Abstract
Senescence is a sequence of biochemical and physiological events that constitute the final stage of development. The identification of genes that alter senescence has practical value and is helpful in revealing pathways that influence senescence. However, the genetic mechanisms of senescence are largely unknown. The leaf of the oresara9 (ore9) mutant of Arabidopsis exhibits increased longevity during age-dependent natural senescence by delaying the onset of various senescence symptoms. It also displays delayed senescence symptoms during hormone-modulated senescence. Map-based cloning of ORE9 identified a 693–amino acid polypeptide containing an F-box motif and 18 leucine-rich repeats. The F-box motif of ORE9 interacts with ASK1 (Arabidopsis Skp1-like 1), a component of the plant SCF complex. These results suggest that ORE9 functions to limit leaf longevity by removing, through ubiquitin-dependent proteolysis, target proteins that are required to delay the leaf senescence program in Arabidopsis.
INTRODUCTION
Leaf development ends with senescence, a process consisting of deterioration events that ultimately lead to death (Noodén, 1988). During senescence, leaf cells experience dramatic changes in metabolism. The most striking phenotypic change is the yellowing of the leaf caused by the preferential breakdown of chlorophyll and chloroplasts (Gut et al., 1987). The loss of the photosynthetic pigment chlorophyll and the breakdown of the structural integrity of the chloroplasts attenuate energy-requiring anabolic events such as photosynthesis and protein synthesis. De novo synthesis of specific proteins also is required for senescence. Although degenerative in nature, leaf senescence is an active process programmed by genetic information. Thus, it is not surprising that there are genetic variants with defects that affect the senescence program. Several genetic loci that alter specific senescence symptoms have been identified in Arabidopsis as well as in a variety of crop plants (Thomas, 1987; Oh et al., 1997). In addition, extensive molecular studies have found even more sets of genes involved in leaf senescence (Davies and Grierson, 1989; Hensel et al., 1993; Buchanan-Wollaston, 1994, 1997; Lohman et al., 1994; Park et al., 1998).
Although senescence occurs in an age-dependent manner in many species (Noodén, 1988), the initiation and progression of senescence can be modulated by a variety of environmental factors such as temperature, mineral deficiency, drought conditions, and pathogen infection. It is known that internal factors such as plant growth regulators, reproduction, and cellular differentiation also influence senescence (Thomas and Stoddart, 1980; Smart, 1994). Among these internal factors, plant hormones have been characterized most thoroughly at the molecular and physiological levels. For example, abscisic acid (ABA) regulates many aspects of plant growth and development, including tolerance to adverse environmental conditions as well as seed maturation and dormancy (Leung et al., 1997). ABA also induces leaf abscission, flower senescence, and leaf senescence (Zeevaart and Creelman, 1988). Ethylene has been known as an endogenous modulator of senescence, including fruit ripening and flower and leaf senescence (Abeles et al., 1988). Indeed, several ethylene-insensitive mutants, etr1, er, ein2, and ein3, were shown to have delayed leaf senescence symptoms (Zacarias and Reid, 1990; Grbić and Bleecker, 1995; Chao et al., 1997; Oh et al., 1997). Jasmonates induce plant defense responses and mechanical wounding (Bell et al., 1995; Reymond and Farmer, 1998). In addition, jasmonates are important cellular regulators of diverse developmental processes, such as flower and fruit development, leaf abscission, and senescence (Creelman et al., 1992). Other previous analyses also showed the effectiveness of ABA, ethylene, and methyl jasmonate (MeJA) in promoting leaf senescence in Arabidopsis (Oh et al., 1996, 1997; Weaver et al., 1998).
The three delayed senescence mutants ore1 (oresara1; oresara means “long living” in Korean), ore3, and ore9 were isolated previously (Oh et al., 1997). These mutants exhibit delayed senescence symptoms in age-dependent in planta leaf senescence as well as in dark-induced senescence. As mentioned above, leaf senescence in plants is affected not just by endogenous developmental factors such as aging and hormones but also by environmental factors. To gain further insight into the molecular basis of leaf senescence in Arabidopsis, we explored the function and molecular nature of the ORE9 gene.
In this study, senescence symptoms induced by age and hormones were examined in the ore9-1 mutant at the physiological and molecular levels. We show that the ore9-1 mutation increases longevity by delaying the onset of various senescence symptoms during age-dependent natural senescence and hormone-induced senescence. This finding suggests that ORE9 may be linked to a common step of senescence induced by ABA, ethylene, and MeJA. It also implies that these senescence-related hormones share a common senescence pathway coupled to ORE9.
