Abstract
Loss of the tumour suppressor BRCA1 results in profound chromosomal instability. The fundamental defect underlying this catastrophic phenotype is not yet known. In vivo, BRCA1 forms a heterodimeric complex with BARD1. Both proteins contain an N-terminal zinc RING-finger domain which confers E3 ubiquitin ligase activity. We have isolated full-length human BRCA1/BARD1 complex and have shown that it has a dual E3 ubiquitin ligase activity. First, it mediates the monoubiquitylation of nucleosome core histones in vitro, including the variant histone H2AX that co-localizes with BRCA1 at sites of DNA damage. Secondly, BRCA1/BARD1 catalyses the formation of multiple polyubiquitin chains on itself. Remarkably, this auto-polyubiquitylation potentiates the E3 ubiquitin ligase activity of the BRCA1/BARD1 complex >20-fold. Even though BRCA1 has been reported to associate with a C-terminal ubiquitin hydrolase, BAP1, this enzyme does not appear to function in the deubiquitylation of the BRCA1/BARD1 complex.
Keywords: BARD1/BRCA1/E3 ligase/ubiquitin
Introduction
Germline mutations in BRCA1 pre-dispose women to early-onset familial breast and ovarian cancers (Miki et al., 1994). In cells, loss of BRCA1 function leads to spontaneous chromosome breakage and sensitivity to DNA damage (Deng and Scott, 2000). BRCA1 is implicated in a variety of cellular processes including transcription, DNA repair and cell cycle progression. Indeed, the BRCA1 protein has been shown to interact directly with a number of proteins involved in these processes, such as RAD51 (Scully et al., 1997b), RNA polymerase II (Scully et al., 1997a) and p53 (Zhang et al., 1998). Another protein known to interact with BRCA1 is BARD1 (Wu et al., 1996). These two proteins co-exist as a heterodimeric complex (Meza et al., 1999; Brzovic et al., 2001) and, in cells that lack BARD1, the stability of BRCA1 is much reduced (Joukov et al., 2001). However, to date, studies of these multiple interactions have yielded little insight into the fundamental molecular function of the BRCA1 protein.
Studies of the amino acid sequence of BRCA1 also revealed little about the protein’s function. Nevertheless, two potentially important motifs were apparent; a BRCT domain, thought to act as a region of protein–protein interaction, and a zinc RING-finger domain (reviewed in Deng and Brodie, 2000). Interestingly, BARD1 too has a zinc RING-finger domain and BRCT domains. Further insight into the function of BRCA1 was gained through the demonstration that RING-finger domains, including that of BRCA1, could confer E3 ubiquitin ligase activity (Lorick et al., 1999).
Ubiquitylation is a form of post-translational modification in which a small, globular 76 amino acid protein, ubiquitin, is covalently attached to protein substrates. Although ubiquitylation has been seen traditionally as a mechanism for targeting proteins for destruction by the proteasome, a wider view is emerging. Indeed, it is now known that there are different forms of ubiquitin modification, which may serve different functions. While some proteins are modified with a single ubiquitin molecule (monoubiquitylation), others have chains of ubiquitin molecules (polyubiquitylation). Furthermore, these chains can take various forms depending on the way the ubiquitin molecules are linked together. It is now clear that ubiquitin modification, in its various forms, plays roles in diverse cellular processes from protein transport to DNA repair (reviewed in Hershko and Ciechanover, 1998; Hicke, 2001; Pickart, 2001; Weissman, 2001).
Ubiquitylation is a multistep process whereby ubiquitin is first activated, in an ATP-dependent process, through the formation of a thiol–ester link between its C-terminal glycine and a cysteine residue on a ubiquitin-activating enzyme (E1). The ubiquitin is then transferred to an ubiquitin-conjugating enzyme (E2), with which it forms a new thiol–ester bond. Finally, ubiquitin is transferred to the ε-amino group of a lysine on its substrate protein in a reaction catalysed by a ubiquitin ligase (E3). There are two subgroups of E3 ligases, the HECT domain ligases such as E6-AP (Huibregtse et al., 1995), and the RING domain ligases (Joazeiro and Weissman, 2000). Whereas HECT domain ligases form ubiquitylated intermediates as part of the transfer of ubiquitin to substrate, RING domain ligases mediate the direct transfer of ubiquitin from the E2 to the substrate.
Mounting evidence supports a role for BRCA1 in the ubiquitylation pathway. First, the DNA damage-induced monoubiquitylation of the Fanconi anaemia D2 protein (FANCD2) is reduced in cells that are defective for BRCA1 (Garcia-Higuera et al., 2001). Furthermore, BRCA1 interacts with a ubiquitin hydrolase enzyme, BAP1 (Jensen et al., 1998). Finally, a number of groups have shown that truncated forms of BRCA1 and BARD1 containing the RING domains have E3 activity in vitro (Lorick et al., 1999; Hashizume et al., 2001; Ruffner et al., 2001; Chen et al., 2002).
One of the major limitations of in vitro systems using RING domain proteins has been the need for defined substrates (Lorick et al., 1999; Hashizume et al., 2001). The lack of specificity in these systems may well be due to the large regions of BRCA1 and BARD1 that are missing in these RING domain proteins. This consideration therefore led us to investigate the biochemical function of the full-length BRCA1/BARD1 complex.
Here, we examine the molecular function of the full-length BRCA1/BARD1 complex in vitro. We describe in detail the biochemical activity and the substrate specificity exhibited by the full-length BRCA1/BARD1 ubiquitin ligase and further show that the activity of the complex can be stimulated by auto-modification.
Results
BRCA1/BARD1 is an E3 mono-ubiquitin ligase
Previous studies have shown that the isolated RING domains of BRCA1 and BARD1 exhibit E3 ligase activity in vitro, promoting formation of polyubiquitin chains (Lorick et al., 1999; Hashizume et al., 2001; Ruffner et al., 2001). Since the great majority of the amino acid sequence is missing in these truncated forms of BRCA1 and BARD1, we wished to investigate the E3 ligase activity of the full-length proteins.
