Abstract
We recently demonstrated that polysome-associated mRNAs that co-isolate with mitochondria encode a subset of mitochondrial proteins, and that the 3′ UTRs of these transcripts are essential for their localization to the vicinity of the organelle. To address the question of the involvement of the mRNA targeting process in mitochondrial biogenesis, we studied the role of ATP2 3′ UTR. An altered ATP2 allele in which the 3′ UTR was replaced by the ADH1 3′ UTR exhibits properties supporting the importance of mRNA localization to the vicinity of mitochondria: (i) the mutated strain presents a respiratory dysfunction; (ii) mito chondrial import of the protein translated from the altered gene is strongly reduced, even though the precursor is addressed to the organelle surface; (iii) systematic deletions of ATP2 3′ UTR revealed a 100 nucleotide element presenting RNA targeting properties. Additionally, when the ATM1 3′ UTR was replaced by the ADH1 3′ UTR, we obtained cells in which ATM1 mRNA is also delocalized, and presenting a respiratory dysfunction. This demonstrates that mRNA localization to the vicinity of mitochondria plays a critical role in organelle biogenesis.
Keywords: co-translational import/mRNA mitochondrial targeting/mitochondria/respiratory deficiency/3′ UTR
Introduction
The biogenesis of mitochondria is a complex cellular process that involves the concerted expression of both nuclear and mitochondrial genomes. It is assumed that mitochondria contain ∼15–20% of cellular proteins (Kumar et al., 2002) and must import several hundreds of polypeptides encoded by the nuclear DNA (Pfanner and Geissler, 2001). Mitochondrial function is essential for the life of mammalian cells, as has been well documented for over 15 years in man (Wallace, 1999). The correct sorting of mitochondrial proteins is the first step to ensure organelle functionality. A classic mechanism for segregating mitochondrial proteins relies on targeting sequences that are either cleavable extensions or within the mature protein. The signal sequences are decoded by a dynamic, multisubunit transport machinery, the TOM and TIM complexes, which directs them to their correct destination within one of the four mitochondrial subcompartments: the outer membrane, the intermembrane space, the inner membrane and the matrix (Pfanner and Geissler, 2001). Alternatively, mounting evidence suggests that the localization of mRNAs to the vicinity of mitochondria is another mechanism employed by cells for mitochondrial protein sorting. In this case, a co-translational phase might assist the import of some precursors (Corral-Debrinski et al., 1999, 2000; Fünfschilling and Rospert, 1999; Gratzer et al., 2000; George et al., 2002). Using yeast DNA microarrays to analyze the mRNA populations associated with free and mitochondrion-bound polysomes, we recently demonstrated that >100 mRNAs encoding mitochondrial proteins localize to mitochondrion-bound polysomes. These mRNAs have 3′ untranslated regions (3′ UTRs) conferring mitochondrion-targeting properties (Marc et al., 2002). We then decided to determine whether the mitochondrial localization of one of these mRNAs is essential for the function of the corresponding protein. ATP2 mRNA localizes exclusively to mitochondrion-bound polysomes, and encodes the β-subunit of the F1-ATP synthase or respiratory chain complex V. The use of an RNA-labeling system to visualize RNA localization in living cells (Beach et al., 1999; Corral-Debrinski et al., 2000) allowed us to demonstrate that the 3′ UTR of ATP2 contains sufficient information to address a reporter RNA to the vicinity of mitochondria. The replacement in vivo, by homologous recombination, of ATP2 3′ UTR with the 3′ UTR of ADH1, a gene encoding a cytoplasmic protein, leads to a respiratory-deficient strain. In these cells, not only is the altered ATP2 mRNA enriched in free cytoplasmic polysomes, but also the Atp2 protein is accumulated as a precursor poorly translocated into the organelle, even though its reaches the outer mitochondrial membrane. The significant decrease in the amount of mature Atp2p inside the mitochondria is responsible for the respiratory deficiency observed, as complementation with a wild-type form of ATP2 completely restores cell respiration. Therefore, ATP2 mRNA delivery to the vicinity of mitochondria is essential for the functionality of the respiratory chain.
In an attempt to generalize the functional importance of 3′ UTRs, we replaced the 3′ UTR of ATM1, a gene coding for an ABC transporter of the inner mitochondrial membrane (Leighton and Schatz, 1995), by the ADH1 3′ UTR and we obtained a cell respiration impairment. In these cells, the altered ATM1 mRNA is over-represented in free cytoplasmic polysomes and the ability to grow on glycerol is restored by transformation with a wild-type ATM1 gene. Thus, the absence of ATM1 or ATP2 3′ UTRs leads to an abnormal cellular distribution of the corresponding transcripts associated with a respiratory deficiency, confirming that mRNA sorting to the vicinity of mitochondria represents a key step to ensure organelle function.
Results
Either the mts or the 3′ UTR of ATP2 is able to address a reporter RNA to the vicinity of mitochondria in living yeast cells
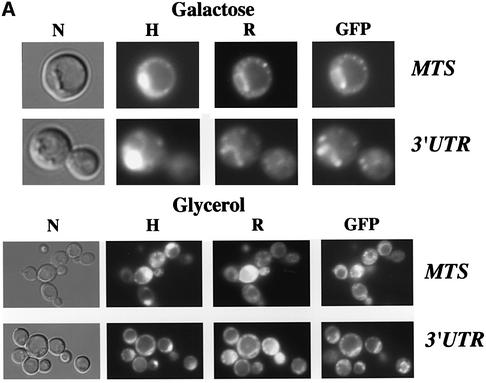
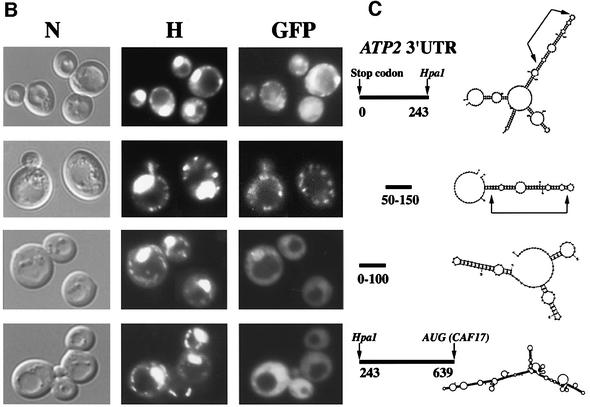
The use of a whole-genome approach to identify mRNAs localized to the vicinity of mitochondria allowed a MLR (mitochondrial localization of mRNA) value of 98 out of 100 to be attributed to the ATP2 transcript, meaning that it localizes to mitochondrion-bound polysomes (Marc et al., 2002). We then decided to visualize RNA in living cells (Beach et al., 1999; Corral-Debrinski et al., 2000) to examine two sequences within ATP2 as possible regulators of RNA sorting: the mitochondrial targeting sequence (mts) and the 3′ UTR. The nucleotide region corresponding to the first 35 amino acids of Atp2p was fused to two CP (coat protein from the MS2 bacteriophage) binding sites in the pIIIA/MS2-2 plasmid, which will direct the synthesis of the reporter RNA. This RNA can be visualized by fluorescence microscopy when cells express the CP–GFP fusion protein. It has been well described in yeast that mitochondria consist of a branched tubular network, the continuity of which depends on a balanced frequency of fusion and fission events (Egner et al., 2002); furthermore, the size and the shape of the mitochondrial compartment depend on the carbon source (Stevens, 1977; Miyakawa et al., 1984). When cells were grown on galactose, we observed small discrete fluorescent spots, which represent mitochondria, as visualized with rhodamine B, a well-characterized mitochondrial probe (Johnson et al., 1980; Chen, 1989). Additionally, reporter RNA localized to mitochondrial nucleoids observed with the Hoechst reagent (Figure 1A). When cells were grown on the non-fermentable source glycerol, the reporter RNA was visualized in branched and more elaborated networks also stained by rhodamine B and Hoechst reagents. Therefore, the region coding for the mts of Atp2 is sufficient to address an RNA to the periphery of mitochondria, as we showed previously for the ATM1 mts (Corral-Debrinski et al., 2000). A region of 639 nucleotides (nt) between the ATP2 stop codon and the CAF17 AUG (CAF17 is contiguous to ATP2 in chromosome X) was inserted in the pIIIA/MS2-2 plasmid. The cellular distribution of this RNA was followed in cells expressing the CP–GFP fusion protein (Figure 1A). Hybrid RNA in these cells was present as small discrete fluorescent spots, which remarkably co-localized with Hoechst staining, representing mitochondrial nucleoids (Miyakawa et al., 1984) When cells were grown on glycerol, we observed the reporter RNA in the typical reticulum structure of mitochondria as visualized with rhodamine B (Figure 1A). When either the mts or the 3′ UTR was placed in the opposite orientation such that the non-coding sequence was transcribed, the reporter RNAs were diffuse in their distribution throughout the cytoplasm (not shown). Thus, information within the mts or the 3′ UTR of ATP2 is sufficient to promote export of the RNA from the nucleus and targeting to the periphery of mitochondria. To map more precisely cis-acting elements within the 639 nt of ATP2 3′ UTR, the distribution of various deletion RNAs was analyzed. Three consensus polyadenylation signals (AAUAAA) are present in this region: the first is situated 5 nt downstream of the stop codon, the second 239 nt farther and the third 302 nt downstream of the stop codon. The hybrid RNA containing the last 396 nt did not localize to the vicinity of mitochondria, and a diffuse staining in the cytoplasm was consistently observed, while the first 243 nt of the 3′ UTR are as efficient as the complete 3′ UTR, allowing the reporter RNA to produce a punctate distribution of fluorescent speckles that localize to mitochondrial structures (Figure 1B). The MFOLD program for RNA secondary structure (Zuker, 1989) predicted that the 250 nucleotide element could fold into a long stem–loop structure formed by four stems separated by asymmetric bulges (Figure 1C). To define further the minimal structural motif in this stem–loop, we examined the targeting properties of the region encompassing the first 150 nt. A reporter RNA with the region 50–150, in which the stem–loop structure was also predicted, presents the proper localization in close contact with mitochondria (Figure 1B). However, the reporter RNA encompassing the 1–100 region showed a diffuse cytoplasmic staining, suggesting that the reporter RNA does not localize to the vicinity of mitochondria (Figure 1B). Interestingly, the folding of this fragment does not predict any particular stable stem–loop structure (Figure 1C). These results demonstrate that specific regions within ATP2 3′ UTR play a crucial role in the mRNA targeting process.