The predicted ORE9 protein consists of a degenerate F-box motif and 18 imperfect leucine-rich repeats (LRRs). The F-box motif of ORE9 interacts physically with the Arabidopsis Skp1-like protein ASK1 in a yeast two-hybrid assay and in an in vitro binding assay. These results suggest that ORE9 functions to limit the longevity of the leaf through ubiquitin-dependent proteolysis and that a novel, senescence-related proteolytic system exists in Arabidopsis.
RESULTS
The ore9-1 Mutant Shows Increased Leaf Longevity during Age-Dependent Senescence
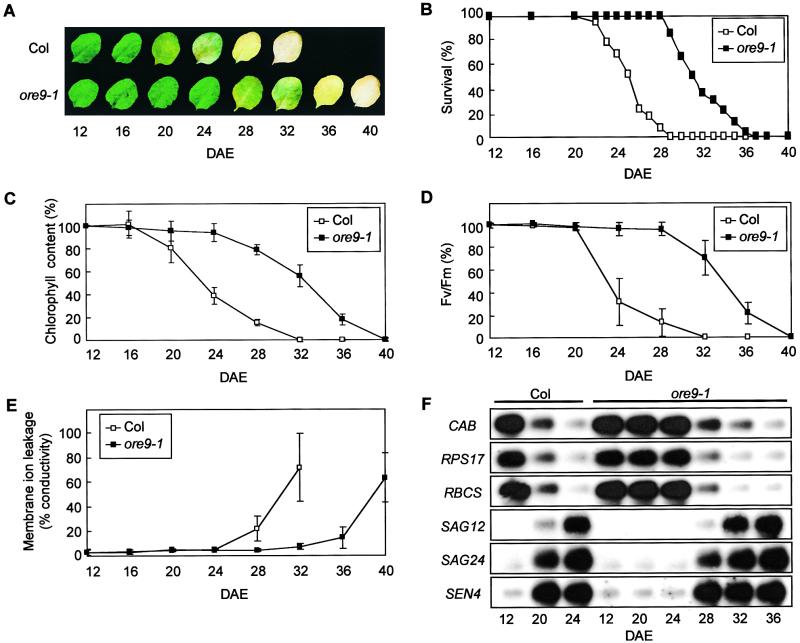
The ore9-1 mutant of Arabidopsis was isolated initially as a delayed leaf senescence mutant. We examined various senescence symptoms during age-dependent natural senescence. We first examined the effect of the ore9-1 mutation on leaf longevity visually (Figures 1A and 1B). The phenotype of individual leaves was followed from the formation of a visually recognizable leaf primordium (1 mm in size). The leaf was considered dead when the entire leaf turned yellow (Grbić and Bleecker, 1995). Leaf yellowing caused by the loss of chlorophyll is a typical symptom of senescence. We observed that the leaf longevity of ore9-1 was extended by ∼27.1%, from 24.7 to 31.4 days after leaf emergence (DAE), when the timing of survival of 50% of leaf populations was compared (Figure 1B).
Figure 1.
Age-Dependent Senescence Symptoms in the ore9-1 Mutation.
(A) The age-dependent senescence phenotype of wild-type (Columbia [Col]) and mutant leaves. Photographs show representative leaves at each time point.
(B) Survival curve. The graph shows the percentage of leaves alive on a given DAE. n = 100.
(C) to (E) Chlorophyll content (C), photochemical efficiency of PSII (D), and membrane ion leakage (E) were examined every 4 days from 12 DAE, when the fourth rosette leaves were just fully grown. Fv/Fm, maximum quantum yield of PSII electron transport (maximum variable fluorescence/maximum yield of fluorescence). Error bars indicate sd; n = 48.
(F) Age-dependent changes of gene expression. Total cellular RNA was extracted at 12, 20, and 24 DAE from wild-type leaves and at 12, 20, 24, 28, 32, and 36 DAE from ore9-1 leaves.
Leaf longevity of the mutant also was assessed by measuring typical senescence-associated physiological markers, such as chlorophyll content, photochemical efficiency of photosystem II (PSII), and membrane ion leakage (Fan et al., 1997; Oh et al., 1997). From 12 DAE on, which is the day the fourth rosette leaves were fully grown, chlorophyll content was measured. At 24 DAE, the leaves of wild-type plants had lost ∼60% of their chlorophyll, whereas yellowing had just started in the ore9-1 mutant (Figure 1C). Delayed senescence symptoms of the mutant also were defined as delays in the decrease in photosynthetic activities and the increase in membrane ion leakage (Figures 1D and 1E). The ore9-1 mutant leaves consistently showed later development of senescence-associated changes in these phenotypes. These results suggest that the ore9-1 mutation causes increased leaf longevity by delaying the onset of various senescence symptoms during age-dependent senescence.
