further into several fragments, the so-called ‘hidden breaks’, upon integration into the 50S ribosomal subunit. Thus, the shifted fragment is most likely part of the plastid 23S rRNA. A probe encompassing the entire 23S rRNA detected this polymorphism in hybridization experiments (Figure 5D). Furthermore, two additional fragments were shifted to larger sizes compared with wild-type 23S rRNA (Figure 5D, arrows). These RNA species most likely represent 3′ unprocessed 23S rRNA molecules, which, despite the absence of 3′ maturation, undergo other processing reactions. Accordingly, all three RNA fragments were ∼100 nucleotides longer than the corresponding wild-type fragments, which correlates with the size of the intergenic region between 23S and 4.5S rRNA (98 nucleotides). We used a probe covering the 3′-terminal 315 nucleotides of the 23S rRNA to test whether the shifted fragments contain the 3′ end of the 23S rRNA. The results depicted in Figure 5E clearly indicate that this is the case for all three fragments.
Fig. 5. Analysis of rRNA expression and processing. (A) Schematic diagram of the organization of the rRNA operon and the location of the probes used for northern hybridizations (black bars). Gray boxes indicate structural RNAs, white boxes indicate introns and lines show intergenic sequences. (B) An ethidium bromide-stained gel after electrophoretic separation of total leaf RNA. The arrow indicates a rRNA fragment, which is shifted in size compared with the wild type (WT). (C–E) Results of northern analyses of three identical membranes with the probes indicated in (A). The probes were specific for 16S rRNA (C), the complete 23S rRNA (D) or the 3′ end (315 nucleotides) of the 23S rRNA (E). The numbers on the right indicate the sizes of detected RNA species (in kb). The arrows in (D) mark fragments, which are shifted in size compared with the wild type.
As observed for mRNAs, over-expression of AtcpPNPase had no effect on 23S rRNA processing. In addition, we did not observe any differences in the processing of 16S rRNA, regardless of the activity of the AtcpPNPase being enhanced or reduced (Figure 5C). This represents a rather surprising finding, since a 300 nucleotide 3′ trailer between the 16S rRNA and the tRNAIle has to be removed to generate the mature 16S rRNA. Apparently, this processing reaction does not depend on AtcpPNPase function.
PNPase activity is dispensable for psaA/psaB/rps14 3′-end maturation but mediates tRNA degradation
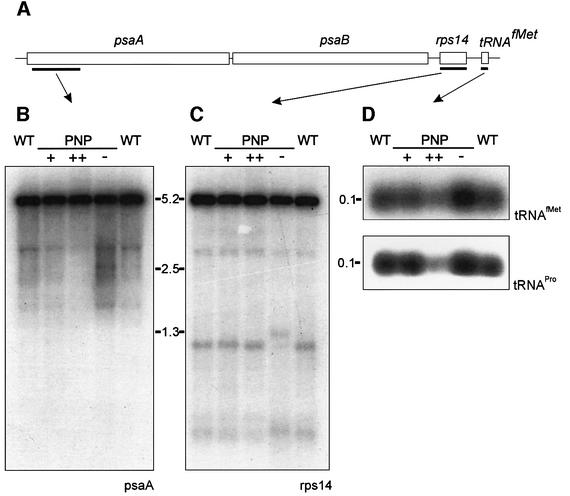
The only apparent difference between the 3′ trailers of the 16S and 23S rRNAs appears to be their mode of generation. The 3′ trailer of the 16S rRNA is most likely generated as a result of efficient RNase P cleavage at the 5′ end of the downstream tRNAIle (Figure 5A). In contrast, the 3′ trailer of the 23S rRNA originates from the release of the 4.5S rRNA from the primary rRNA transcript. Although the enzymatic activity mediating the 5′-end processing of the 4.5S rRNA is currently unknown, it is likely to be conveyed by another, RNase P-independent, endonucleolytic cleavage. To test whether the observed PNPase independence of 3′ trimming of RNase P processed transcripts represents a general phenomenon in plastid RNA metabolism, we chose to investigate processing of the psaA/psaB/rps14/tRNAfMet operon. This operon represents an example of a polycistronic RNA harboring a tRNA at its 3′ end (Figure 6A; Meurer et al., 1998). Northern analyses with both psaA- and rps14-specific probes detected a single major RNA species of ∼5.2 kb in wild-type, PNP+/++ and PNP– plants. This transcript appears to be devoid of tRNAfMet, since tRNA-specific probes did not hybridize with this band but instead detected only the mature tRNAfMet (Figure 6D). Notably, no difference in either length or amount of transcript was observed for the psaA/psaB/rps14 mRNA in all plant lines analyzed. Since the resolution of the northern hybridization might not have allowed the detection of minor differences in length of the psaA/psaB/rps14 transcript, we also used S1 nuclease protection assays to exactly determine the 3′ end of this mRNA. This analysis revealed an identical mature rps14 3′ end encompassing ∼60 nucleotides 3′ UTR in PNP–, PNP++ and wild-type plants (data not shown). The consistently observed PNPase independence of the 3′-end maturation of both 16S rRNA and psaA/psaB/rps14 strongly suggests that 3′ trailers resulting from RNase P cleavage are subject to a distinct maturation pathway.