Fig. 1. Imaging of fluorescent RNA in living yeast cells. The RNA-labeling system (Beach et al., 1999) was used to determine the addressing information included in both the 3′ UTR of the ATP2 gene and the sequence encoding the first 35 amino acids of Atp2p representing its mitochondrial targeting sequence (MTS). Co-expression of CP–GFP plasmid and reporter RNAs leads to formation of a GFP-labeled RNA, which was visualized using fluorescence microscopy techniques. (A) The ATP2 3′ UTR of 639 bp long or the sequence corresponding to Atp2p’s mts of 105 bp long were cloned in the pIIIA/MS2-2 plasmid. Cells had grown either in 2% galactose or 2% glycerol medium and were visualized at early log phase. GFP indicates the green RNA labeling, H the Hoechst staining and R the rhodamine B labeling; cells were also photographed with Nomarski optics (N). (B) Several lengths of the ATP2 3′ UTR were inserted into the pIIIA/MS2-2 plasmid. Cells expressing each reporter RNA and the CP–GFP protein were grown in 2% galactose medium and visualized after Hoechst staining (H). Green RNA labeling is indicated (GFP) and cells were also photographed with Nomarski optics (N). (C) The MFOLD program for RNA secondary structure (Zuker, 1989) was applied to predict the stem–loop structures that can be formed in the different ATP2-3′UTR fragments tested in the RNA-labeling system. The arrows indicate the stable 70-nt-long stem–loop structure.
In vivo substitution of the ATP2 3′ UTR by the ADH1 3′ UTR leads to respiratory-deficient cells associated with an abnormal localization of the ATP2 mRNA
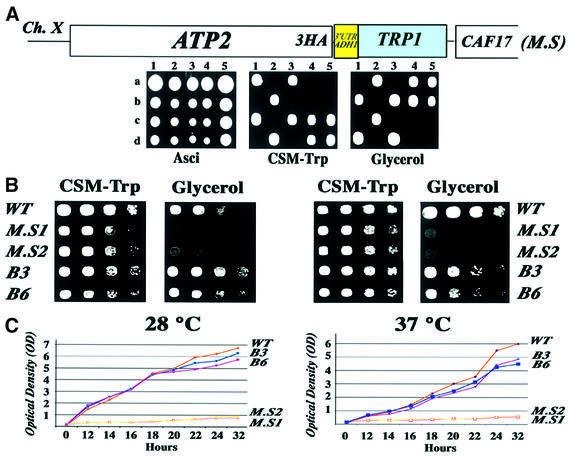
The localization of ATP2 mRNA to the vicinity of mitochondria seems to be dependent on at least two regions within the transcript: the mts and the 3′ UTR, as we have previously reported for ATM1 (Corral-Debrinski et al., 2000). We can predict that the in vivo modification of either of these elements could alter the final destination of the corresponding transcript. The 3′ UTR of ATP2 possesses the same targeting properties as the ATM1 3′ UTR (Figure 1) and is certainly involved in the localization of ATP2 mRNA to the vicinity of the mitochondria (Corral-Debrinski et al., 2000; Marc et al., 2002). In order to assess the biological role of the ATP2 mRNA targeting process, we modified the corresponding chromosomal allele by homologous recombination. Heterozygous diploids of the BMA64 strain were transformed with a PCR product of the pFA6a-3HA-TRP1 plasmid (Longtine et al., 1998), which was designed to recombine within the 3′ region of the chromosomal ATP2 allele. After dissection of each tetrad, two TRP+ spores were obtained, and were called ATP2-3′UTRADH1 cells. In these haploid cells, the C-terminus of the ATP2 ORF is fused to three tandem repeats of the influenza virus hemagglutin epitope (3HA), followed by the 3′ UTR of ADH1, a gene coding for a cytoplasmic protein (Figure 2A). These cells are unable to form colonies on media containing the non-fermentable carbon source glycerol at both 28 and 37°C. They also have a reduced growth rate in minimum synthetic medium (Figure 2B). The ability to grow on liquid glycerol medium was measured in wild-type and ATP2-3′UTRADH1 cells. As shown in Figure 2C, the growth rate of the mutated strains, MS1 and MS2, is dramatically reduced at both 28 and 37°C as compared with wild-type cells (WT). To exclude the possibility that the respiratory-deficient phenotype is due to the addition of the three HA epitopes at the C-terminus of Atp2p, we obtained BMA64 haploid cells, which express a HA-tagged Atp2 protein associated to the ATP2 3′ UTR. These cells, called B3 and B6, are able to grow on glycerol at both 28 and 37°C (Figure 2B and C); thus, the chromosomal ATP2 allele with a C-terminal triple HA tag functions like the wild-type allele. This result indicates that the respiratory chain deficiency of the ATP2-3′UTRADH1 strain is due to the absence of the ATP2 3′ UTR.
Fig. 2. In vivo substitution of the ATP2 3′ UTR by the ADH1 3′ UTR leads to a respiratory chain dysfunction. (A) The pFA6a-3HA1-TRP1 plasmid (Longtine et al., 1998) was used to obtain a modified strain (MS) from the diploid BMA64 strain called ATP2-3′UTRADH1 in which the C-terminus of ATP2 was fused to three tandem repeats of the HA epitope followed by the 3′ UTR of the ADH1 gene, encoding a cytoplasmic protein. After dissection of tetrads, each TRP+ spore was tested for its ability to grow on medium with the non-fermentable carbon source glycerol. (B) The ability to grow on glycerol plates was examined for two independent haploid TRP+ cells (M.S1, M.S2), for cells in which ATP2 locus was modified to obtain the synthesis of a protein with three tamdem repeats of the HA epitope while conserving its own 3′ UTR (B3, B6), and for wild-type BMA64 cells transformed with the pFL45 plasmid, allowing them to grow in the absence of tryptophan (WT). Cells were grown on complete synthetic medium containing 2% glucose and devoid of tryptophan to an OD of 2. They were serially diluted (1:5) and spotted either on complete synthetic medium devoid of tryptophan (CSM-Trp) or on 2% glycerol medium (Glycerol). The plates were then incubated for 2 days at both 28 and 37°C. (C) The growth rate of MS1 and MS2 cells was measured on liquid glycerol medium and compared with wild-type cells (WT) and with cells expressing the HA-tagged version of Atp2p (B3 and B6) either at 28 or 37°C. Cells were grown overnight on complete synthetic medium devoid of tryptophan and containing 2% glucose. The quantity of cells corresponding to an OD of 0.2 was diluted in 40 ml of glycerol medium. OD measurements were performed every 2 h.