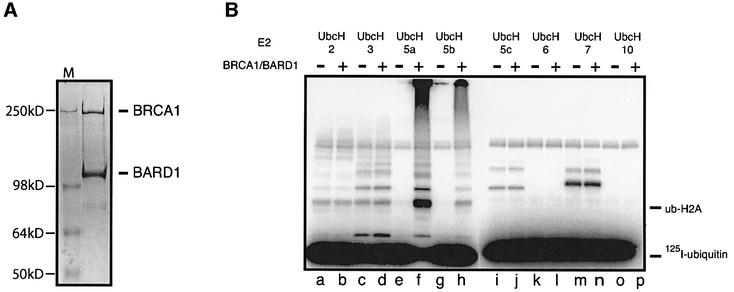
Until now, detailed biochemical studies of BRCA1 and BARD1 have been hampered by difficulty in isolating full-length proteins (Paull et al., 2001). Nevertheless, by co-expressing full-length human BRCA1 and BARD1 in insect cells, we were able to purify these proteins together as a complex. Analysis of the purified complex by SDS–PAGE demonstrated stoichiometric amounts of the BRCA1 and BARD1 proteins, consistent with the formation of a heterodimer (Figure 1A).
Fig. 1. BRCA1/BARD1 has E3 monoubiquitin ligase activity in vitro. (A) A Coomassie-stained gel of the full-length BRCA1/BARD1 complex. His-tagged BRCA1 and BARD1 proteins were co-expressed and co-purified as a complex from Sf9 insect cells as described in Materials and methods. A 20 µl aliquot of the purified complex was analysed by SDS–PAGE using a 6% Tris–glycine gel and visualized by staining with Simply Blue Safe Stain (Invitrogen). Molecular weight markers (M) and the BRCA1 and BARD1 proteins are indicated. (B) BRCA1/BARD1 complex exhibits ubiquitin ligase activity in the presence of the E2 enzyme, UbcH5a. Ubiquitin E3 ligase activity was investigated by incubating histone H2A substrate with BRCA1/BARD1 complex in the presence of E1 enzyme and a variety of E2 enzymes (as indicated). Reactions were carried out with a 30–60 min pre-incubation step as described in Materials and methods. 125I-Labelled products were analysed by SDS–PAGE and visualized using a PhosphorImager. [125I]ubiquitin substrate and 125I-ubiquitylated H2A products (Ub-H2A) are indicated.
To determine whether the BRCA1/BARD1 complex has E3 ligase activity, we examined its ability to modify a substrate with 125I-labelled ubiquitin ([125I]Ub) in the presence of ATP, an E1 enzyme and a range of E2 enzymes. Although genetic evidence suggests the FANCD2 protein as a likely substrate for BRCA1/BARD1-dependent ubiquitylation in vivo, we were unable to obtain sufficient quantities of this protein to investigate this in vitro. Instead, we used histone proteins as substrates. Not only are histone proteins monoubiquityl ated in vivo, but the variant histone, H2AX, co-localizes with BRCA1 at sites of DNA damage (Paull et al., 2001).
Incubation of BRCA1/BARD1 complex with histone H2A and [125I]Ub gave rise to an 125I-labelled product that migrated with an apparent Mr of 23 kDa after SDS–PAGE, consistent with the addition of a single [125I]Ub molecule to histone H2A (Figure 1B). Mass spectrometry and western blot analysis with antibodies against H2A and ubiquitin (data not shown) confirmed this product as ubiquitylated-H2A (Ub-H2A).
Although BRCA1/BARD1-dependent ubiquitylation demonstrated a clear preference for the E2 enzyme UbcH5a, very low levels of Ub-H2A were also detected with UbcH5b. In contrast Ub-H2A product was not detected with UbcH6, UbcH7 and UbcH10 (Figure 1B, lanes l, n and p). While Ub-H2A was also detected with the E2s UbcH2 and UbcH3, these reactions were not dependent on BRCA1/BARD1 (Figure 1B, lanes a–d) and were considerably less efficient than with UbcH5a. This is consistent with studies in yeast, which have shown that the E2 enzymes RAD6 and CDC34 (homologues of UbcH2 and UbcH3, respectively) can ubiquitylate histones H2A and H2B independently of an E3 enzyme (Robzyk et al., 2000).
Although in our assay BRCA1/BARD1-dependent modification of H2A was primarily monoubiquitylation, low levels of diubiquitylated H2A were also detected. This probably results from a minor contamination of the [125I]Ub with diubiquitin, rather than by successive additions of single ubiquitin molecules. Western blot analysis confirmed the absence of polyubiquitylated H2A products (data not shown).
The RING domains
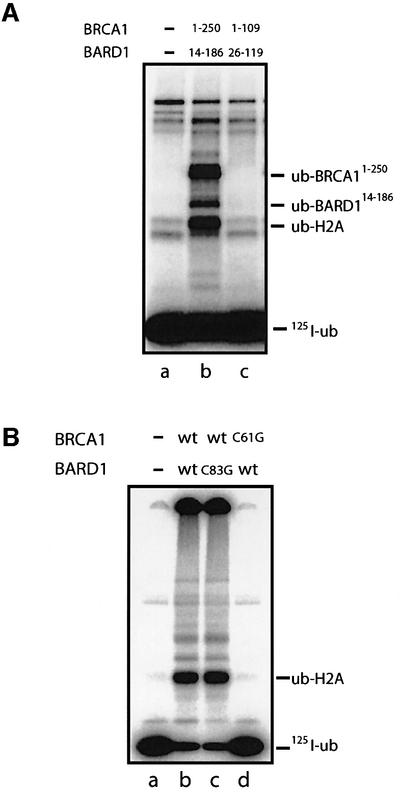
BRCA1 and BARD1 are known to associate through their RING domains. Indeed the minimal interacting RING domain proteins, BRCA11–109 and BARD126–119, have been shown to form a stable RING–RING heterodimeric complex in vitro (Brzovic et al., 2001). Nevertheless, we found that these proteins were defective for E3 activity (Figure 2A). In contrast, larger RING-containing proteins, BRCA11–250 and BARD114–186, which contain additional flanking sequences, efficiently ubiquitylated H2A when present together in the reaction (Figure 2A, lane b). Clearly the RING domains of BRCA1 and BARD1, while necessary, are not sufficient for E3 activity, and sequences outside the RING domains make an important contribution to this function.