Leaf senescence is accompanied by decreased expression of genes related to photosynthesis and protein synthesis and increased expression of senescence-associated genes (SAGs) (Nam, 1997). To determine the effect of ore9-1 on gene expression, we examined the expression patterns of these genes during leaf development (Figure 1F). In wild-type leaves, the expression of genes involved in photosynthesis (chlorophyll a/b binding protein [CAB] and ribulose bisphosphate carboxylase small subunit [RBCS]) and protein synthesis (chloroplast ribosomal protein S17 [RPS17]) declined in an age-dependent manner. However, in ore9-1 leaves, their expression was maintained at much higher levels until 24 DAE and started to decrease at 28 DAE. Expression of the SAG12, SAG24, and SEN4 genes increased dramatically along with leaf age in wild-type plants. However, in the ore9-1 mutant, expression of SAG12 was not detectable during the same age span and increased only at 28 DAE. In the case of SAG24 and SEN4, their expression was maintained at low levels until 24 DAE and was induced at 28 DAE. This result further showed that ore9-1 delays the age-dependent senescence process not only in overall physiology but also with regard to specific molecular events.
The ore9-1 Mutant Shows Delayed Senescence Symptoms in ABA-, MeJA-, and Ethylene-Induced Senescence
Leaf senescence is regarded as a developmentally programmed event. However, leaf senescence can be modulated by a range of plant hormones, such as ABA, MeJA, and ethylene (Weidhase et al., 1987; Zeevaart and Creelman, 1988; Aharoni, 1989). This phenomenon has been suggested to be a mechanism to coordinate the senescence process with various physiological events, such as the stress responses governed by these hormones (Nam, 1997).
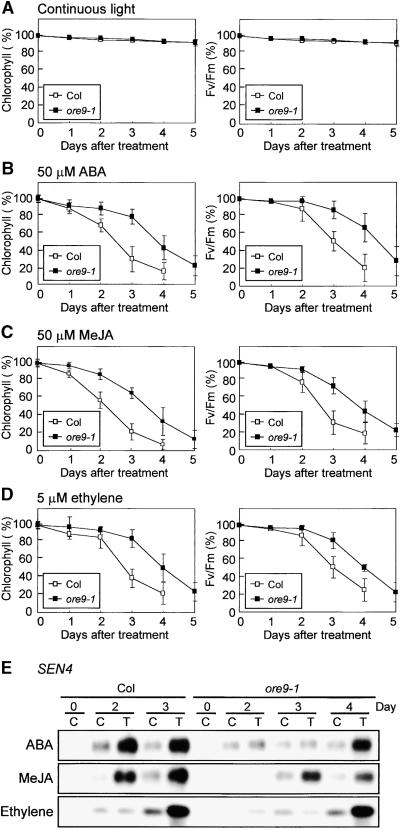
Before examining the hormone-induced senescence symptoms, the effects of the incubation of a detached leaf in continuous light were first assessed as a control experiment. As shown in Figure 2A, detached leaves gradually lost chlorophyll content and photochemical efficiency during the incubation period of 5 days. After incubation for 5 days in the light, the remaining chlorophyll content of wild-type and ore9-1 leaves was 84 and 87%, respectively, and photochemical efficiency was reduced to 92 and 93%, respectively. This result indicated that the change in the two parameters was not significantly different in wild-type and mutant leaves in this condition.
Figure 2.
Delay of Leaf Senescence in the ore9-1 Mutant during Senescence Accelerated by Plant Hormones.
The change of chlorophyll content and photochemical efficiency in continuous light (A), ABA-induced senescence (B), MeJA-induced senescence (C), and ethylene-induced senescence (D). (E) shows the change in expression of the SEN4 gene. Fv/Fm, maximum quantum yield of PSII electron transport (maximum variable fluorescence/maximum yield of fluorescence). Error bars indicate sd; n = 24. Total cellular RNA was extracted from control (C) and hormone-treated (T) leaves.
To determine whether ORE9 is involved in the senescence process induced by these hormones, we examined the senescence symptoms of the ore9-1 mutant after treatment with the plant hormones. Upon treatment with ABA, the chlorophyll content of wild-type leaves decreased rapidly to 69% after 2 days and to 27% after 3 days, whereas the ore9-1 mutant leaves retained 89 and 78% of their chlorophyll, respectively (Figure 2B) at the same times. The decrease in photochemical efficiency showed a trend similar to that of chlorophyll content. Expression of the SEN4 gene in the wild-type leaves increased from 2 days after treatment. In ore9-1 leaves, its expression was induced at 4 days after treatment (Figure 2E).