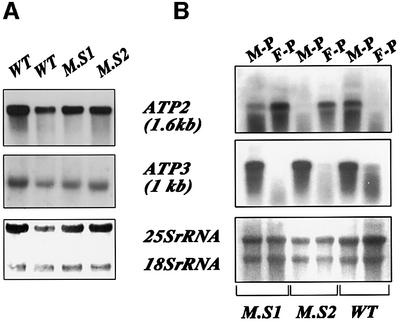
To determine whether this respiration dysfunction is the consequence of an abnormal localization of ATP2-3′UTRADH1 mRNA, fractionation experiments were performed to obtain free and mitochondrion-bound polysomes from wild-type and ATP2-3′UTRADH1 cells. RNAs were purified and subjected to northern blot analysis (Figure 3B). As expected, in wild-type cells, ATP2 and ATP3 transcripts (Marc et al., 2002) were remarkably enriched in mitochondrion-bound polysomes (Figure 3B, M-P). In ATP2-3′UTRADH1 cells, while ATP3 mRNA was enriched in mitochondrion-bound polysomes, the distribution of ATP2-3′UTRADH1 transcript was dramatically affected. Indeed, the mRNA predominantly localized to free cytoplasmic polysomes (Figure 3B, F-P), and very low amount of the overall ATP2 mRNA level was detected in mitochondrion-bound polysomes. The steady-state levels of ATP2 mRNA were similar in all the cells examined, as shown in northern blot experiments performed with total cellular RNA preparations (Figure 3A), indicating that the respiratory deficiency of the ATP2-3′UTRADH1 strain is not due to the instability of the RNA transcribed from the altered ATP2 allele. Therefore, the presence of ATP2 3′UTR is required for the accurate subcellular localization of the transcript in vivo.
Fig. 3. ATP2-3′UTRADH1 mRNA steady-state levels and subcellular localization in the ATP2-3′UTRADH1 strain. (A) Total RNAs were purified from ATP2-3′UTRADH1 cells (M.S1 and M.S2) and wild-type cells (WT), and subjected to northern blot analysis using successively ATP2 and ATP3 ORFs as radiolabeled probes. (B) Mitochondrion-bound polysomes (M-P) and free cytoplasmic polysomes (F-P) were purified from ATP2-3′UTRADH1 cells (M.S1 and M.S2) and wild-type cells (WT). Eight micrograms of RNA extracted from each polysomal population were separated on formaldehyde–agarose gels, subjected to northern blot analysis and hybridized successively with ATP2 and ATP3 probes. Autoradiograms shown in (A) and (B) represent an exposure time of 6 h at –80°C with Amersham intensifying screens. Methylene blue staining of the filters prior to hybridization is shown at the bottom.
In the ATP2-3′UTRADH1 strain, Atp2 precursor is not efficiently translocated into the mitochondria
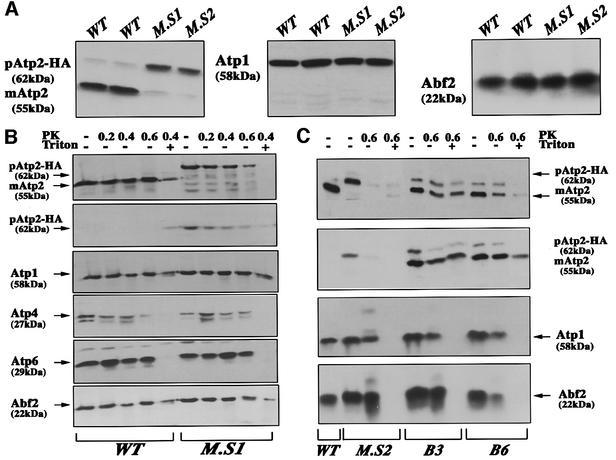
To determine whether the Atp2 protein is produced at normal levels in the ATP2-3′UTRADH1 strain, we performed immunoblotting analyses using mitochondrial purifications. Two independent clones were examined; all the cells tested produced similar amounts of Atp2p. In ATP2-3′UTRADH1 cells (M.S1 and M.S2), very little of the mature form of the protein was detected: the majority of the fusion protein was present as the full-length precursor form of ∼62 kDa. In contrast, in wild-type cells (WT) almost all the Atp2p detected had an apparent mol. wt of ∼55 kDa, which corresponds to the mature form, in which the first 34 amino acids were cleaved (Bedwell et al., 1987) (Figure 4A). These data indicate that in the ATP2-3′UTRADH1 strain (M.S1), the mitochondrial import of Atp2p was impaired. To confirm the defect in Atp2p mitochondrial translocation, purified mitochondria from wild-type and ATP2-3′UTRADH1 cells were treated with proteinase K (PK) and subjected to western blot analyses (Figure 4B). The results for four independent experiments unambiguously showed that the mitochondrial import of Atp2p was significantly reduced in the ATP2-3′UTRADH1 strain. Nearly all the Atp2 protein signal in these cells was the full-length precursor form of ∼62 kDa. This protein was also detected with a specific anti-HA antibody, indicating that it is synthesized from the altered ATP2 allele (Figure 4B). Furthermore, we observed that in mitochondria purified from ATP2-3′UTRADH1 cells the Atp2 precursor was sensitive to proteolysis by PK, while in wild-type cells the majority of Atp2p was less accessible to proteolysis. Indeed, when PK concentrations used were >0.4 mg/ml, the protein produced from the altered ATP2 allele was almost entirely digested, whereas a significant amount of Atp2p produced in wild-type cells remained protease insensitive (Figure 4B), confirming that the majority of Atp2p produced from the altered allele is blocked on the surface of mitochondria. To examine the levels of other complex V proteins in the ATP2-3′UTRADH1 strain, western blots were performed using anti-Atp1p, anti-Atp4p and anti-Atp6p antibodies. The overall amounts of these proteins were not changed in the ATP2-3′UTRADH1 strain, suggesting that their mitochondrial import was not affected (Figure 4B). This also holds true for Abf2p, a mitochondrial DNA binding protein, the level of which was not affected in the mutated strain (Figure 4B). Additionally, we examined mitochondrial preparations of cells containing the chromosomal ATP2 allele with a C-terminal triple HA tag and the ATP2 3′UTR (B3 and B6). The fusion Atp2p is successfully translocated inside the mitochondria. Both the precursor and the mature form of the fusion protein were detected at the expected molecular weights when anti-Atp2p or anti-HA antibodies were used (Figure 4C), thus confirming that the addition of three HA epitopes at the C-terminus of Atp2p did not affect protein mitochondrial import, cleavage of the mts and assembly of the mature protein into complex V. This result is in agreement with the ability of these cells to grow on glycerol medium (Figure 2B and C). We can envisage that the modified form of ATP2 mRNA, which mainly localizes to free cytoplasmic polysomes, leads to the synthesis of a protein which, although recognized by the mitochondrial import machinery, is not successfully translocated into the organelle.
Fig. 4. Amount of Atp2 protein and localization in the ATP2-3′UTRADH1 strain. (A) Mitochondria were prepared from wild-type cells (WT) as from two independent clones of the ATP2-3′UTRADH1 strain (M.S1 and M.S2). SDS–PAGE was performed with 30 µg of proteins and analyzed by western blotting using antibodies against Atp2, Atp1 and Abf2 proteins.The precursor of Atp2p migrated at an approximate mol. wt of 62 kDa, while the mature protein migrated at ∼55 kDa. (B) To determine the precise cellular localization of Atp2p, mitochondria were treated for 30 min at 0°C with PK at 0.2, 0.4 or 0.6 mg/ml (PK); the PK digestion at 0.4 mg/ml was also performed in combination with 1% Triton X-100 (Triton). The following antibodies were used: polyclonal antibodies against Atp2p, Atp1p, Atp4p, Atp6p, Abf2p and monoclonal antibody 12CA5 against the HA epitope (second from the top). The precursor of Atp2p migrated at an approximate mol. wt of 62 kDa, while the mature protein migrated at ∼55 kDa. In wild-type mitochondria, no signal was detected with the 12CA5 antibody, thus confirming the identity of the revealed 62 kDa polypeptide with the protein expressed from the altered ATP2 allele. Moreover, the HA-tagged version of the Atp2 precursor was almost entirely digested by a 0.6 mg/ml concentration of PK, while the mature form of the protein in wild-type cells was more protected against digestion. These data indicate that Atp2 protein synthesized from the altered allele is present at the mitochondrial surface, but its mitochondrial import is not efficient. (C) To determine the cellular localization of a HA-tagged version of Atp2, mitochondria were purified from B3 and B6 cells and subjected to western blotting after digestion with 0.6 mg/ml PK in the presence or absence of 1% Triton X-100. As controls, CW04 strain (WT) and ATP2-3′UTRADH1 strain (MS2) were used. Antibodies against Atp2, Atp1 and Abf2 proteins and against the HA epitope (second from the top) were used successively. Even though we were able to detect more of the HA-tagged Atp2p precursor in B3 and B6 cells than in wild-type cells, the amount of the mature Atp2p insensitive to externally added protease in B3 and B6 cells was significantly increased as compared with that observed in the MS2 strain, thus confirming that the impairment of Atp2p mitochondrial import observed in the ATP2-3′UTRADH1 strain is not due to the presence of the three HA epitopes at the C-terminus of the protein.