Fig. 6. Expression analysis of the psaA operon and chloroplast tRNAs. (A) Schematic diagram of the psaA operon and location of the probes used for northern analyses (black bars). (B–D) Results from northern hybridizations of four identical membranes to the probes indicated in (A). The probes were specific for the coding regions of psaA (B), rps14 (C) and tRNAfMet (D, upper), respectively. The faint hybridization signals detected in (B) and (C) do not correspond to monocistronic processing products and most likely represent degradation intermediates. Oligonucleotide probes complementary to nucleotides 35–74 were used for expression analyses of tRNAPro (D, lower). The numbers indicate the sizes of the detected RNA species (in kb).
In contrast to the northern analyses of the psaA/psaB/rps14 transcript, expression analysis of tRNAfMet revealed significant differences in the steady-state level of this RNA, which appeared to correlate with PNPase activity (Figure 6D). Despite the observed PNPase resistance of mature mRNA species, over-expression of AtcpPNPase led to a significant decrease in tRNAfMet amounts (∼70% of the amount in wild-type plants), and this tRNA accumulated to an even higher extent in PNP– plants (∼130% of the amount in wild-type plants). As for the processing of 23S rRNA, an in vivo function of PNPase in tRNA degradation has not been demonstrated so far. To address the potential general relevance of this surprising finding, we performed additional northern analyses to determine the steady-state levels of tRNAPro. Again, we observed the very same PNPase dependence of tRNA accumulation as for tRNAfMet (Figure 6D). Since over-expression of PNPase led to reduced tRNA amounts and depletion of PNPase activity led to enhanced tRNA accumulation, we conclude that AtcpPNPase functions as a genuine tRNA degrading enzyme in vivo. Taken together, our findings extend the function of PNPase towards all major classes of plastid RNAs.
PNPase activity and the extent of plastid mRNA polyadenylation are inversely correlated
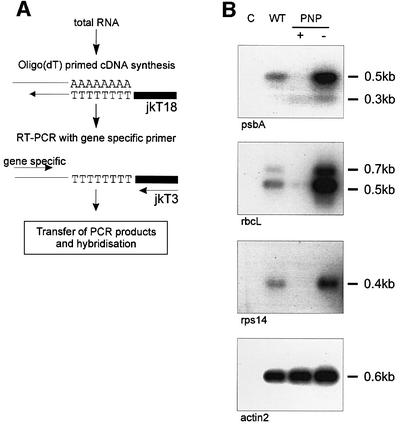
Recent studies have identified polyadenylation of bacterial and plant organellar mRNAs as a mechanism effectively triggering the 3′→5′ degradation of otherwise stable transcripts (Sarkar, 1997; Hayes et al., 1999). The low abundance of polyadenylated mRNAs in the steady-state RNA population of wild-type cells has hampered studies on RNA polyadenylation in chloroplasts. Research on bacterial polyadenylation has circumvented this obstacle by the use of appropriate exonuclease mutants (Sarkar, 1997; Coburn and Mackie, 1999). Likewise, the transgenic lines generated during this study provide a suitable experimental system to address fundamental aspects of polyadenylation in chloroplasts in vivo. We therefore pursued a PCR-based approach to explore the correlation of PNPase activity and polyadenylation and the enzymatic basis for mRNA polyadenylation. To this end, RNA from wild-type, PNPase over-expressing and PNPase co-suppressed plants was reverse transcribed