Fig. 2. The RING domains of BRCA1 and BARD1 are necessary but not sufficient for E3 activity. (A) Sequences outside the minimal interacting RING domains of BRCA1 and BARD1 are required for ubiquitin ligase activity. Ubiquitin ligase activity of two RING domain complexes, BRCA11–109/BARD126–119 and BRCA11–250/BARD114–186, was investigated. Reactions were incubated for 1.5 h at 37°C as described in Materials and methods. Ubiquitylated H2A product (Ub-H2A) and ubiquitylated BRCA1 and BARD1 products (Ub-BRCA11–250 and Ub-BARD114–186) are indicated. (B) Mutation of a key metal-binding residue in BRCA1 but not BARD1 abolishes the E3 activity of BRCA1/BARD1. Mutant complexes BRCA1(C61G)/BARD1 and BRCA1/BARD1(C83G) were examined for E3 activity as described in Figure 1. Each reaction contained 0.1 µg of BRCA1/BARD1 complex. Ubiquitylated H2A (Ub-H2A) product is indicated.
Many cancer-predisposing missense mutations in BRCA1 are found in key metal-binding residues of its RING domain (Couch and Weber, 1996). Previous studies have shown that the E3 activity of the RING domain proteins is abolished by some of these mutations (Hashizume et al., 2001; Ruffner et al., 2001). Since these RING domain proteins contained only small regions of the BRCA1 protein, it was important to examine these mutations in the context of the full-length complex. One such mutant, BRCA1(C61G), was examined in a complex with wild-type BARD1 and found to be completely defective for ubiquitin ligase activity (Figure 2B, lane d). This provides definitive proof that a single amino acid substitution in BRCA1, known to confer a pre-disposition to breast cancer, results directly in the loss of BRCA1/BARD1 ubiquitin ligase function in vitro.
The RING domains of BRCA1 and BARD1 do not, however, play equivalent roles in the ubiquitin ligase function of the BRCA1/BARD1 complex. Whereas BRCA1(C61G) protein was defective in E3 ligase function, the BARD1(C83G) mutant protein, which has an equivalent RING substitution in BARD1, exhibited wild-type activity (Figure 2B, lane c).
Substrate specificity in vitro
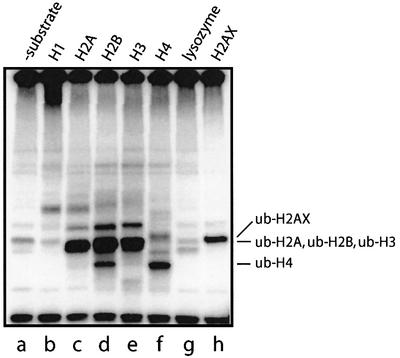
Histone proteins are known to be monoubiquitylated in vivo. Whereas in yeast histones H2A and H2B are ubiquitylated by the RAD6 protein (Robzyk et al., 2000), it remains unclear which enzymes carry out ubiquitylation of histones in mammalian cells. Intriguingly, BRCA1 co-localizes with the histone variant H2AX at sites of DNA damage (Paull et al., 2000). Since H2AX is also known to be ubiquitylated in vivo, we examined whether it, and other histone proteins, were substrates for BRCA1/BARD1-dependent ubiquitylation in vitro. Strikingly, as well as H2AX, the nucleosome core histones H2A, H2B, H3 and H4 were all ubiquitylated efficiently by BRCA1/BARD1, while the linker histone H1 was not (Figure 3). Additionally, denatured lysozyme, which has been used as a generic substrate for ubiquitylation by other E3 ligases, was not ubiquitylated by BRCA1/BARD1 (Figure 3, lane g). Hence, in vitro the ubiquitin ligase function of the BRCA1/BARD1 complex exhibits defined substrate specificity, with a strong preference for nucleosome core histones.
Fig. 3. Substrate specificity of the BRCA1/BARD1 complex. A 0.1 µg aliquot of BRCA1/BARD1 was incubated with 1 µg of the indicated substrate as described in Materials and methods. Lysozyme (lane g) was denatured previously, by heating to 100°C for 15 min in a waterbath. 125I-Labelled products were analysed by SDS–PAGE and visualized with a Molecular Dynamics Typhoon PhosphorImager.
Auto-ubiquitylation of BRCA1/BARD1
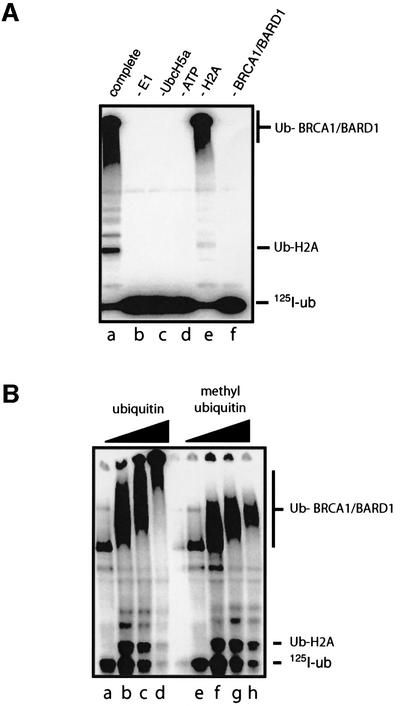
As well as ubiquitylated histones, we detected an additional 125I-ubiquitylated product that migrated near the well after SDS–PAGE (Figure 1B, lane f, and Figure 4A, lanes a and e). Western blot analysis revealed that this high molecular weight product comprised ubiquitylated forms of BRCA1 and BARD1 (Ub-BRCA1/BARD1) (data not shown), indicating that the BRCA1/BARD1 complex promotes its own ubiquitylation. Since Ub-BRCA1/BARD1 was detected in the absence of H2A, this modification was not simply a byproduct of the ubiquitylation of histones (Figure 4A, lane e).