During treatment with MeJA, the leaves showed severely accelerated senescence symptoms (Figure 2C). After 3 days of treatment, the chlorophyll content of the wild-type leaves was reduced to 21% but that of mutant leaves was 66%. After treatment with MeJA, the expression of SEN4 was strongly induced from 2 days in the wild type and from 3 days in the mutant (Figure 2E). After treatment with ethylene for 3 days, the chlorophyll content and photochemical efficiency of the wild-type leaves decreased to 39 and 52%, respectively, whereas those of the ore9-1 mutant leaves were 85 and 82%, respectively (Figure 2D). Expression of SEN4 increased at 3 days in wild type but at 4 days in the mutant after treatment with ethylene (Figure 2E). These data showed that ORE9 plays an important role in the senescence process modulated by these hormones as well as in age-dependent senescence. This in turn suggests that age-dependent and hormone-modulated senescence processes use at least one common regulator, ORE9.
Responses of ore9-1 Seedlings to ABA, Ethylene, and MeJA Stimulation
The delayed senescence symptoms observed in the ore9-1 mutant during hormone-induced senescence could be explained by a defect in hormone perception in the mutant. To test this hypothesis, seedling responses to hormones were assayed (data not shown). One well-known physiological role of ABA is the regulation of seed dormancy. The seed dormancy of wild-type and ore9-1 seed was analyzed on B5 medium with or without 10 μM ABA. The control seedlings (no ABA) of wild type and the ore9-1 mutant germinated normally, and the cotyledons opened and expanded under the continuous light conditions used. Both wild-type and ore9-1 seedlings plated on ABA-containing medium barely germinated and did not show the phenotype of growth.
When seedlings from many different dicotyledonous species are grown in the dark in the presence of ethylene, they adopt a striking morphology referred to as the triple response. The seedlings of wild type and ore9-1 showed the distinctive phenotypes of the triple response in 1-aminocyclopropane-1-carboxylic acid (ACC)-containing medium. The regulation of root formation is a well-known function of jasmonate and its derivatives (Staswick et al., 1992). Upon treatment with 10 μM MeJA, the root growth of wild-type and ore9-1 seedlings was reduced and inhibited, whereas hypocotyls and cotyledons of the MeJA-treated seedlings had the same growth patterns as the untreated control seedlings. These results indicate that the ore9-1 mutation does not cause defects in the known signal transduction pathway that underlies the perception of and the response to these hormones.
Isolation of the ORE9 Gene
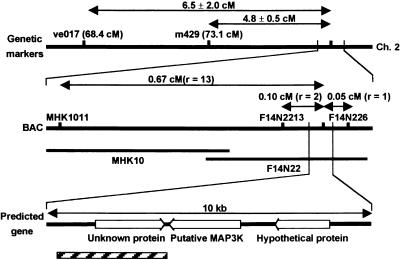
Genetic mapping, using cleaved amplified polymorphic sequence (CAPS) markers, placed ORE9 4.8 ± 0.5 centimorgans from the m429 locus on chromosome 2. Further mapping located ORE9 on bacterial artificial chromosome clone F14N22. We generated CAPS markers using the genomic sequences of the bacterial artificial chromosome clone. Two CAPS markers designated F14N226 and F14N2213 had one and two recombinant chromosomes, respectively, in 984 T2 progeny. These mapping data located ORE9 at approximately 0.05 and 0.1 centimorgan from these markers, respectively (Figure 3). In a 10-kb region around the map location of the ORE9 gene, we found three predicted open reading frames (ORFs). We then compared the wild-type and mutant nucleotide sequences of these ORFs. A single base pair change in the ore9-1 mutant, a C-to-T substitution, was identified in one of the predicted ORFs. This mutation resulted in the early termination of translation.
Figure 3.
Map-Based Cloning of ORE9.
The numbers of recombination events between the CAPS markers and the ORE9 locus (r) are shown. The hatched bar represents the 4.5-kb fragment used for a complementation experiment. Ch. 2, chromosome 2; BAC, bacterial artificial chromosome; cM, centimorgan; MAP3K, mitogen-activated protein kinase kinase kinase.
To confirm that the mutated ORF corresponds to the ORE9 gene, we performed a complementation experiment. The 4.5-kb genomic fragment containing the wild-type sequence of the mutated ORF was sufficient to complement the delayed senescence phenotype in the ore9-1 mutant (Table 1). We also generated transgenic plants that express the ORE9 gene in the antisense direction. Among the 28 transgenic lines we examined visually, five independent lines showed delayed leaf senescence (data not shown). These results confirmed that the identified ORF sequence is the ORE9 gene. ORE9 is a single copy gene in Arabidopsis, as determined by genomic DNA blot analysis (data not shown).