using an oligo(dT)18 primer with an anchor sequence. Subsequent semi-quantitative RT–PCR analysis followed the procedure described for the initial detection of polyadenylated chloroplast mRNAs (Kudla et al., 1996). The amount of PCR products in each reaction was analyzed by DNA blotting and hybridization to a gene-specific probe (Figure 7A). We initially analyzed polyadenylation of the psbA transcript. As shown in Figure 7B, polyadenyl ated psbA mRNA is detectable in wild-type plants after 20 cycles of amplification. In contrast, RT–PCR analyses of exonuclease-deficient PNP– plants revealed a much higher amount of polyadenylated transcripts. In these lines, strong psbA-specific signals already became detectable after 20 cycles of amplification. Intriguingly, this approach detected hardly any polyadenylated psbA mRNAs in exonuclease over-expressing PNP+ plants in these analyses. To confirm that these observations were not limited to the psbA mRNA, we next analyzed polyadenylation of rbcL and rps14 transcripts. As shown in Figure 7B, these analyses revealed similar results, indicating reduced polyadenylation in PNP+ lines and enhanced mRNA polyadenylation in exonuclease-deficient PNP– lines. Taken together, these data strongly suggest that polyadenylated mRNAs provide the preferred substrates for PNPase-mediated degradation. Moreover, the fact that polyadenylated transcripts accumulate to even higher amounts in PNP– plants is suggestive of a function of PNPase as poly(A) mRNA 3′→5′ degrading exonuclease in vivo. Finally, the observed enhanced accumulation of polyadenylated transcripts in PNPase-deficient plants strongly suggests the function of an enzyme distinct from PNPase as the major poly(A) polymerase in higher plant chloroplasts.
Fig. 7. PNPase activity and the extent of chloroplast mRNA polyadenyl ation are inversely correlated. (A) Scheme of the amplification and detection of polyadenylated mRNAs. (B) Southern blot analyses of RT–PCR products containing poly(A) sequences after 20 cycles of amplification. Equal amounts of RNA from wild-type (WT), PNPase over-expressing (+) and PNPase co-suppressed (–) plants were used for cDNA synthesis. A primer pair specifically amplifying the nuclear- encoded and constitutively expressed actin2 transcript was used as a control. C, RNA without reverse transcription.
Discussion
Previous studies had identified a 100RNP from spinach chloroplasts as a plant counterpart to the bacterial exoribonuclease PNPase (Hayes et al., 1996). Subse quent in vitro studies of this enzyme suggested a function in 3′-end maturation of mRNAs and in polyadenylation-mediated mRNA decay (Kudla et al., 1996; Lisitsky et al., 1997). The work described here provides the first comprehensive in vivo analyses of PNPase function in processing and degradation of different classes of RNAs in chloroplasts. In the course of this work, we have shown that PNPase activity in chloroplasts is required for efficient mRNA 3′-end processing of mRNAs but is insufficient to mediate mRNA degradation in vivo.