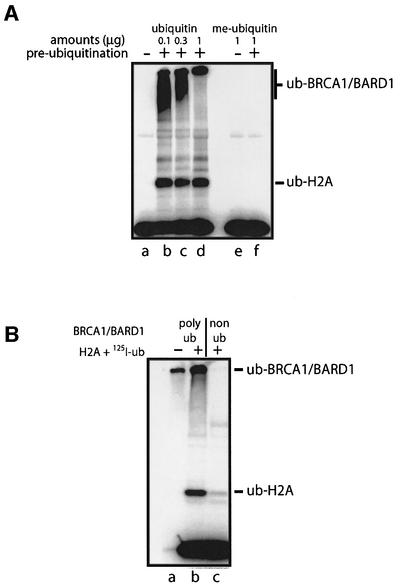
Fig. 4. Auto-ubiquitylation of BRCA1/BARD1 complex involves addition of multiple polyubiquitin chains. (A) BRCA1/BARD1-dependent ubiquitylation of H2A requires all the key reaction components. Reactions were carried out as described in Materials and methods, but omitting the indicated component from the reaction. The complete reaction is shown in lane a. Ubiquitylated BRCA1/BARD1 com plex (Ub-BRCA1/BARD1) and ubiquitylated H2A are indicated. (B) Ubiquitylation of the BRCA1/BARD1 complex in the presence of unmodified ubiquitin (lanes a–d) or methyl ubiquitin (lanes e–h). Reactions were carried out as outlined in Materials and methods using 0, 0.1, 0.3 or 1 µg (lanes a and e, b and f, c and g, and d and h) of ubiquitin or methyl ubiquitin as indicated. Reactions were performed for 60 min. Products were analysed by SDS–PAGE on a 4–20% Tris–glycine gel. Ubiquitylated H2A (Ub-H2A) and auto-ubiquitylated BRCA1/BARD1 products (Ub-BRCA1/BARD1) are indicated.
The large molecular weight of the ubiquitylated BRCA1 and BARD1 proteins suggested they were modified with multiple ubiquitin molecules. While this might arise through the attachment of polyubiquitin chains, it might also arise through modification with single ubiquitin molecules at multiple lysine residues on these large proteins. To investigate these possibilities, we used methyl ubiquitin, which cannot form polyubiquitin chains. When BRCA1/BARD1 was incubated with increasing concentrations of methyl ubiquitin, the Ub-BRCA1/BARD1 products were increasingly retarded during SDS–PAGE, consistent with ubiquitylation at multiple sites (Figure 4B, lanes e–h). At the highest concentrations of methyl ubiquitin, this retardation was limited, presumably as the sites of ubiquitylation became saturated (Figure 4B, lane h). In contrast, modification with ubiquitin, which permits formation of extended chains, gave rise to products that were retarded as far as the well, suggesting that they were much larger (Figure 4B, lanes a–d). From this, we infer that auto-ubiquitylation of BRCA1/BARD1 probably involves the formation of polyubiquitin chains at multiple sites on each protein. Hence, BRCA1/BARD1 catalyses two distinct forms of modification; monoubiquitylation of heterologous substrates and polyubiquitylation of itself.
Auto-ubiquitylation stimulates the ubiquitin ligase activity of BRCA1/BARD1
Although polyubiquitylation is most often regarded as a signal for protein degradation by the proteasome, it has also been shown to stimulate the activity of at least one E3 ligase, TRAF-6 (Wang et al., 2001). We therefore investigated whether, at low concentrations of BRCA1/BARD1 complex, polyubiquitylation might stimulate E3 ligase function similarly. To this end, BRCA1/BARD1 was pre-incubated under conditions that permitted its polyubiquitylation. The E3 activity of this ubiquitylated complex was then measured by the addition of histone H2A substrate and [125I]Ub. In control reactions, BRCA1/BARD1 was omitted from the pre-incubation step and then added along with the H2A substrate and [125I]Ub. Analysis of 125I-labelled products by SDS–PAGE clearly demonstrated that ubiquitylation of H2A was more efficient with the polyubiquitylated form of the BRCA1/BARD1 complex (Figure 5A). Furthermore, stimulation was a property of polyubiquitin chains since it was not observed with BRCA1/BARD1 complexes that had been pre-ubiquityl ated with methyl ubiquitin (Figure 5A, lanes e and f).
Fig. 5. Ubiquitylation of BRCA1/BARD1 complex potentiates its E3 ligase activity. (A) A 0.025 µg aliquot of BRCA1/BARD1 was pre-incubated at 37°C with the indicated amounts of ubiquitin or methyl ubiquitin in conditions permissible for auto-ubiquitylation (see Materials and methods). After 2 h, 1 µg of H2A substrate, excess [125I]ubiquitin and ATP were added to each reaction, followed by a further 20 min incubation at 37°C. Reactions were stopped by the addition of SDS loading dye and boiling for 2 min (+pre-incubation). In control reactions, BRCA1/BARD1 was omitted from the pre-incubation step and added instead with H2A and [125I]ubiquitin (–pre-incubation). Products were analysed by SDS–PAGE and visualized using a PhosphorImager. The ubiquitylated H2A and BRCA1/BARD1 products are indicated. (B) Ubiquitylation of H2A by polyubiquitylated and non-polyubiquitylated BRCA1/BARD1 complex. Polyubiquitylated and non-ubiquitylated forms of the BRCA1/BARD1 complex were isolated as described in Materials and methods. E3 activity was examined in fresh reactions as described in Figure 1, and analysed by SDS–PAGE. Ubiquitylated BRCA1/BARD1 complexes and ubiquitylated H2A are indicated.
To confirm this result, we purified polyubiquitylated and non-ubiquitylated forms of BRCA1/BARD1 using nickel-NTA resin. These isolated complexes were washed and added to fresh reactions to measure their E3 activity with H2A. Once again, ubiquitylation of H2A was >20-fold more efficient with polyubiquitylated BRCA1/BARD1 complex than with the non-ubiquitylated form (Figure 5B). Clearly, polyubiquitylation of the BRCA1/BARD1 complex stimulates its E3 function.