Table 1.
Complementation Test of the ore9-1 Mutanta
| Hygromycin Resistance |
Phenotype
|
|||||
|---|---|---|---|---|---|---|
| Line | HygR | HygS | χ2 | + | − | χ2 |
| Col | NA | 25 | 0 | |||
| ore9-1 | NA | 0 | 25 | |||
| T2
| ||||||
| ore9-1/ORE9-a | 215 | 68 | 0.143 (P > 0.5) | 56 | 19 | 0.004 (P > 0.9) |
| ore9-1/ORE9-b | 231 | 73 | 0.158 (P > 0.5) | 53 | 18 | 0.005 (P > 0.9) |
| ore9-1/ORE9-c | 276 | 14 | 1.001 (P > 0.1) | 67 | 6 | 0.483 (P > 0.1) |
a The χ2 values are for an expected ratio of 3:1 (lines a and b) or 15:1 (line c). HygR, resistant to hygromycin; HygS, sensitive to hygromycin; +, wild type; −, delayed senescence; NA, not applicable.
The ORE9 Protein Contains an F-Box Motif and 18 LRRs
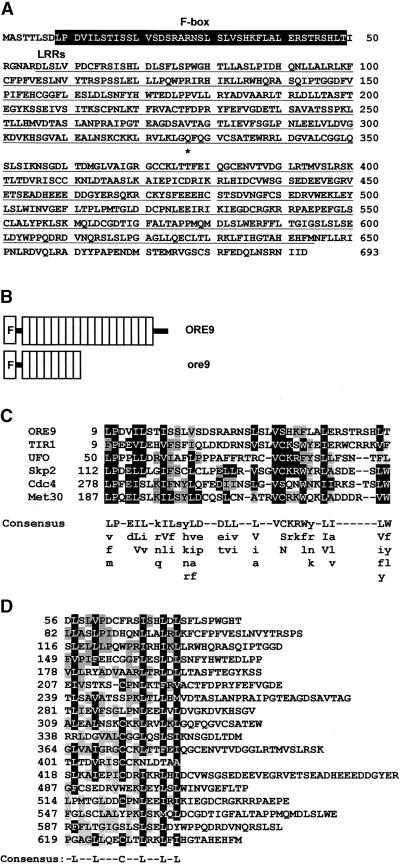
Comparison of the ORE9 cDNA with the genomic sequence revealed that the ORE9 gene has no introns. The ORE9 cDNA sequence predicts an ORF coding for a 693–amino acid protein. The predicted ORE9 protein has a degenerate F-box motif and 18 imperfect LRRs (Figure 4). A database search with the predicted polypeptide sequence showed that ORE9 is related to other LRR-containing F-box proteins: Arabidopsis TIR1 (48.4% similarity), yeast Cdc4 (47.8% similarity), yeast Met30 (44.4% similarity), and human Skp2 (38.2% similarity).
Figure 4.
Molecular Analysis of the ORE9 Gene.
(A) Predicted amino acid sequence of the ORE9 gene. The F-box and the LRRs are noted by a black box and by underlining, respectively. The mutated residue in the ore9-1 mutation is indicated by an asterisk.
(B) Scheme of the predicted structures of the ORE9 and ore9 proteins. The F-box and the LRRs are noted by the letter F and by white boxes, respectively.
(C) Alignment of the F-box motifs from various proteins and an F-box consensus sequence generated from an alignment of 38 F-box–containing proteins (Patton et al., 1998). Highly conserved residues are capitalized.
(D) Alignment of the 18 LRRs in ORE9. Highly conserved residues are boxed in black; gray shading indicates functionally conservative substitutions in more than half of the comparisons.
The F-box is a 40–amino acid residue domain composed of highly degenerate hydrophobic sequences. It is a component of the ubiquitin E3 ligase complex (SCF complex). In the case of yeast, the complex is composed of an F-box protein, Skp1, and Cdc53. The SCF complexes are known to ubiquitinate specific target substrates and lead to subsequent proteolytic degradation by the 26S proteasome in many organisms (Craig and Tyers, 1999; Callis and Vierstra, 2000). ORE9 has an F-box, suggesting a role in ubiquitinating proteins targeted for degradation.