Northern analyses of psbA and rbcL transcripts revealed that an increase in chloroplast PNPase does not result in an accelerated decay of mature mRNAs. Previous in vitro studies suggested that the stem–loop structure located in the 3′ UTR is able to impede but not prevent 3′ exoribonucleolytic degradation by PNPase (Hayes et al., 1996). Our findings that the amount of mature mRNAs does not decrease in PNPase over-expressing plants indicate that this RNA secondary structure exhibits a very effective protective function against exonucleolytic digestion by PNPase in vivo. These results also suggest that, in vivo, PNPase activity is not sufficient to mediate the decay of mature mRNAs and that other degradation-initiating events, like polyadenylation and/or endoribo nucleolytic cleavage upstream of the stem–loop structure, are irreplaceable and rate limiting. An additional possible cause for the observed differences between in vivo and in vitro studies could be the complexation of chloroplast transcripts with certain RNA-binding proteins (RNPs) in vivo but not in in vitro analyses of synthetic transcripts. Consequently, these RNPs could only function in planta as impeding factors for AtcpPNPase (Hayes et al., 1999). Such RNA–protein interactions could also provide the explanation for another discrepancy with the results of previous in vitro studies that were suggestive of a two-step mechanism generating the mature 3′ end of mRNAs. In this model, 3′-end processing would be initiated by an endonucleolytic cleavage ∼15 nucleotides downstream of the stem–loop structure and completed through a PNPase-mediated further exonucleolytic trimming towards this secondary structure (Hayes et al., 1996). Our demon stration that PNPase-deficient plants accumulate large amounts of 3′ unprocessed RNAs, which are extended by 200–300 nucleotides at their 3′ terminus, questions the relevance of this model for the situation in vivo. Again, a likely explanation for the observed difference is the use of ‘naked’ RNA in in vitro studies, which could expose potential cleavage sites in these assays that we effectively masked by proteins in vivo. In any case, our findings described here significantly extend our understanding of 3′-end processing in chloroplasts and underline the importance of in vivo studies. Another interesting outcome of our investigations is that, despite the reduced accumulation of mature rbcL and psbA mRNAs, the total (mature plus unprocessed) amount of transcripts did not change significantly. Studies of exonuclease-deficient mutants from E.coli revealed that the loss of PNPase function could be compensated by the remaining hydrolytic RNase II activity and that only pnp/rnb double mutants are not viable (Donovan and Kushner, 1986). A similar scenario involving additional chloroplast-localized exoribonuclease(s) might explain our findings. Even though we currently cannot completely rule out a contribution of residual PNPase activity to the mRNA turnover in co-suppressed plants, the function of another exoribonuclease appears to be more likely. In line with this hypothesis, we have detected an additional hydrolytic exoribonuclease activity in Arabidopsis chloroplasts (O.Batistic and J.Kudla, unpublished results).
Most interestingly, our expression studies revealed evidence for PNPase-independent 3′-end maturation of at least the psaA/psaB/rps14 and 16S rRNA transcripts. The 3′ termini of both precursor RNAs most likely result from an RNase P-mediated processing of the tRNAs located downstream. In this regard, it is tempting to speculate that the initial mode of 3′-end precursor generation might determine the subsequent processing events and thus contribute to a ‘channeling’ of specific RNAs into specific maturation pathways.
Despite considerable efforts to understand the processing of 23S rRNA in chloroplasts, the enzymes responsible for generating the mature termini could not be identified. This work provides evidence for an essential function of PNPase in the formation of the 3′ end of 23S rRNA in chloroplasts. In addition, this finding reveals extensive differences to the well characterized 23S rRNA 3′-end maturation in E.coli (Li et al., 1999) and points to an alternative mechanism of rRNA processing in plastids. Previous studies in E.coli uncovered a two-step mechanism consisting of an initial RNase III cleavage 7–9 residues downstream of the mature 23S rRNA 3′ end and subsequent trimming of the remaining single-stranded overhang by RNase T (Li et al., 1999). The chloroplast 23S rRNA precursor transcript harbors an additional 98 residues at its 3′ end and is most likely generated by a yet unknown endoribouclease mediating the release of the 4.5S RNA. Our data demonstrate that the final 3′-end processing of this 98 nucleotide tail of the 23S rRNA precursor is solely accomplished through exonucleolytic trimming by PNPase.
Both the nine nucleotide extended 23S rRNA precursor from E.coli and the 98 nucleotide extended 23S rRNA precursor from chloroplasts are effectively incorporated into ribosomes. Previous studies in E.coli demonstrated a more efficient 3′-end processing of ribosome-integrated 23S rRNA than of free 23S rRNA molecules (Li et al., 1999). Taken together, these data may suggest that, in general, the ribosome association of 23S rRNA precursor molecules, which is accompanied by major changes of RNA tertiary structure, facilitates their final maturation. This, together with our finding that the 3′-end processing of mRNAs also does not represent a prerequisite for their ribosome association, might be suggestive for a not yet unraveled importance of 3′-end maturation for the proper regulation of RNA metabolism in chloroplasts.