Deubiquitylation
The ubiquitylation of many proteins can be counter balanced by the action of deubiquitylating enzymes. Interestingly, BRCA1 is known to associate in vivo with a C-terminal ubiquitin hydrolase enzyme, BAP1 (Jensen et al., 1998). Therefore, we wished to determine whether BAP1 could regulate BRCA1/BARD1 E3 activity through deubiquitylation. To this end, recombinant BAP1 was expressed and purified from Escherichia coli and shown to have ubiquitin hydrolase activity on an artificial substrate Ub-AMC (Figure 6A). As expected, the mutant protein BAP1(C91S), which carries an active site mutation, was unable to hydrolyse Ub-AMC. However, when wild-type BAP1 was incubated with the polyubiquitylated form of BRCA1/BARD1, we observed no deubiquitylation of the complex (Figure 6B). On the contrary, isopeptidase T, a C-terminal ubiquitin hydrolase involved in the recycling of ubiquitin, deubiquitylated BRCA1/BARD1 very efficiently, releasing [125I]Ub. While we currently do not rule out the possibility that in the presence of additional factors BAP1 might function to deubiquitylate BRCA1/BARD1, we think it unlikely that this is its primary role.
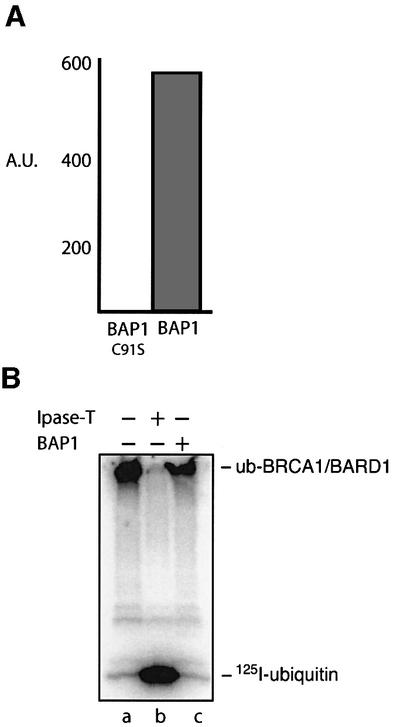
Fig. 6. BAP1 does not deubiquitylate BRCA1/BARD1 complex. (A) Recombinant BAP1 has C-terminal ubiquitin hydrolase activity. Recombinant BAP1 and the mutated protein BAP1(C91S) were incubated with ubiquitin-AMC as described in Materials and methods. Ubiquitin hydrolysis was measured by fluorescence using a Perkin Elmer Luminescence Spectrometer with excitation wavelength of 380 nm and emission wavelength of 460 nm. Fluorescence is plotted as arbitrary units (A.U.) (B) The 125I-polyubiquitylated form of BRCA1/BARD1 was isolated using nickel-NTA resin, washed in buffer and incubated with 100 nM BAP1 protein or 100 nM isopeptidase T as described in Materials and methods. Products were analysed by SDS–PAGE and deubiquitylation determined by the release of [125I]ubiquitin.
Discussion
Although BRCA1 has been implicated in a variety of cellular processes, its molecular function remains unclear. Data presented here and elsewhere provide compelling evidence that at least one biochemical function of BRCA1 is as an E3 ubiquitin ligase.
As with other E3s, BRCA1/BARD1-dependent ubiquitylation requires E1 and E2 enzymes, as well as ATP. In particular, BRCA1/BARD1 demonstrated a preference for the E2, UbcH5a. In contrast to earlier studies (Lorick et al., 1999; Ruffner et al., 2001), but in agreement with Hashizume et al. (2001), we find that E3 function requires both the BRCA1 and BARD1 proteins. Furthermore, while truncated forms of BRCA1 and BARD1 could be purified separately and mixed to reconstitute E3 activity, the full-length proteins were only active when co-expressed and purified as a complex (C.J.Vandenberg, unpublished data).
The purified complex contains equimolar amounts of the BRCA1 and BARD1 proteins, consistent with earlier reports that they exist as a heterodimer in vivo (Brzovic et al., 2001). Within the complex, BRCA1 and BARD1 have been shown to interact specifically through their RING domains. However, even though the minimal RING domain proteins BRCA11–109 and BARD126–119 form a stable heterodimeric complex in vitro, they do not exhibit E3 activity. On the contrary, the larger RING domain proteins BRCA11–250 and BARD114–186 do have E3 activity, indicating that sequences which lie outside the RING domain are important for E3 function. Although these flanking regions may contribute to the structure of the complex helping it maintain an active conformation, it is also possible that these additional sequences contain important catalytic residues necessary for ubiquitin transfer.
The metal-binding residues of the BRCA1 RING domain are clearly important for ubiquitin transfer. A single amino acid substitution in BRCA1(C61G) abolishes the E3 activity of the BRCA1(C61G)/BARD1 complex in vitro. Although similar observations were reported previously, these studies used severely truncated forms of the BRCA1 protein (Hashizume et al., 2001; Ruffner et al., 2001). We can now be confident that this single amino acid substitution, which confers predisposition to breast and ovarian cancer, is by itself sufficient to cause loss of BRCA1/BARD1 ubiquitin ligase function. Interestingly, an equivalent mutation in the metal-binding site of the BARD1 RING domain does not affect E3 activity. It is likely, therefore, that the catalytic residues for ubiquitin transfer reside specifically in the RING domain of BRCA1.
The BRCA1/BARD1 complex exhibits a dual specificity, catalysing two distinct types of ubiquitin chain modification; monoubiquitylation of heterologous substrate and polyubiquitylation of itself. Currently, very little is known about why modification of some lysine residues is limited to a single ubiquitin molecule while others become modified with ubiquitin chains. In our reactions, BRCA1/BARD1-dependent monoubiquitylation and polyubiquitylation exhibit similar requirements for E1, UbcH5a and ATP. Rather, the major determining factor for mono- versus polyubiquitylation by BRCA1/BARD1 appears to be the substrate. Perhaps as more substrates are identified and their sites of ubiquitylation determined, it will be possible to define specific consensus sequences for sites of monoubiquitylation and polyubiquitylation. Alternatively, steric factors associated with particular substrates may limit modification to a single ubiquitin molecule.