The expression of a wild-type version of ATP2 rescues the respiratory deficiency of the ATP2-3′UTRADH1 strain
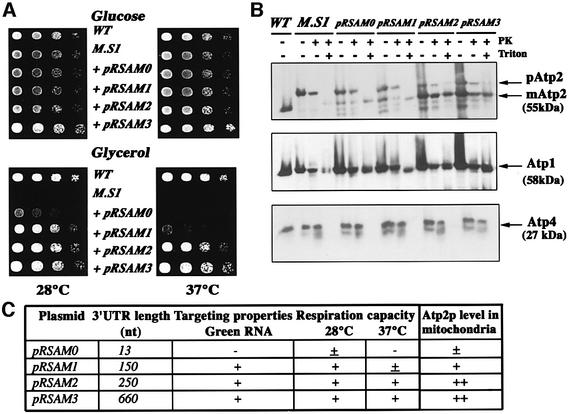
The above results establish a link between the respiratory deficiency and the decrease in Atp2p levels in the inner mitochondrial membrane of ATP2-3′UTRADH1 cells. To fully confirm the role of ATP2 mRNA localization in respiratory chain dysfunction, we transformed the modified strain with several low-copy-number plasmids containing the complete ATP2 ORF and different lengths of the ATP2 3′ UTR. A complete rescue of the respiratory capacity was obtained when ATP2-3′UTRADH1 cells were transformed with the pRSAM3 plasmid, which contains the full-length ATP2 3′ UTR. These cells are able to grow on glycerol medium at both 28 and 37°C as efficiently as wild-type cells (Figure 5A). The expression of pRSAM0 in the mutated strain led to a very low rate of growth on glycerol medium at 28°C, and these cells were unable to grow at 37°C. In this plasmid, the stop codon of Atp2p is followed by only 13 nt (Figure 5A). When the expression of ATP2 is directed from pRSAM1, which contains 150 nt of the ATP2 3′ UTR, cell growth on glycerol was improved at 28°C; however, the cell growth rate remained low at 37°C. The pRSAM2 encompassing 250 nt of the ATP2 3′ UTR allowed a full rescue of cell respiration; cells expressing ATP2 from this plasmid were able to respire at both 28 and 37°C, and their growth rate was identical to that observed in both wild-type cells and cells transformed with pRSAM3 (Figure 5A).
Fig. 5. Complementation experiments of the ATP2-3′UTRADH1 strain. (A) The ATP2-3′UTRADH1 strain (M.S1) was transformed with an array of low-copy-number plasmids expressing the ATP2 ORF in combination with different lengths of the ATP2 3′ UTR (pRSAM0-pRSAM3); as a control, the ATP2-3′UTRADH1 strain transformed with the empty pRS416 vector was used (M.S1). The ability of the transformed cells to grow on glycerol plates was determined at both 28 and 37°C. Cells were grown on complete synthetic medium containing 2% glucose and devoid of tryptophan to an OD of 2. They were serially diluted (1:5) and spotted either on complete synthetic medium devoid of tryptophan and containing 2% glucose (Glucose) or on 2% glycerol medium (Glycerol). The plates were then incubated for 2 days. Wild-type cells transformed with pRS416 and pFL45 (WT) were used as a positive control, since they are able to grow on glycerol and in the absence of tryptophan and uracile. (B) To compare the amount of Atp2p produced from each pRSAM plasmid, mitochondria from wild-type cells (WT) and transformed ATP2-3′UTRADH1 cells (M.S1) were analyzed by western blotting using antibodies against Atp2, Atp1 and Atp4 proteins. The precursor of Atp2p migrated at an approximate mol. wt of 62 kDa, while the mature protein migrated at the expected size of ∼55 kDa. (C) The table summarizes the size of the 3′ UTR in each tested plasmid, their ability to rescue cell respiration and the targeting properties determined in Figure 1. A clear correlation can be drawn between the length of the 3′ UTR, the level of the mature protein inside mitochondria (Figure 5B), the phenotypic complementation and the ability of the sequence to target a reporter RNA to the vicinity of mitochondria.
To determine the amount and the localization of Atp2p synthesized from the different plasmids, mitochondria were purified from each transformed strain. Figure 5B shows that the full-length precursor of Atp2p was produced at similar levels in all the cells examined, a majority of the signal might correspond to the protein synthesized from the altered ATP2 allele. The differences detected concerned the amounts of Atp2p mature form; indeed, very low levels of the protein were detected in cells transformed with pRSAM0. Similar amounts of the mature form of Atp2p were detected in wild-type cells and in cells expressing pRSAM2 or pRSAM3. Interestingly, the level of the protein was ∼4-fold lower in cells expressing pRSAM1 than in cells expressing pRSAM2 or pRSAM3, which might explain the reduced respiratory capacity of these cells at 37°C (Figure 5A). These data suggest that ATP2 mRNAs transcribed from pRSAM2 and pRSAM3 plasmids were enriched in mitochondrion-bound polysomes. This specific targeting is followed by the complete mitochondrial translocation of Atp2p, which leads to the recovery of respiratory chain function.
In vivo substitution of the ATM1 3′ UTR by the ADH1 3′ UTR also leads to respiratory-deficient cells due to an abnormal cellular localization of the ATM1 mRNA
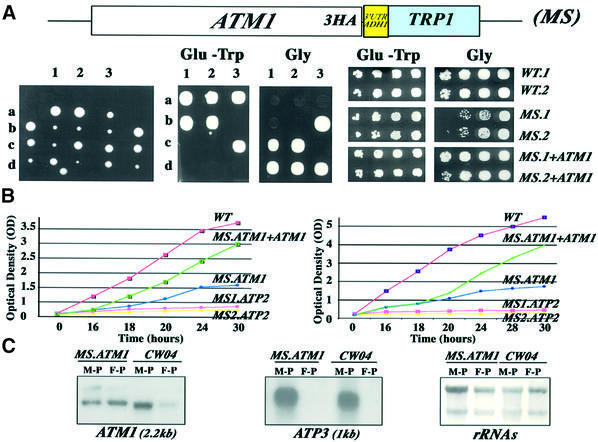
To generalize the biological importance of the 3′ UTR for mRNA localization to the vicinity of mitochondria and, as a consequence, for the functionality of the organelle, we replaced in vivo the 3′ UTR of ATM1 by the 3′ UTR of ADH1 and examined the phenotype of the mutated strain obtained, called ATM1-3′UTRADH1. As in the case of ATP2, ATM1-3′UTRADH1 (MS) cells grew poorly on the non-fermentable source glycerol (Figure 6A). Mitochondrial function was restored when the mutated strain was transformed with a low-copy-number vector directing the expression of a wild-type version of ATM1 (Figure 6A, MS+ATM1). ATM1 disruption leads to a respiratory deficiency and atm1 cells spontaneously become ρ– because of the instability of their mitochondrial genome; indeed, few of these cells are still able to grow on glycerol several days after plating (Leighton and Schatz, 1995). To examine whether the in vivo replacement of ATM1 3′ UTR by ADH1 3′ UTR leads to ρ– cells, we determined the ability of the ATM1-3′UTRADH1 strain to grow on liquid glycerol medium after transformation with the wild-type ATM1 gene. Figure 6B confirms the inability of ATM1-3′UTRADH1 cells to grow on non-fermentable carbon sources, although this impairment was less severe than that measured in the ATP2-3′UTRADH1 cells (MS.ATP2). When ATM1-3′UTRADH1 cells expressed a functional ATM1 gene, a significant recovery of respiratory function was observed even if the growth rate measured is lower than that measured in wild-type cells. Therefore, the absence of ATM1 3′ UTR renders the cells partially ρ–, as described for the ATM1 disruption. As a step towards determining the biological role of ATM1 3′ UTR on the respiratory deficiency of ATM1-3′UTRADH1 cells, we purified mitochondrion-bound polysomes and free cytoplasmic polysomes from ATM1-3′UTRADH1 cells. Our previous work clearly demonstrated that ATM1 mRNA localizes to the vicinity of mitochondria (Corral-Debrinski et al., 2000). Figure 6C confirms that in wild-type cells ATM1 transcript mainly localized to mitochondrion-bound polysomes, and ATM1 mRNA is almost undetectable on free cytoplasmic polysomes fractions. In ATM1-3′UTRADH1 cells, ATM1 transcript was enriched in free cytoplasmic polysomes, and a reduction in the overall signal of ATM1 mRNA on mitochondrion-bound polysomes was consistently measured. ATP3 mRNA in ATM1-3′UTRADH1 cells presented the proper localization to mitochondrion-bound polysomes (Figure 6C). Thus, perturbation on ATM1 mRNA targeting to the vicinity of mitochondria in ATM1-3′UTRADH1 cells is associated to the absence of the 3′ UTR in the ATM1 altered allele and might be involved in the impairment of mitochondrial function observed in these cells.