Investigations in E.coli have greatly advanced our understanding of bacterial tRNA processing and degradation (Li and Deutscher, 1996; Li et al., 1999; Ow and Kushner, 2002). However, whether or not PNPase participates in these processes in bacteria in vivo remains unclear (Li et al., 1999). tRNA metabolism in chloroplasts is poorly understood. Our findings that PNPase over-expression significantly reduced the steady-state levels of tRNAs and, conversely, PNPase-deficient plants exhibited enhanced accumulation of tRNAs, assign for the first time a direct contribution of PNPase to the regulation of tRNA turnover. This finding is rather surprising, since previous studies predominantly performed in E.coli have established that highly structured RNAs impede exonucleolytic digestion by PNPase (Symmons et al., 2002). Currently, one cannot clearly distinguish whether unique enzymatic properties of the chloroplast PNPase are responsible for this function or whether the concerted action together with other proteins enables the turnover of tRNAs. However, the latter scenario appears to be more likely, since the in vitro characterization of chloroplast PNPase activities and substrate preferences revealed no significant differences when compared with the bacterial enzyme (Hayes et al., 1996; Lisitsky et al., 1997). The fact that the affected organism can tolerate a significant reduction of tRNA amounts might point to an inherent cellular back-up system provided by a relative oversupply of functional tRNAs. Accordingly, similar observations have recently been reported in E.coli (Li and Deutscher, 1996).
Recent studies have established that polyadenylation of RNAs in bacteria and plant organelles renders transcripts susceptible for rapid decay. Several in vitro studies suggested an enhanced affinity of PNPase towards polyadenylated transcripts as an efficient molecular mechanism mediating the selective and rapid degradation of mRNAs marked by poly(A) tails (Hayes et al., 1996; Lisitsky et al., 1997). Although facile biochemical separation of PNPase and poly(A) polymerase activities from spinach chloroplasts had been reported earlier (Lisitsky et al., 1996), recent in vitro studies suggested a role for the spinach PNPase as a bifunctional enzyme mediating both polyadenylation and 3′→5′ degradation (Yehudai-Resheff et al., 2001). Our present investigations have revealed an increased amount of polyadenylated mRNAs in PNPase-deficient plants and, conversely, a strong reduction of polyadenylated transcript in plants over-expressing this exoribonuclease. These data suggest a pivotal role for PNPase in degrading polyadenylated transcripts. Moreover, the fact that polyadenylated transcripts accumulate in PNPase-deficient plants makes it very unlikely that this enzyme fulfills a poly(A) polymerase function in vivo. Instead, we propose that, as in E.coli, a prokaryotic-type poly(A) polymerase conveys this transcript modification in chloroplasts. A comprehensive analysis of candidate genes identified in the Arabidopsis genome is currently under way (J.Kilian and J.Kudla, unpublished results).
A general conclusion emerging from this work is the notion that PNPase is involved in the metabolism of all classes of chloroplast RNAs and therefore plays a much more fundamental role in RNA processing and decay than previously anticipated. The availability of the complete Arabidopsis genome sequence and the rapidly increasing amount of ‘knock out’ mutant lines (including three novel potential loss-of-function alleles for the PNPase; M.Walter and J.Kudla, unpublished results) will facilitate the identification and functional analyses of additional general processing enzymes from chloroplasts in the near future.
Materials and methods
General methods and plant material
Molecular biology techniques were performed using standard protocols (Sambrook and Russell, 2001). Total RNA was isolated using the Trifast solution (Peqlab). A list of primers used in the course of this work can be obtained upon request. UV cross-linking, cDNA synthesis and PCR for detection of polyadenylated transcripts were performed as described previously (Kudla et al., 1996).
Arabidopsis thaliana cv. Columbia plants were cultivated at 21°C on soil in a growth chamber under an 8/16 h light/dark cycle. After growth for 5–6 weeks, leaves were harvested for experimental analysis.
Cloning, sequence analysis, plasmid construction and plant transformation
The partial AtcpPNPase cDNA was amplified by PCR from Arabidopsis cDNA with a degenerated primer pair and cloned into the PCRII TA vector as described by the manufacturer (Invitrogen). Library screens, cDNA sequencing and computational sequence analyses were performed as described previously (Kudla et al., 1999). The AtcpPNPase cDNA sequence was submitted to the DDBJ/EMBL/GenBank databases (accession No. AF450480).