The potentiation of BRCA1/BARD1-dependent E3 activity as a consequence of auto-ubiquitylation is intriguing since it suggests that BRCA1/BARD1 is regulated through a positive feedback mechanism. Indeed, this effect is quite large, with the polyubiquitylated form of the complex exhibiting a >20-fold increase in E3 activity compared with the unmodified form. In vivo stimulation of BRCA1/BARD1 E3 activity might be important to deliver a rapid pulse of BRCA1-dependent ubiquitylation of specific substrates, for example in response to DNA damage. However, one can also envisage the need for a mechanism to limit this positive feedback loop. This could be achieved, for example, through destruction of the polyubiquitylated BRCA1/BARD1 complex by the proteasome. Alternatively, the BRCA1/BARD1 complex might be down-regulated through the action of a deubiquitylating enzyme such as a C-terminal ubiquitin hydrolase. However, although BRCA1 associates with the C-terminal ubiquitin hydrolase, BAP1, in vivo (Jensen et al., 1998), this protein does not deubiquitylate BRCA1/BARD1 in vitro. In contrast, isopeptidase T, another C-terminal ubiquitin hydrolase associated with ubiquitin recycling and regulation of the proteasome, very efficiently deubiquitylated BRCA1 in vitro. Hence, while it is possible that BAP1 deubiquitylates BRCA1/BARD1 under different conditions or with additional cofactors, it is more likely that this enzyme acts on other substrates.
Activation of the BRCA1/BARD1 complex is dependent on modification with polyubiquitin chains and was not observed with complexes modified with methyl ubiquitin. We currently envisage two mechanisms through which activation of E3 function might occur. First, ubiquitylation at one or more sites could produce allosteric changes in the BRCA1/BARD1 complex that directly increase its E3 activity. Alternatively, polyubiquitin chains themselves might increase the affinity of the complex for its substrate or for UbcH5a, thereby increasing the rate of ubiquitin transfer. Recently, another RING E3 ligase, TRAF6, which acts in the IκB signalling pathway, was shown to be activated by auto-ubiquitylation (Wang et al., 2001). Although the mechanism of TRAF6 activation is not fully understood, evidence suggests that it is mediated through variant polyubiquitin chains that are linked through Lys63 residues, rather than Lys48. These mechanisms currently are being investigated.
Although polyubiquitylation is known to play a critical role in targeting proteins for degradation by the proteasome, much less is known about monoubiquitylation. Nevertheless, this post-translational modification seems to play an important role in regulating the location and activity of cellular proteins (reviewed in Hicke, 2001). In yeast, for example, monoubiquitylation provides a signal for proteins located in the plasma membrane to be internalized for entry into the endocytic pathway. In retroviruses, monoubiquitylation of the GAG protein plays an important role in viral budding.
The major genetic evidence supporting ubiquitin ligase function for BRCA1 in vivo comes from studies on the FANCD2 protein. Whereas in wild-type cells the FANCD2 protein co-localizes with BRCA1 in nuclear foci and becomes monoubiquitylated in response to DNA damage, HCC1937 cells, which encode a mutated form of BRCA1, are largely defective for both monoubiquitylation of FANCD2 and foci formation (Garcia-Higuera et al., 2001). However, since HCC1937 retains low levels of monoubiquitylated FANCD2, it remains to be demonstrated whether BRCA1 is the only E3 ligase able to ubiquitylate this protein. Furthermore, the role of FANCD2 ubiquitylation in genomic stability and the part played by the other Fanconi anaemia proteins in this process also remain unclear.
In vitro, nucleosome core histones are ubiquitylated very efficiently by the BRCA1/BARD1 complex. What then is the role of BRCA1/BARD1-dependent ubiquitylation of histones in vivo? In yeast, disruption of histone ubiquitylation leads to defects in mitotic cell growth, meiosis and DNA repair (Martini et al., 2002; Robzyk et al., 2000). In addition, recent data have demonstrated that ubiquitylation of histone H2B also plays a crucial role in gene silencing (Sun and Allis, 2002). Although it is possible that BRCA1 and BARD1 are involved in any or all of these processes in mammalian cells, it is difficult to draw direct parallels since yeast do not encode homologues of these proteins. Moreover, in yeast, ubiquitylation of histones is performed by RAD6, an E2 enzyme (Robzyk et al., 2000), rather than an E3. Whilst mammalian cells have two homologues of the yeast RAD6 protein (HR6A and HR6B) (Koken et al., 1992), it is not clear whether these proteins are involved in a general pathway for histone ubiquitylation. It is entirely possible, however, that more than one pathway exists for ubiquitylation of histones in mammals and that BRCA1/BARD1 might act specifically during the cell cycle or after DNA damage.
In mammalian cells, BRCA1 co-localizes in nuclear foci with the phosphorylated form of the variant histone H2AX (known as γH2AX), at the sites of DNA breaks (Paull et al., 2000). Since H2AX is a substrate for BRCA1/BARD1-dependent ubiquitylation in vitro, it is tempting to speculate that these nuclear foci are sites of active H2AX ubiquitylation by BRCA1/BARD1 and that modification occurs specifically at sites of DNA damage. Although the exact function of H2AX is not well understood, mice lacking H2AX exhibit sensitivity to DNA damage and profound chromosomal instability (Celeste et al., 2002). Generally, histone proteins are required for compaction of nuclear DNA into chromatin, and their modification is thought to loosen this compaction. Therefore, one might envisage that ubiquitylation of γH2AX by BRCA1/BARD1 at DNA breaks modulates local chromatin packaging to facilitate the action of DNA repair enzymes. It is noteworthy, therefore, that the DNA repair complex MRE11/RAD50/NBS1 also co-localizes with γH2AX and BRCA1 at DNA breaks (Zhong et al., 1999; Paull et al., 2000), further supporting a link with DNA repair.