ORE9 Interacts Physically with ASK1
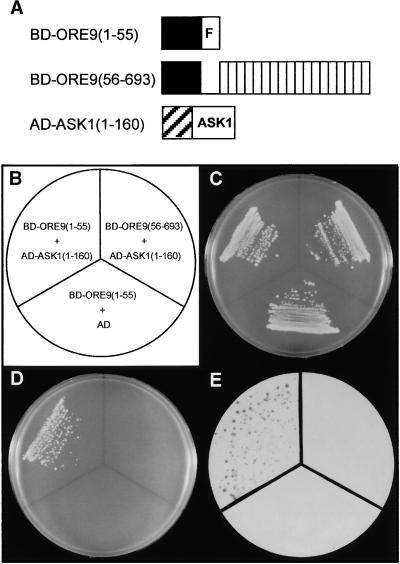
To examine the possibility that ORE9 might function as an F-box protein to form an SCF complex, we tested whether ORE9 could interact with an Arabidopsis protein homologous with yeast Skp1. We first performed yeast two-hybrid analysis using the Arabidopsis homolog of Skp1, ASK1 (Figure 5). The result showed that the F-box domain of ORE9 (amino acid residues 1 to 55) could interact with ASK1. The other part of ORE9 (amino acid residues 56 to 693) did not interact with ASK1 (Figures 5D and 5E). These data indicate that the F-box region of ORE9 is both necessary and sufficient for interaction with ASK1.
Figure 5.
Interaction of ORE9 and the Arabidopsis Homolog of Yeast Skp1, ASK1, in a Yeast Two-Hybrid Experiment.
(A) Scheme of the ORE9 and ASK1 constructs used in the yeast two-hybrid analysis. Solid and hatched boxes represent the GAL4 binding domain (BD) and the GAL4 activation domain (AD), respectively. The F-box and the LRRs are noted by the letter F and by white boxes, respectively.
(B) Plasmid pairs used in the yeast two-hybrid experiment.
(C) and (D) Yeast cells transformed with the plasmid pairs were cultured either on control synthetic minimal medium containing 2% dextrose but lacking tryptophan and leucine (C) or on viability test medium (synthetic minimal medium lacking tryptophan, leucine, and histidine and containing 2 mM 3-amino-1,2,4-triazole) (D).
(E) β-Galactosidase activities of the transformants.
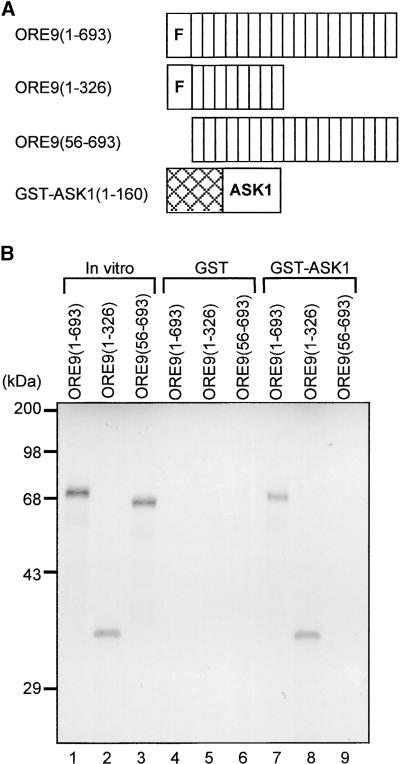
An in vitro binding assay was performed to confirm the direct interaction between ORE9 and ASK1. The assay was performed using a glutathione S-transferase (GST)-ASK1 fusion protein and 35S-labeled ORE9 derivatives generated by in vitro translation (Figure 6A). The full length ORE9 protein (amino acid residues 1 to 693) and the mutant protein derived from the ore9-1 mutant (amino acid residues 1 to 326) were coprecipitated with the GST-ASK1 fusion protein. The ORE9 fragment without the F-box (amino acid residues 56 to 693) did not bind to the GST-ASK1 fusion protein (Figure 6B). These data, together with the results of the yeast two-hybrid experiment, show that ORE9 can interact with ASK1. These results suggest that ORE9 may form the SCFORE9 complex.
Figure 6.
Interaction of ORE9 and ASK1 in an in Vitro Binding Assay.
(A) The ORE9 and ASK1 constructs used in the in vitro binding analysis. The cross-hatched box represents the GST protein. The F-box and the LRRs are noted by the letter F and by white boxes, respectively.
(B) In vitro binding experiment for analysis of the ORE9-ASK1 interaction. The in vitro translation products of ORE9(1-693), ORE9(1-326), and ORE9(56-693) are shown in lanes 1, 2, and 3, respectively. The 35S-labeled proteins were incubated with resin-bound GST (lanes 4, 5, and 6) or GST-ASK1 (lanes 7, 8, and 9).