For construction of the plasmid used for plant transformation, the Arabidopsis PNPase cDNA sequence was amplified with Pwo-Polymerase (Peqlab) with primers designed to amplify the complete coding region and to introduce the FLAG tag sequence and appropriate restriction sites for cloning into the target vectors. The amplification products were cloned into the desired plasmids and verified by sequencing of both DNA strands. Over-expression of the recombinant AtcpPNPase was performed with the pET24b plasmid system (Novagen). Plant transformation constructs were based on the vectors pRTΩNot and pGPTV-BAR/AscI (Figure 2). Arabidopsis thaliana plants were transformed by vacuum infiltration using the A.tumefaciens strain GV3101::pMP90. Seeds were planted in soil and selected for BASTA resistance by spraying the seedlings with a 0.1% BASTA solution containing 0.1% Tween-20.
Isolation and analysis of chloroplasts, thylakoid membranes and proteins
Leaves (30–40 g) were used to isolate chloroplasts according to Somerville et al. (1981). To isolate chloroplast proteins, the pellet was resuspended in an equal volume of 2× SDS loading buffer containing 0.1% Triton X-100 and denatured for 5 min at 95°C. Insoluble material was removed by centrifugation for 15 min at 15 000 g.
Thylakoid membranes were isolated according to the protocol of Machold et al. (1979), resuspended in 2× SDS loading buffer and incubated for 30 min at 40°C. Insoluble material was removed by centrifugation.
To isolate total leaf proteins, 100 mg of leaf material were extracted as described previously (Martinez-Garcia et al., 1999). Immunoblotting was performed using standard protocols. For detection, the ECLplus kit (Amersham) was applied.
Isolation of polysomal RNA
Polysomal RNA was isolated from 700 mg of leaf material following the procedure described by Barkan (1993). Fractions were collected according to Ruf et al. (1997). From top to bottom, the volumes of the fractions were 150, 700, 750, 900 and 900 µl. RNA was extracted from all fractions with phenol/chloroform and precipitated with isopropanol after the addition of 10 µg of glycogen. RNA pellets were resuspended in 20 µl of water and samples of 5 µl were subsequently analyzed by northern blot hybridization.
Acknowledgments
Acknowledgements
We thank Dr R.Bock for valuable discussions and critical reading of the manuscript. We also thank Dr K.Harter for advice with the FLAG tag system and Dr R.Bock and Dr M.Hippler for providing antibodies. The initial cloning of a partial AtcpPNPase cDNA was supported by a fellowship to J.Kudla (DFG Ku 931/1-3) as postdoctoral fellow in the laboratory of Dr W.Gruissem at UC Berkeley. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to J.Kudla (DFG Ku 931/2-1, 2-2).
References
- Baginsky S. et al. (2001) Chloroplast PNPase exists as a homo-multimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA, 7, 1464–1475. [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1993) Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell, 5, 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis A.J., Vanzo,N.F. and Raynal,L.C. (1999) mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet., 15, 24–28. [DOI] [PubMed] [Google Scholar]
- Coburn G.A. and Mackie,G.A. (1999) Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol., 62, 55–108. [DOI] [PubMed] [Google Scholar]
- Decker C.J. (1998) The exosome: a versatile RNA processing machine. Curr. Biol., 8, R238–R240. [DOI] [PubMed] [Google Scholar]
- Donovan W.P. and Kushner,S.R. (1986) Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl Acad. Sci. USA, 83, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drager R.G., Higgs,D.C., Kindle,K.L. and Stern,D.B. (1999) 5′ to 3′ exoribonucleolytic activity is a normal component of chloroplast mRNA decay pathways. Plant J., 19, 521–531. [DOI] [PubMed] [Google Scholar]
- Garcia-Mena J., Das,A., Sanchez-Trujillo,A., Portier,C. and Montanez,C. (1999) A novel mutation in the KH domain of polynucleotide phosphorylase affects autoregulation and mRNA decay in Escherichia coli. Mol. Microbiol., 33, 235–248. [DOI] [PubMed] [Google Scholar]
- Hayes R., Kudla,J., Schuster,G., Gabay,L., Maliga,P. and Gruissem,W. (1996) Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J., 15, 1132–1141. [PMC free article] [PubMed] [Google Scholar]