For a long time, BRCA1 has been described largely in terms of the catastrophic phenotype that accompanies its loss of function, and its large number of molecular interactions. The demonstration that BRCA1 functions as a E3 ubiquitin ligase in vitro is certainly in keeping with its role as a key regulator for maintaining genomic stability, and explains how it can influence a diverse range of cellular processes. Furthermore, the development of an in vitro system, using full-length BRCA1/BARD1 complex, provides an invaluable tool for dissecting the biochemical mechanism of BRCA1-dependent ubiquitylation. The goal now is to identify substrates for the ubiquitin ligase function of BRCA1/BARD1 and to determine at a biochemical level the consequences of their modification. In this way, we hope to define the molecular pathways through which BRCA1 contributes to maintenance of genomic stability and gain insight into how defects in these pathways lead eventually to malignant transformation.
Materials and methods
Proteins
Histones H2A, H2B, H3 and H4 were purchased from Roche. Lysozyme was purchased from Sigma. Rabbit reticulocyte E1 enzyme, E2 enzymes, isopeptidase T and Ub-AMC were purchased from Boston Biochem.
Purification of BRCA1 and BARD1
Full-length human BRCA1 and BARD1 (gift from R.Baer) were tagged with His12 and FLAG and cloned into pDEST8 using the Gateway cloning system (Invitrogen). Recombinant baculovirus was generated using the Bac-to-Bac baculovirus expression system (Invitrogen). Viruses vBRCA1-FLH12 and vBARD1-FLH12 were co-infected in Sf9 insect cells for 48 h, after which cells were harvested by centrifugation at 1500 × g for 5 min in a Sorvall RT7. Cleared cell lysates were prepared from fresh cells by sonication in buffer A (20 mM Tris–HCl pH 7.5, 500 mM KCl, 10% glycerol, 0.1% Triton X-100, 2 mM 2-mercapto ethanol) containing 5 mM imidazole and complete protease inhibitors (Roche), followed by ultracentrifugation at 55 000 r.p.m. for 1 h. The lysate was loaded onto a HiTrap Ni2+ chelating column (Pharmacia) and washed with 10 column volumes of buffer A (5 mM imidazole) followed by 10 column volumes of buffer A, containing 60 mM imidazole. Proteins were eluted in 10 ml of buffer A containing 750 mM imidazole, and diluted 2-fold by addition of buffer B [20 mM Tris–HCl pH 7.5, 10% glycerol, 0.1% Triton X-100, 2 mM dithiothreitol (DTT)] to give a final concentration of 250 mM KCl. The Ni2+ eluate was loaded onto a 1 ml heparin column and washed with buffer B containing 250 mM KCl. BRCA1/BARD1 was eluted with a 250 mM–1 M KCl gradient. Peak protein fractions, which eluted at ∼800 mM KCl, were dialysed against storage buffer (25 mM Tris–HCl pH 7.5, 10% glycerol and 2 mM DTT) containing 150 mM KCl. Aliquots were frozen on liquid nitrogen and stored at –80°C.
Bacterially expressed proteins His6-BAP1, His6-BRCA1 (1–250) and (1–109), and His6-BARD (14–186) and (26–119) were purified using Ni-NTA resin, (Qiagen) as described above for BRCA1/BARD1.
H2AX purification
Full-length human H2AX was cloned into the vector pOPTH (gift from O.Perisic). His-tagged H2AX protein was expressed in bacterial strain BL21 (DE3), after induction with 0.4 mM isopropyl-β-d-thiogalacto pyranoside (IPTG) at 37°C. Cells were harvested and H2AX was purified as described for BRCA1/BARD1 with elution off heparin at 1 M KCl. Protein-containing fractions were dialysed against storage buffer containing 250 mM KCl, frozen on liquid nitrogen and stored at –80°C.
Ubiquitylation assays
Unless stated otherwise, reactions (10 µl) contained 50 ng of purified BARD1/BRCA1, 300 ng of E1 and 300 ng of UbcH5a (Boston Biochem), 1 µg of ubiquitin (Sigma), 0.5 µg of [125I]Ub (Amersham) and 1 µg of the indicated substrate, in buffer (50 mM Tris–HCl pH 7.5, 2.5 mM MgCl2, 15 mM KCl, 0.7 mM DTT, 0.01% Triton, 1% glycerol and 2 mM ATP). Reactions were incubated for 30–60 min at 37°C and stopped by addition of SDS sample buffer containing 1 mM DTT. Samples were heated at 100°C for 5 min and the products resolved by SDS–PAGE. 125I-Labelled products were visualized using the Molecular Dynamics Typhoon PhosphorImager and ImageQuaNT software.
Where indicated, BRCA1/BARD1 was first pre-incubated under the conditions outlined above but without H2A substrate and [125I]Ub, which were added at a later time. Reactions with methyl ubiquitin (Sigma) were carried out similarly, using the concentrations of methyl ubiquitin stated in the figure legends.
Deubiquitylation assays
BAP1 or BAP1(C91S) (both at 100 nM) were incubated with Ub-AMC (1 µM) in reaction buffer (50 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM DTT). Reactions were performed as described in Dang et al. (1998) using excitation and emission wavelengths of 380 and 460 nm, respectively. Hydrolysis of Ub-AMC was measured by fluorescence using a Perkin Elmer Luminescence Spectrometer and quantified in arbitrary units. Deubiquitylation of 125I-polyubiquitylated BRCA1/BARD1 complex was carried out using 250 ng of BRCA1/BARD1 complex which had been ubiquitylated with [125I]Ub as described above and then purified using Ni2+-NTA resin (Qiagen). Nickel–agarose beads containing polyubiquitylated complex were washed with 25 mM HEPES pH 7.0 and added to 10 µl of deubiquitylation reaction buffer. Then, 100 nM BAP1 or isopeptidase T was added as indicated and incubation carried out at 37°C for 30 min. Products were analysed by SDS–PAGE using a 14% Tris–glycine gel (Invitrogen) and 125I-labelled products visualized using a PhosphorImager.
Acknowledgments
Acknowledgements
We wish to thank Dr Richard Baer for the kind gift of BRCA1 and BARD1 cDNAs, and Dr Olga Perisic for plasmid pOPTH. We also thank our colleagues at the Laboratory of Molecular Biology for advice and valuable discussions. C.J.V. is supported by a grant from the Association for International Cancer Research, St Andrews, Scotland, to K.H.