DISCUSSION
ORE9 May Function Upstream in the Regulatory Cascade of Senescence Pathways
Senescence is a developmental event that leads to the death of a cell, an organ, or an organism upon aging. The aging process that results in senescence and limits longevity is a ubiquitous biological phenomenon in most organisms. It is now clear that, at least in part, the longevity of many organisms is controlled genetically. Plants, especially annual plants, exhibit distinctive aging and senescence processes. However, the systematic study of the genetics of longevity mutations began only recently using the leaf of Arabidopsis as a model system (Grbić and Bleecker, 1995; Oh et al., 1997).
We observed that the leaf longevity of ore9-1 was extended by ∼27.1% (Figure 1B). The delay in the leaf senescence of the mutant was demonstrated by measuring changes in typical senescence-associated physiological markers such as chlorophyll content, photochemical efficiency, and membrane ion leakage (Figures 1C to 1E). The findings suggest that ORE9 may function normally as a positive regulator of leaf senescence in Arabidopsis, limiting the longevity of the leaf. Because the ore9-1 mutation affects a wide variety of senescence symptoms, ORE9 may function upstream in the regulatory cascade of senescence pathways.
Leaf senescence is delayed in the ore9-1 mutant, but ultimately it does occur. The progression of leaf senescence observed in the mutant can be explained if senescence proceeds through several parallel pathways. Accepting the notion that senescence is important for the fitness of plants, the strategy of having genetically redundant pathways would be advantageous for plants to promote the fidelity of the senescence process (Thomas, 1993; Nam, 1997). In support of this notion, double mutant analysis among ore1, ore3, and ore9 suggests that ORE1, ORE3, and ORE9 may not function on a single senescence pathway but may act in overlapping pathways (data not shown).
Three Senescence-Related Hormones, ABA, MeJA, and Ethylene, Share a Common Senescence Pathway Involving ORE9
Although leaf senescence is a developmentally programmed event, the initiation and progression of senescence can be influenced by a range of hormones, such as ABA, MeJA, and ethylene. These plant hormones have diverse effects on leaf senescence, affecting parameters such as the onset, progression, and termination of leaf senescence. The question was raised whether the signals and processes induced by these hormones share a common senescence pathway(s). The ore9-1 mutant was chosen to investigate this question in the context of hormone-induced senescence symptoms.
The senescence symptoms we examined were loss of chlorophyll content and photochemical efficiency and induction of SEN4 gene expression. The ore9-1 mutant exhibited a delay in the appearance of these senescence symptoms induced by the three hormones ABA, ethylene, and MeJA. These studies revealed that the ore9-1 mutation is linked to a common step in the ABA-, ethylene-, and MeJA-induced senescence processes. This finding implies that ORE9 is required for the proper progression of leaf senescence induced by ABA, ethylene, and MeJA. Although the ore9-1 mutant showed delayed hormone-induced senescence, this mutation did not cause defects in the known signal transduction pathways that underlie the perception of and the response to these hormones. This was determined by the demonstration of normal hormone effects at the seedling stage. Therefore, three senescence-related hormones, ABA, ethylene, and MeJA, share at least one common pathway that involves ORE9 and is involved in the hormonal induction of senescence.
Protein Degradation during Leaf Senescence
Leaf senescence involves massive structural and functional disintegration of cells through the activation of various hydrolytic activities (Noodén, 1988; Smart, 1994). These hydrolytic activities are required to degrade the nutrients that accumulate during the vegetative growth phase and, subsequently, to mobilize the nutrients. The degradation of protein is probably the most significant breakdown process. The spectrum of genes involved in protein turnover, such as cysteine protease– and aspartic protease–like genes, is being expanded continuously.
The ubiquitin pathway for targeted protein degradation is important in the elimination of abnormal cytoplasmic proteins and in the rapid turnover of short-lived proteins. The identification of an E2-type ubiquitin carrier protein gene that shows increased expression during leaf senescence in Nicotiana sylvestris (Genschik et al., 1994) and the increased expression of a polyubiquitin gene, SEN3, in senescing leaves of Arabidopsis (Park et al., 1998) suggest that ubiquitin-dependent proteolysis occurs during leaf senescence. However, it appears unlikely, based on the structure of the genes of the ubiquitin pathway, that any of these proteins could be involved in the degradation of chlorophyll proteins (Vierstra, 1993). Therefore, it has been suggested that ubiquitin-mediated protein degradation may not be involved in the massive breakdown of proteins during senescence but may have a role in degrading specific proteins. Our study of the function of ORE9 provides evidence that leaf senescence involves the ubiquitin-mediated degradation of specific proteins.