- Hayes R., Kudla,J. and Gruissem,W. (1999) Degrading chloroplast mRNA: the role of polyadenylation. Trends Biochem. Sci., 24, 199–202. [DOI] [PubMed] [Google Scholar]
- Hilleren P., McCarthy,T., Rosbash,M., Parker,R. and Jensen,T.H. (2001) Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature, 413, 538–542. [DOI] [PubMed] [Google Scholar]
- Kudla J., Hayes,R. and Gruissem,W. (1996) Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J., 15, 7137–7146. [PMC free article] [PubMed] [Google Scholar]
- Kudla J., Xu,Q., Harter,K., Gruissem,W. and Luan,S. (1999) Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl Acad. Sci. USA, 96, 4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. and Deutscher,M.P. (1996) Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell, 86, 503–512. [DOI] [PubMed] [Google Scholar]
- Li Z., Pandit,S. and Deutscher,M.P. (1999) Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA, 5, 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K. and Link,G. (1997) Chloroplast endoribonuclease p54 involved in RNA 3′-end processing is regulated by phosphorylation and redox state. Nucleic Acids Res., 25, 2403–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky I., Klaff,P. and Schuster,G. (1996) Addition of destabilizing poly(A)-rich sequences to endonuclease cleavage sites during the degradation of chloroplast mRNA. Proc. Natl Acad. Sci. USA, 93, 13398–13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky I., Kotler,A. and Schuster,G. (1997) The mechanism of preferential degradation of polyadenylated RNA in the chloroplast. The exoribonuclease 100RNP/polynucleotide phosphorylase displays high binding affinity for poly(A) sequence. J. Biol. Chem., 272, 17648–17653. [DOI] [PubMed] [Google Scholar]
- Machold O., Simpson,D.J. and Moller,B.L. (1979) Chlorophyll-proteins of thylakoids from wild type and mutants of barley (Hordeum vulgare L.). Carlsberg Res. Commun., 44, 235–254. [Google Scholar]
- Martinez-Garcia J.F., Monte,E. and Quail,P.H. (1999) A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J., 20, 251–257. [DOI] [PubMed] [Google Scholar]
- Meurer J., Meierhoff,K. and Westhoff,P. (1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta, 198, 385–396. [DOI] [PubMed] [Google Scholar]
- Meurer J., Grevelding,C., Westhoff,P. and Reiss,B. (1998) The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol. Gen. Genet., 258, 342–351. [DOI] [PubMed] [Google Scholar]
- Mitchell P. and Tollervey,D. (2000) Musing on the structural organization of the exosome complex. Nat. Struct. Biol., 7, 843–846. [DOI] [PubMed] [Google Scholar]
- Ow M.C. and Kushner,S.R. (2002) Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev., 16, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier P. and Arraiano,C.M. (2000) Degradation of mRNA in bacteria: emergence of ubiquitous features. BioEssays, 22, 235–244. [DOI] [PubMed] [Google Scholar]
- Rochaix J.D. (1996) Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol. Biol., 32, 327–341. [DOI] [PubMed] [Google Scholar]
- Rott R., Levy,H., Drager,R.G., Stern,D.B. and Schuster,G. (1998) 3′-processed mRNA is preferentially translated in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol., 18, 4605–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S., Kössel,H. and Bock,R. (1997) Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol., 139, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J. and Russell,D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sarkar N. (1997) Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem., 66, 173–197. [DOI] [PubMed] [Google Scholar]
- Somerville C.R., Somerville,S.C. and Ogren,W.L. (1981) Isolation of photosynthetically active protoplasts and chloroplasts from Arabidopsis thaliana. Plant Sci. Lett., 21, 89–96. [Google Scholar]
- Symmons M.F., Williams,M.G., Luisi,B.F., Jones,G.H. and Carpousis,A.J. (2002) Running rings around RNA: a superfamily of phosphate-dependent RNases. Trends Biochem. Sci., 27, 11–18. [DOI] [PubMed] [Google Scholar]
- van Hoof A. and Parker,R. (1999) The exosome: a proteasome for RNA? Cell, 99, 347–350. [DOI] [PubMed] [Google Scholar]
- Yehudai-Resheff S., Hirsh,M. and Schuster,G. (2001) Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol., 21, 5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]