Fig. 6. In vivo substitution of the ATM1 3′ UTR by the ADH1 3′ UTR also leads to a respiratory chain dysfunction. (A) The pFA6a-3HA1-TRP1 plasmid (Longtine et al., 1998) was used to obtain a modified strain (MS) from the diploid BMA64 strain called ATM1-3′UTRADH1 in which the C-terminus of ATM1, the gene encoding an ABC transporter of the inner mitochondrial membrane (Leighton and Schatz, 1995), was fused to three tandem repeats of the HA epitope followed by the 3′ UTR of the ADH1 gene, encoding a cytoplasmic protein. After dissection of tetrads, each TRP+ spore was tested for its ability to grow on medium with the non-fermentable carbon source glycerol. Three tetrads are shown (1–3), all the Trp+ cells grew poorly on glycerol at 28°C. Two independent clones (MS.1 and MS.2) were transformed with a wild-type version of the ATM1 gene obtained by gap repair and their ability to grow on glycerol was compared with that of wild-type cells transformed with the pFL45 plasmid and with that of the original MS.1 and MS.2 cells. Cells were grown on complete synthetic medium containing 2% glucose and devoid of tryptophan to an OD of 2. They were serially diluted (1:5) and spotted either on complete synthetic medium devoid of tryptophan (Glu-Trp) or on 2% glycerol medium (Glycerol). The plates were then incubated for 2 days at 28°C. (B) The growth rate of ATM1-3′UTRADH1 (MS.ATM1) cells was measured on liquid glycerol medium and compared with wild-type cells (WT), and with cells expressing the wild-type ATM1 gene (MS.ATM1+ATM1) and the ATP2-3′UTRADH1 strain (MS1.ATP2 and MS2.ATP2) either at 28 or 37 °C. Cells were grown overnight on complete synthetic medium devoid of tryptophan and containing 2% glucose. The quantity of cells corresponding to an OD of 0.2 was diluted in 40 ml of glycerol medium. OD measurements were performed every 2 h. (C) To determine the subcellular localization of the modified ATM1 mRNA in the ATM1-3′UTRADH1 strain, mitochondrion-bound polysomes (M-P) and free cytoplasmic polysomes (F-P) were purified from ATM1-3′UTRADH1 cells (MS.ATM1) and wild-type cells (WT). Eight micrograms of RNA extracted from each polysomal population were separated on formaldehyde–agarose gels, subjected to northern blot analysis and hybridized successively with ATM1 and ATP3 probes. Autoradiograms shown represent an exposure time of 16 h for ATM1 and of 6 h for ATP2 at –80°C with Amersham intensifying screens. Methylene blue staining of the filters prior hybridization is also shown.
Discussion
Recently, a genomic scale analysis allowed us to show that >100 mRNAs encoding mitochondrial proteins localize to the vicinity of mitochondria in Saccharomyces cerevisiae (Marc et al., 2002). The main goal of our research was to determine whether this process is involved in organelle biogenesis. To undertake this project, we generated a mutated strain in which the 3′ UTR of the ATP2 gene encoding the β-subunit of the F1-ATP synthase was replaced by the 3′ UTR of ADH1, a gene coding for a cytoplasmic protein. First, we demonstrated that the 3′ UTR of ATP2 acts in vivo as a localizing element, which allows the targeting of a reporter RNA to the close proximity of mitochondria. Secondly, we showed that in the mutated strain the altered ATP2 mRNA was remarkably over-represented in free cytoplasmic polysomes. Finally, these cells present a deficiency of cell respiration. Interestingly, we observed a significant decrease in the mitochondrial import of Atp2p synthesized from the altered ATP2 allele. It is then tempting to envisage that impairment of ATP2 mRNA sorting could be responsible for the respiratory chain dysfunction observed. To further confirm the impact on mitochondrial biogenesis that could have caused the perturbation of mRNA targeting, we examined cells in which the 3′ UTR of ATM1, another essential mitochondrial gene, was replaced by the ADH1 3′ UTR. Cells present a respiratory dysfunction and, as has been described for ATM1 disruption, some of them become ρ–. We showed that this phenomenon is correlated to the poor sorting of the altered ATM1 mRNA to the vicinity of the mitochondria.
Therefore, this study constitutes the first demonstration that mRNA sorting to the vicinity of mitochondria represents a crucial step for the functionality of the organelle.
Mitochondrial mRNA targeting process is essential for protein function
Recent studies in the squid giant axon demonstrated that the supply of proteins for mitochondria localized to the distal compartments of these large asymmetric neurons is obtained by local synthesis, thus establishing an intimate link between translation and translocation to ensure the viability of the organelle located in these neuronal domains (Gioio et al., 2001). Sorting of mRNAs encoding mitochondrial proteins could then be a key step to ensure the functionality of the corresponding polypeptides inside the organelle. Specific mRNA localization in somatic cells often involves 3′ UTR sequences (Jansen, 2001), as we demonstrated for the ATM1 transcript (Corral-Debrinski et al., 2000) and other genes with as yet unattributed functions in yeast (Marc et al., 2002).
These data prompted us to examine two important points: (i) the minimal size of the localization element present in the 3′ UTR of a mitochondrial gene; (ii) the impact on protein function of mRNA targeting impairment. For this purpose, we took ATP2 as a model system. ATP2 encodes the β-subunit of the F1-ATP synthase. The visualization of RNA in living cells allowed us to show that both the sequence encoding the first 35 amino acids of Atp2p and the ATP2 3′ UTR possess cis-acting information that guides mRNA to the mitochondrial periphery. A 243 nt stretch following Atp2p stop codon is able to address an RNA to the vicinity of mitochondria. A deletion of 50 nt both at the 5′ and 3′ of this sequence did not alter its properties; therefore, a minimal localization element within ATP2 3′ UTR is 100 nt in length and encompasses the sequence between nt 50 and 150 downstream of the stop codon. The MFOLD program for RNA secondary structure (Zuker, 1989) predicted that this region folds into an apparently stable long stem–loop structure formed by four stems separated by asymmetrical bulges. To determine whether this motif can be found in other mRNAs encoding mitochondrial proteins, we examined 3′ UTRs of the top 25 highest and lowest MLR values (Marc et al., 2002) and compared them with 3′ UTRs for 25 cytoplasmic proteins. We found that this structure is found three times more frequently in mRNAs with high MLR values than in mRNAs with low MLR values or mRNAs for cytoplasmic proteins. Moreover, a similar structure can be found in the 3′ UTR of ATP5b, human homolog of ATP2, which is strongly enriched in mitochondrion-bound polysomes purified from HeLa cells (data not shown).
Since the ATP2 3′ UTR acts as a localizing element, we decided to replace it with ADH1 3′ UTR in the chromosomal ATP2 gene to assess the effect on mRNA sorting and protein function. We obtained cells unable to respire; when a wild-type ATP2 gene was expressed, respiration function was restored, confirming that the chromosomal modification is responsible for the phenotype observed. In these cells, ATP2 mRNA was poorly localized to mitochondrion-bound polysomes. Therefore, the in vivo impairment of ATP2 mRNA sorting is associated with a cell respiration defect, which is reminiscent of that observed in the null ATP2 mutant (Chen and Clark, 1999).