References
- Brzovic P.S., Rajagopal,P., Hoyt,D.W., King,M.C. and Klevit,R.E. (2001) Structure of a BRCA1–BARD1 heterodimeric RING–RING complex. Nat. Struct. Biol., 8, 833–837. [DOI] [PubMed] [Google Scholar]
- Celeste A. et al. (2002) Genomic instability in mice lacking histone H2AX. Science, 296, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Kleiman,F.E., Manley,J.L., Ouchi,T. and Pan,Z.Q. (2002) Auto-ubiquitination of the BRCA1/BARD1 RING ubiquitin ligase. J. Biol. Chem., 277, 22085–22092. [DOI] [PubMed] [Google Scholar]
- Couch F.J. and Weber,B.L. (1996) Mutations and polymorphisms in the familial early-onset breast cancer (BRCA1) gene. Breast Cancer Information Core. Hum. Mutat., 8, 8–18. [DOI] [PubMed] [Google Scholar]
- Dang L.C., Melandri,F.D. and Stein,R.L. (1998) Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry, 37, 1868–1879. [DOI] [PubMed] [Google Scholar]
- Deng C.X. and Brodie,S.G. (2000) Roles of BRCA1 and its interacting proteins. Bioessays, 22, 728–737. [DOI] [PubMed] [Google Scholar]
- Deng C.X. and Scott,F. (2000) Role of the tumor suppressor gene Brca1 in genetic stability and mammary gland tumor formation. Oncogene, 19, 1059–1064. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I., Taniguchi,T., Ganesan,S., Meyn,M.S., Timmers,C., Hejna,J., Grompe,M. and D’Andrea,A.D. (2001) Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell, 7, 249–262. [DOI] [PubMed] [Google Scholar]
- Hashizume R., Fukuda,M., Maeda,I., Nishikawa,H., Oyake,D., Yabuki,Y., Ogata,H. and Ohta,T. (2001) The RING heterodimer BRCA1–BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem., 276, 14537–14540. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hicke L. (2001) Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell. Biol., 2, 195–201. [DOI] [PubMed] [Google Scholar]
- Huibregtse J.M., Scheffner,M., Beaudenon,S. and Howley,P.M. (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl Acad. Sci. USA, 92, 2563–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen D.E. et al. (1998) BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene, 16, 1097–1112. [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A. and Weissman,A.M. (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell, 102, 549–552. [DOI] [PubMed] [Google Scholar]
- Joukov V., Chen,J., Fox,E.A., Green,J.B. and Livingston,D.M. (2001) Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl Acad. Sci. USA, 98, 12078–12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koken M.H., Smit,E.M., Jaspers-Dekker,I., Oostra,B.A., Hagemeijer,A., Bootsma,D. and Hoeijmakers,J.H. (1992) Localization of two human homologs, HHR6A and HHR6B, of the yeast DNA repair gene RAD6 to chromosomes Xq24–q25 and 5q23–q31. Genomics, 12, 447–453. [DOI] [PubMed] [Google Scholar]
- Lorick K.L., Jensen,J.P., Fang,S., Ong,A.M., Hatakeyama,S. and Weissman,A.M. (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl Acad. Sci. USA, 96, 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E.M., Keeney,S. and Osley,M.A. (2002) A role for histone H2B during repair of UV-induced DNA damage in Saccharomyces cerevisiae. Genetics, 160, 1375–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza J.E., Brzovic,P.S., King,M.C. and Klevit,R.E. (1999) Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J. Biol. Chem., 274, 5659–5665. [DOI] [PubMed] [Google Scholar]
- Miki Y. et al. (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science, 266, 66–71. [DOI] [PubMed] [Google Scholar]
- Paull T.T., Rogakou,E.P., Yamazaki,V., Kirchgessner,C.U., Gellert,M. and Bonner,W.M. (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol., 10, 886–895. [DOI] [PubMed] [Google Scholar]
- Paull T.T., Cortez,D., Bowers,B., Elledge,S.J. and Gellert,M. (2001) From the cover: direct DNA binding by Brca1. Proc. Natl Acad. Sci. USA, 98, 6086–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem., 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Robzyk K., Recht,J. and Osley,M.A. (2000) Rad6-dependent ubiquitination of histone H2B in yeast. Science, 287, 501–504. [DOI] [PubMed] [Google Scholar]
- Ruffner H., Joazeiro,C.A., Hemmati,D., Hunter,T. and Verma,I.M. (2001) Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl Acad. Sci. USA, 98, 5134–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Anderson,S.F., Chao,D.M., Wei,W., Ye,L., Young,R.A., Livingston,D.M. and Parvin,J.D. (1997a) BRCA1 is a component of the RNA polymerase II holoenzyme. Proc. Natl Acad. Sci. USA, 94, 5605–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Chen,J., Plug,A., Xiao,Y., Weaver,D., Feunteun,J., Ashley,T. and Livingston,D.M. (1997b) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell, 88, 265–275. [DOI] [PubMed] [Google Scholar]
- Sun Z.W. and Allis,C.D. (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature, 418, 104–108. [DOI] [PubMed] [Google Scholar]
- Wang C., Deng,L., Hong,M., Akkaraju,G.R., Inoue,J. and Chen,Z.J. (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature, 412, 346–351. [DOI] [PubMed] [Google Scholar]
- Weissman A.M. (2001) Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell. Biol., 2, 169–178. [DOI] [PubMed] [Google Scholar]
- Wu L.C. et al. (1996) Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet., 14, 430–440. [DOI] [PubMed] [Google Scholar]
- Zhang H., Somasundaram,K., Peng,Y., Tian,H., Bi,D., Weber,B.L. and El-Deiry,W.S. (1998) BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene, 16, 1713–1721. [DOI] [PubMed] [Google Scholar]
- Zhong Q., Chen,C.F., Li,S., Chen,Y., Wang,C.C., Xiao,J., Chen,P.L., Sharp,Z.D. and Lee,W.H. (1999) Association of BRCA1 with the hRad50–hMre11–p95 complex and the DNA damage response. Science, 285, 747–750. [DOI] [PubMed] [Google Scholar]