Remarkably, studies conducted with the mutated strain allowed us to establish some connections between mitochondrial mRNA targeting, translation and protein import. Indeed, we observed that a critical amount of Atp2p inside the mitochondria is necessary to fully rescue the respiratory deficiency of the mutated strain. A plasmid containing the ATP2 ORF associated to a minimal 100 nt localizing element only partially rescued cell respiration, although this element was able to target a reporter RNA in the vicinity of mitochondria. The amount of Atp2p inside mitochondria in these cells was obviously not sufficient to obtain a functional respiratory chain. Therefore, this element is necessary, but not sufficient, to promote and/or maintain the precursor in an import-competent conformation. When the ATP2-3′UTRADH1 strain was transformed with a plasmid in which ATP2 ORF was fused to the 250 nt element, respiration ability was totally recovered. This strongly suggests that the physiologically functional ATP2 3′ UTR is 250 nt in length. We can hypothesize that: (i) this sequence might interact with the 5′ region of the transcript, allowing the cross-talk of both mRNA ends involved in the recruitment of the translational machinery; (ii) its presence might be necessary for the assembly of the ribonucleoprotein complex, and thus for protein translation and membrane translocation; and (iii) as in the case of the yeast ASH1 mRNA (Gonzalez et al., 1999) or of the oligodendrocyte MBP mRNA (Bassell et al., 1999), redundant localizing elements are required for the delivery, anchoring and translation activation of the corresponding transcripts.
Connections between mRNA sorting, mRNA translation and mitochondrial protein import
Whether in vivo mitochondrial protein import occurs co-translationally or post-translationally is still not known. It has been shown by the utilization of a homologous cell-free mitochondrial import system derived from yeast that, in vivo, the import of several mitochondrial proteins was very tightly coupled to their synthesis (Fujiki and Verner, 1993). Additionally, ribosome-attached nascent chains can initiate import into mitochondria in a homologous in vitro system (Fünfschilling and Rospert, 1999). It is noteworthy that the nascent polypeptide-associated complex (NAC) is required for the effective targeting of ribosome nascent chain complexes to mitochondria (George et al., 1998, 2000). Therefore, in mitochondria, a co-translational import mechanism may actually be required for some precursors, and this may be initiated by mRNA localization to the mitochondrial periphery, a phenomenon that we have observed for ∼100 mitochondria-destined proteins in yeast (Marc et al., 2002).
The results reported here clearly demonstrated an intimate link between ATP2 mRNA targeting to the vicinity of mitochondria and translocation of Atp2p inside the organelle. Indeed, ATP2-3′UTRADH1 cells, in which the altered ATP2 mRNA is mostly found in free cytoplasmic polysomes, accumulate the majority of the full-length Atp2 precursor presumably in the cytosol. Even though the protein was able to interact with the mitochondrial import machinery, its productive import did not occur. This defect is restricted to Atp2p, since Atp1p, Atp4p, Atp6p and Abf2p were successfully translocated inside the organelle. Moreover, the inability of Atp2p produced from the altered allele to be imported to the mitochondria is not the consequence of the triple HA epitope present at the C-terminus of the protein. Hence, the respiratory deficiency of ATP2-3′UTRADH1 cells is the direct consequence of the absence of the ATP2 3′ UTR.
The β-subunit of the F1-ATP synthase complex has been extensively used to study the process of mitochondrial protein import in mammalian cells. High-resolution in situ hybridization experiments in rat hepatocytes have revealed that β-F1-ATPase mRNA localized as clusters in close contact with the outer mitochondrial membrane (Egea et al., 1997). These structures are translationally active, indicating that the import of the corresponding precursor requires the sorting of its mRNA to mitochondrial proximity (Ricart et al., 1997). More recently, the biological role of the N-terminal region and the 3′ UTR of β-F1-ATP synthase mRNA in the assembly of the ribonucleoprotein complex was demonstrated in rat hepatocytes (Ricart et al., 2002). In yeast, it has been reported that Atp2p can be efficiently imported in vitro (Maccecchini et al., 1979) and that this can hold true in vivo. However, several observations could argue in favor of a co-translational mitochondrial insertion. (i) Under normal steady-state growth conditions, precursor forms of mitochondrial proteins, especially Atp2p, are below the limits of detection (Ades and Butow, 1980). Only under special conditions, when antibiotics were used to abolish the membrane potential across the mitochondrial inner membrane, were precursor forms observed (Reid and Schatz, 1982). (ii) When a denatured form of the mitochondrial F1-ATPase β-subunit precursor was prepared either in vivo or in vitro, its import into mitochondria did not occur even when targeting signals for its mitochondrial import were present (Ohta and Schatz, 1984). (iii) F1 β-precursor is able to form tetramers in solution, in vitro synthesized tetramers are not competent for mitochondrial import indicating the requirement of an import-competent structure to allow a complete mitochondrial translocation of the protein (Chen and Douglas, 1987). (iv) The N-terminal region of Atp2p functions as an intramolecular chaperone, as the absence of a redundant signal within this sequence reduces the ability of the protein to be imported in a post-translational manner (Hajek et al., 1997). Since we demonstrated here that the N-terminal region of the ATP2 transcript is necessary for its association with mitochondrion-bound polysomes, we can hypothesize that the above results could be explained by the requirement of ATP2 mRNA mitochondrial targeting to obtain an efficient mitochondrial import of the precursor. We can consider an attractive model: in vivo ATP2 mRNA molecules translated in free cytoplasmic polysomes give rise to precursors lacking the import-competent structure required for an efficient mitochondrial membrane translocation. In an attempt to reconcile our data with post-translational import of accumulated precursors in experiments conducted with spheroplasts (Reid and Schatz, 1982), we can suggest that Atp2 precursors obtained under CCCP treatment were able to interact with the import machinery or/and chaperone-associated activity, which maintained them in an ‘import-competent’ conformation, while in our mutated strain, Atp2 precursor was strictly translated in free cytoplasmic polysomes away from the import machinery, thus presenting a conformation incompatible with mitochondrial import.
Concluding remarks
In conclusion, we present here for the first time, to our knowledge, compelling evidence that for two mitochondria-destined proteins, Atp2p and Atm1p, mRNA-specific delivery and translation at the vicinity of mitochondria represent a key step to ensure the functionality of corresponding polypeptides inside the organelle. Our previous report showing that at least 100 mRNAs encoding mitochondrial proteins are consistently found in close contact with mitochondria (Marc et al., 2002) allows us to hypothesize that this phenomenon is essential to ensure organelle assembly. The functional implication of this mRNA distribution in mitochondrial biogenesis deserves to be explored, since its alteration could be involved in a wide spectrum of degenerative diseases in man.
Material and methods
Strains used
Saccharomyces cerevisiae strains used in this study were CW04, which is isonuclear with the strain W303–1B (MATa, leu2-3, ura 3-1, trp1-1, ade1-2, his3-11, can1-100) (Conde and Fink, 1976) and the diploid BMA64 strain (a/α, ura3-1/ura3-1, trp1-Δ2/trp1-Δ2, leu2-3,112/leu2-3,112, his3-11/his3-11, ade2-1/ade2-1, can1-100/can1-100). The ATP2-3′UTRADH1 and ATM1-3′UTRADH1 strains were obtained by homologous recombination of diploid BMA64 cells using a PCR-mediated technique. DNA for yeast transformation was generated by PCR using as template the pFA6a-3HA-TRP1 plasmid (Longtine et al., 1998), the recombinant Tth DNA polymerase XL (Perkin-Elmer Cetus) and the following primers: for ATP2, the forward primer 5′-ATTGAAGATGTTGTTGCTAAAGC TGAAAAGTTAGCCGCTGAAGCCGCTGAAGCCAACCGGATCC CCGGGTTAATTAA-3′ and the reverse primer 5′-TATCTTTTT TTTCCCATCATCCTTATTGTTCTACCTTATTGTTCTACCTTCGG TATTTCAAGAATTCGAGCTCGTTTAAAC-3′; and for ATM1, the forward primer 5′-GAAGATCTGGATCATTTAGAGAATGAACTAA AAGACCAGGCAAGAACTACGGATCCCCGGGTTAATTAA-3′ and the reverse primer 5′-CGGGTCATTGAAAAACCGCGGAGAAA GATAATAACTTTTCTTTTTTCCGAATTCGAGCTCGTTTAAAC-3′.
In mutated strains, the modified genes expressed triple-HA-tagged proteins and the genuine 3′ UTRs were substituted by the ADH1 3′ UTR, which is 237 bp long. To determine whether a triple-HA-tagged Atp2 protein is functional, we replaced the ADH1 3′ UTR by the 662 bp long ATP2 3′ UTR in the pFA6a-3HA-TRP1 plasmid (Longtine et al., 1998), using the BglII and AscI unique sites and a PCR product of 662 bp long corresponding to the full-length ATP2 3′ UTR. This product was obtained using the same set of primers used for fluorescence microscopy experiments, but adding at the 5′-end the compatible BglII and AscI recognition sequences. The modified pFA6a-3HA-TRP1 plasmid was used to prepare a PCR product, which allows a single-step modification of the chromosomal ATP2 gene on the haploid BMA64 strain (a, ura3-1/ura3-1, trp1-Δ2/trp1-Δ2, leu2-3, 112/leu2-3, 112, his3-11/his3-11, ade2-1/ade2-1, can1-100/can1-100).
Cells were grown at 28°C in rich medium (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose and 30 mg/ml adenine), galactose-rich medium [1% Bacto yeast extract, 2% Bacto peptone, 2% galactose, 0.1% KH2PO4, 0.12% (NH4)2SO4] or synthetic medium containing either 2% glucose, 2% glycerol or 2% galactose supplemented with the appropriate nutritional requirements. For solid medium, 2% agar was added. Yeast cells were transformed using a simplified lithium method (Gietz et al., 1992).
Plasmid construction
For DNA manipulations and cloning, Escherichia coli strain TG1 was used. To obtain the wild-type version of ATM1, we used a linearized chimeric construct containing both the promoter and the 3′ UTR regions of ATM1 (Corral-Debrinski et al., 2000) to obtain, by gap repair, the complete gene.
The construction of the pRSAM plasmid series was carried out as follows. Plasmid p344SB2-5 (a kind gift of Dr D.Mueller) was cut using restriction endonucleases SalI and HindIII. The 1.8 kb fragment containing the ATP2 promoter, the complete ATP2 ORF and a sequence of 13 nt after the stop codon was recovered and inserted into a pRS416 vector, the plasmid obtained was named pRSAM0. For pRSAM3, p344SB2-5 was cut using SalI and EcoRI restriction endonucleases. The 2.5 kb fragment containing ATP2 promoter, ORF and the full-length 3′ UTR (662 bp) was recovered and inserted into pRS416. Other plasmids in the pRSAM series were constructed using the remaining pRSAM0 XmaI and EcoRI cloning sites just behind the ATP2 stop codon. 3′ UTRs were amplified with 25 cycle PCR amplification using rTth DNA polymerase XL (Perkin-Elmer Cetus) and yeast genomic DNA as template. For pRSAM1, the primers used were 5′-TCCCCCCGGGGGGATGAAT GAATATCGTAACATAATAATAC-3′ and 5′-CCGGAATTCCGGA GCTTAAACCAAGGGAAGCAAAATTTG-3′. For pRSAM2, the primers used were 5′-TCCCCCCGGGGGGACTTTATTTACTTACAG TTGGGAGTAAA-3′ and 5′-CCGGAATTCCGGAGCTTAAACCAAG GGAAGCAAAATTTG-3′.
The PCR products were purified using Nucleospin Extract kit (Macherey Nagel), digested with XmaI and EcoRI, and then inserted into pRSAM0 vector.
Plasmids for the ‘Green-RNA’ labeling system were prepared as described previously (Corral-Debrinski et al., 2000; Marc et al., 2002).
The full-length ATP2 3′ UTR was generated by PCR using as template yeast genomic DNA, the recombinant Tth DNA polymerase XL (Perkin-Elmer Cetus), the forward primer 5′-TCCCCCGGGG GATAGAAGAAATAAAGCTTAAACCAAGGG-3′ and the reverse primer 5′-TCCCCCGGGGGACATGTCCAGTGGGAAAGCGAGGC AAGA-3′. The 662 bp fragment was purified, digested with SmaI and cloned in the pIIIA/MS2-2 plasmid, generously provided by K.Bloom (Beach et al., 1999). For the sequence coding the first 35 amino acids of Atp2p, we used the following primers: 5′-TCCCCCGGGGGA AGAATGGTTTTGCCAAGACTATATACT-3′ and 5′-TCCCCCGGG GGATGAGGCCATACATCTTTTCCACGAAGT-3′. The 3′ UTR fragments of 243 and 396 bp long were obtained by the cloning of respective HphI digestion products of the full-length 3′ UTR. Finally, for the 50–150 and 0–100 fragments, specific oligonucleotides, representing both strands (the 0 position representing the ATP2 stop codon) were purchased, annealed and directly cloned in the SmaI site of the pIIIA/MS2-2 plasmid.
Purification of free and mitochondrion-bound polysomes
Free and mitochondrion-bound polysomes were purified as described previously (Corral-Debrinski et al., 2000; Marc et al., 2002). The full protocol can be found at the following web address: http://www.biologie.ens.fr/fr/genetiqu/puces/publications/mitoloc/MARC_mito_purif_protocol.pdf
RNA extractions
RNA extraction from mitochondrion-bound polysomes, free cytoplasmic polysomes and whole cells was performed using a hot-phenol method (Schmitt et al., 1990) or RNAesy Protect MIDI kit (Qiagen). For whole-cell extraction, cells were grown in synthetic medium (BIO101 Inc.) containing 2% galactose, 0.5% glucose and appropriate auxotrophy requirements to an OD600 of 1. For northern blots, 8 µg (subcellular polysome fractions) or 15 µg (whole cells) of RNA were separated by electrophoresis through denaturing formaldehyde–agarose gels; after transfer, nylon membranes were stained with methylene blue solution (Sambrook et al., 1989). Hybridizations and washings were performed as described previously (Corral-Debrinski et al., 2000).
Purification of mitochondria
Cells were grown in synthetic medium with 2% galactose and 0.5% glucose. Mitochondria were purified by successive centrifugations as described previously (Corral-Debrinski et al., 1999). Protein concentrations were measured using the Bradford dye-binding assay. To determine whether Atp2p was translocated into the organelle, 30 µg of mitochondrial proteins were treated with various concentrations (0.2–0.6 mg/ml) of PK at 0°C for 30 min. As a control for protease accessibility, PK digestion was performed in the presence of 1% Triton X-100, a detergent that disrupts both mitochondrial membranes. Samples were then subjected to SDS–PAGE and western blot analysis.
SDS–PAGE and western blot analysis
Thirty micrograms of proteins from mitochondria were resolved by 11% SDS–PAGE and transferred to nitrocellulose. Filters were probed with the following antibodies: rabbit monoclonal antibodies against the Atp2p (1:100 000), Atp1 (1:100 000), Atp4 (1:10 000) and Atp6 (1:10 000), a kind gift of Dr J.Velours, mouse monoclonal antibody 12CA5 against the HA1 epitope (1:5000) and rabbit polyclonal antibody against Abf2p (1:10 000). Immunoreactive bands were visualized with either anti-mouse or anti-rabbit IgGs coupled to horseradish peroxidase followed by ECL detection (Amersham International) according to the manufacturer’s instructions.
Growth and respiration assays
Cells were grown in synthetic medium containing 2% glucose to an OD600 of 2. They were then serially diluted (1:5) and spotted either on synthetic medium containing 2% glucose supplemented with the appropriate nutritional requirements or on 2% glycerol medium (1% yeast extract, 2% peptone, 2% glycerol, 10% sodium phosphate buffer pH 6.2). Plates were incubated at 28 or 37°C for 48 h. For growth assay in liquid glycerol medium, cells were grown overnight in rich medium until they reached stationary phase. The equivalent of 0.2 OD600 of cells were diluted in 40 ml of glycerol medium and OD was measured every 2 h. The wild-type strains were transformed with the pFL45 plasmid, allowing them to grow in a medium without tryptophan. For the experiments performed with the ATP2-3′UTRADH1 strain transformed with different pRSAM plasmids, the wild-type strain was also transformed with the pFL44 plasmid to allow it to grow in medium without uracil.
Fluorescence microscopy
The CP of bacteriophage MS2 was fused to GFP and expressed from the plasmid pCP-GFP, a generous gift of D.Beach and K.Bloom (Beach et al., 1999). pCP-GFP is a low-copy HIS3-selectable plasmid that produced CP–GFP regulated by the MET25 promoter. Cells grown in the presence of methionine produced no detectable CP–GFP protein product. To induce CP–GFP production, cells were switched to a medium without methionine for 2 h. To obtain reporter RNA, we used the pIIIA/MS2-2 plasmid (Beach et al., 1999), which contains two tandem copies of the CP binding site. This plasmid can express a transcript tagged by the two tandem copies of the CP binding site, using the RNaseP promoter; this constitutive RNA polymerase III promoter maintains RNA levels throughout the cell cycle. The single SmaI site was used to introduce the nucleotide sequences with potential targeting properties. In all the cases, the binding site precedes the sequence examined. We studied both possible orientations for each inserted sequence. The cells expressing the constructions where the inserted sequence was placed in the opposite orientation, such that the non-coding sequence was transcribed, were examined by fluorescence microscopy. These cells, like the one expressing the empty pIIIA/MS2-2 plasmid, showed a low amount of fluorescence distributed throughout the cytoplasm and excluded only from the vacuole (data not shown), as shown previously (Beach et al., 1999). Living cells expressing the green RNAs were observed by fluorescence microscopy with a chilled CCD camera (Hamamatsu Photonics, Hamamatsu City, Japan) as described previously (Corral-Debrinski et al., 1999, 2000). For all microscopic observations, cells were harvested in early log phase. Mitochondrial structures and mitochondrial DNA were visualized using rhodamine B at 0.5 nM (Molecular Probes) and the Hoechst reagent (1 nM).
Acknowledgments
Acknowledgements
We are grateful to S.Karniely for useful discussions and comments on the manuscript. We thank Dr K.Bloom for the gift of plasmids required for the RNA-labeling system, Dr D.M.Mueller for the gift of p344SB2-5 plasmid and Dr J.Velours for the gift of the specific anti-Atp2, anti-Atp1, anti-Atp4 and anti-Atp6 antibodies. This work was supported by funds from the CNRS (UMR8541), the Ecole Normale Supérieure and the French association against myopathies (AFM, grant MNM 2000). A.M. is the recipient of a CNRS BDI fellowship.
References
- Ades I.Z. and Butow,R.A. (1980) The transport of proteins into yeast mitochondria. Kinetics and pools. J. Biol. Chem., 255, 9925–9935. [PubMed] [Google Scholar]
- Bassell G.J., Oleynikov,Y. and Singer,R.H. (1999) The travels of mRNAs through all cells large and small. FASEB J., 13, 447–454. [DOI] [PubMed] [Google Scholar]
- Beach D.L., Salmon,E.D. and Bloom,K. (1999) Localization and anchoring of mRNA in budding yeast. Curr. Biol., 9, 569–578. [DOI] [PubMed] [Google Scholar]
- Bedwell D.M., Klionsky,D.J. and Emr,S.D. (1987) The yeast F1-ATPase β subunit precursor contains functionally redundant mitochondrial protein import information. Mol. Cell. Biol., 7, 4038–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.B. (1989) Fluorescent labeling of mitochondria. Methods Cell Biol., 29, 103–123. [DOI] [PubMed] [Google Scholar]
- Chen W.-J. and Douglas,M.G. (1987) The role of protein structure in the mitochondrial import pathway. Analysis of the soluble F1-ATPase β-subunit precursor. J. Biol. Chem., 262, 15598–15604. [PubMed] [Google Scholar]
- Chen X.J. and Clark,G.D. (1999) α and β subunits of F1-ATPase are required for survival of petite mutants in Sacharomyces cerevisiae. Mol. Gen. Genet., 262, 898–908. [DOI] [PubMed] [Google Scholar]
- Conde J. and Fink,G. (1976) A mutant of S. cerevisiae defective for nuclear fusion. Proc. Natl Acad. Sci. USA, 73, 3651–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Debrinski M., Belgareh,N., Blugeon,C., Claros,M.G., Doye,V. and Jacq,C. (1999) Overexpression of yeast karyopherin Pse1p/Kap121p stimulates the mitochondrial import of hydrophobic proteins in vivo. Mol. Microbiol., 31, 1499–1511. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M., Blugeon,C. and Jacq,C. (2000) In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol., 20, 7881–7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea G., Izquierdo,J.M., Martin,R.C.S. and Cuezva,J.M. (1997) mRNA encoding the β-subunit of the mitochondrial F1-ATPase is a localized mRNA in rat hepatocytes. Biochem. J., 322, 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner A., Jakobs,S. and Hell,S.W. (2002) Fast 100-nm resolution three-dimentional microscope reveals structural plasticity of mitochondria in live yeast. Proc. Natl Acad. Sci. USA, 99, 3370–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki M. and Verner,K. (1993) Coupling of cytosolic protein synthesis and mitochondrial protein import in yeast. J. Biol. Chem., 268, 1914–1920. [PubMed] [Google Scholar]
- Fünfschilling U. and Rospert,S. (1999) Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell, 10, 3289–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R., Beddoe,T., Landl,K. and Lithgow,T. (1998) The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc. Natl Acad. Sci. USA, 95, 2296–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R., Walsh,P., Beddoe,T. and Lithgow,T. (2002) The nascent polyptide-associated complex (NAC) promotes interaction of ribosome with the mitochondrial surface in vivo. FEBS Lett., 516, 213–216. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic. Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioio A.E., Eyman,M., Zhang,H., Lavina,Z.S., Giuditta,A. and Kaplan,B.B. (2001) Local synthesis of nuclear-encoded mito chondrial proteins in the presynaptic nerve terminal. J. Neurosci. Res., 64, 447–453. [DOI] [PubMed] [Google Scholar]
- Gonzalez I., Buonomo,S.B.C., Nasmyth,K. and Ahsen,U.v. (1999) ASH1 mRNA localization in yeats involves multiple secondary elements and Ash1 protein translation. Curr. Biol., 9, 337–340. [DOI] [PubMed] [Google Scholar]
- Gratzer S., Beilharz,T., Beddoe,T., Henry,M.F. and Lithgow,T. (2000) The mitochondrial protein targeting suppressor (mts1) mutation maps to the mRNA-binding domain of Npl3p and affects translation on cytoplasmic polysomes. Mol. Microbiol., 35, 1277–1285. [DOI] [PubMed] [Google Scholar]
- Hajek P., Koh,J.Y., Jones,L. and Bedwell,D.M. (1997) The amino terminus of the F1-ATPase β-subunit precursor functions as an intramolecular chaperone to facilitate mitochondrial protein import. Mol. Cell. Biol., 17, 7169–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.-P. (2001) mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol., 2, 247–256. [DOI] [PubMed] [Google Scholar]
- Johnson L.V., Walsh,M.L. and Chen,L.B. (1980) Localization of mitochondria in living cells with rhodamine 123. Proc. Natl Acad. Sci. USA, 77, 990–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. et al. (2002) Subcellular localization of the yeast proteome. Genes Dev., 16, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton J. and Schatz,G. (1995) An ABC transporter in the mitochondrial inner membrane is required for normal growth of yeast. EMBO J., 14, 188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A.,III, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Maccecchini M.-L., Rudin,Y., Blobel,G. and Schatz,G. (1979) Import of proteins into mitochondria: precursor forms of the extra-mitochondrially made F1-ATPase subunits in yeast. Proc. Natl Acad. Sci. USA, 76, 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc P., Margeot,A., Devaux,F., Blugeon,C., Corral-Debrinski,M. and Jacq,C. (2002) Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO rep., 3, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa I., Aoi,H., Sando,N. and Kuroiwa,T. (1984) Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast, Saccharomyces cerevisiae. J. Cell Sci., 66, 21–38. [DOI] [PubMed] [Google Scholar]
- Ohta S. and Schatz,G. (1984) A purified precursor polypeptide requires a cytosolic protein fraction for import into mitochodnria. EMBO J., 3, 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N. and Geissler,A. (2001) Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol., 2, 339–349. [DOI] [PubMed] [Google Scholar]
- Reid G.A. and Schatz,G. (1982) Import of proteins into mitochondria. Yeast cells grown in the presence of carbonyl cyanide m-chlorophenylhydrazone accumulate massive amounts of some mitochondrial precursor polypeptides. J. Biol. Chem., 257, 13056–13060. [PubMed] [Google Scholar]
- Ricart J., Egea,G., Izquierdo,J.M., Martin,C.S. and Cuezva,J.M. (1997) Subcellular structure containing mRNA for β subunit of mitochondrial H+ ATP synthase in rat hepatocytes is trnaslationnaly active. Biochem. J., 324, 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricart J., Izquierdo,J.M., Di Liegro,C.M. and Cuezva,J.M. (2002) Assembly of the ribonucleoprotein complex containing the mRNA of the β-subunit of the mitochondrial H+-ATP synthase requires the participation of two distal cis-acting elements and a complex set of cellular trans-acting proteins. Biochem. J., 365, 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook K.J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schmitt M.E., Brown,T.A. and Trumpower,B.L. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B.J. (1977) Variation in number and volume of the mitochondria in yeast according to growth conditions. A study based on serial sectioning and computer graphics reconstitution. Biol. Cell, 28, 37–56. [Google Scholar]
- Wallace D.C. (1999) Mitochondrial diseases in man and mouse. Science, 283, 1482–1488. [DOI] [PubMed] [Google Scholar]
- Zuker M. (1989) On finding all suboptimal foldings of an RNA molecule. Science, 244, 48–52. [DOI] [PubMed] [Google Scholar]