ORE9 May Function as an F-Box Protein of an SCF Complex
The F-box is found in a diverse array of regulatory proteins, including the Skp2 protein of humans and the Cdc4 and Grr1 proteins of yeast (Bai et al., 1996). F-box proteins have been identified in plants and found to function in the regulation of floral organ identity (UFO), jasmonic acid–regulated defense (COI1), auxin response (TIR1), and control of the circadian clock (ZTL and FKF1) (Ruegger et al., 1998; Xin et al., 1998; Samach et al., 1999; Nelson et al., 2000; Somers et al., 2000). Here we demonstrated that an F-box protein is involved in the control of leaf senescence. The ORE9 derivative, which does not contain the F-box domain, failed to interact with ASK1 in a yeast two-hybrid assay and in an in vitro binding assay (Figures 5 and 6). This finding suggests that the F-box motif may be essential for ORE9 function in leaf senescence.
The ASK1 gene was isolated in two-hybrid screens using TIR1 and UFO F-box proteins as bait (Gray et al., 1999; Samach et al., 1999). At present, 10 ASK genes have been identified in the Arabidopsis genome, suggesting that some specificity may exist in the interaction between F-box and ASK proteins. The presence of a related ASK protein also suggests that functional redundancy may exist among ASK proteins. Categorization and characterization of ASK proteins that interact with ORE9 therefore will help us to understand the mechanism of leaf senescence through ubiquitin-mediated proteolysis.
How Does the Activity of ORE9 Influence Leaf Senescence?
Why is leaf senescence delayed in the ore9-1 mutant? The F-box motif is required to form the SCF complex. The remaining LRR or WD40 repeat region in F-box–containing proteins functions in substrate recognition for subsequent ubiquitination. We propose that ORE9 is involved in the degradation of substrate protein(s) through ubiquitin-mediated proteolysis. In addition, we suggest that the substrate is a key negative regulatory molecule of senescence, such as a transcriptional repressor of SAGs. In the ore9-1 mutants, the substrate(s) may not be recruited and degraded effectively because of the defect in the LRR domain. Consequent accumulation of the negative regulators may prolong the down regulation of SAGs, thereby extending the longevity of the leaf. Alternatively, or in addition, ORE9 may function as a receptor for the selective degradation of self-maintenance proteins. In this case, the continuous presence of these proteins may inhibit the initiation of senescence in the mutant. The ore9-1 mutant displays defects in several senescence pathways. This finding suggests that the SCFORE9 complex may have multiple substrates, some that integrate hormone signaling with leaf senescence and others that control leaf longevity.
Investigation of gene function through the targeted substrates of ORE9 and associated pathways should lead to a better understanding of the mechanisms relevant to leaf senescence in plants. The identification of ORE9, a positive regulator of leaf senescence, as a possible component of an SCF complex provides important information that enhances our understanding of the molecular mechanism of longevity control in plants and in other systems as well.
METHODS
Plants and Other Materials
Arabidopsis thaliana plants were grown in an environmentally controlled growth room (Korea Instruments, Seoul, Korea) at 22°C with a 16-hr-light/8-hr-dark cycle. All experiments were performed with the fourth rosette leaves. Leaf samples were obtained by cutting leaves at the approximate middle of the petioles with a sharp scalpel to minimize wounding effects.
Hormone Treatment
For hormone treatment, leaves at 12 days after leaf emergence (DAE) were detached and floated on 3 mM Mes buffer (pH 5.8) solution in the presence or absence of 50 μM abscisic acid (ABA; Sigma) or 50 μM methyl jasmonate (MeJA; Sigma). In the case of ethylene treatment, detached leaves were incubated in a glass box containing ethylene gas (5 μM). All hormonal treatments were performed at 22°C under continuous lighting.
Hormonal sensitivity was tested by examining seedling responses to each hormone. Wild-type and mutant seedlings were grown as follows. Seed were surface-sterilized for 10 min in 1% sodium hypochlorite and rinsed with sterilized distilled water. Sterilized seed were then plated on B5 medium (Gibco BRL) containing 0.8% agar (type M; Sigma) with or without the addition of 1-aminocyclopropane-1-carboxylic acid (ACC; 10 μM; Sigma), ABA (10 μM), or MeJA (10 μM). Seed plated on B5 medium containing ACC were incubated in darkness, and seed plated on ABA- or MeJA-containing B5 medium were kept under continuous illumination. After 5 days of incubation, the ACC-treated seedlings grown continuously in the dark were assayed. The ABA- and MeJA-treated seedlings grown under continuous light were assayed after 8 days of treatment.
Measurement of Chlorophyll Content, Photochemical Efficiency, and Ion Leakage
Chlorophyll was extracted from individual leaves by boiling the leaves in 95% ethanol at 80°C. Chlorophyll concentration per fresh weight of leaf was calculated as described by Lichtenthaler (1987). The photochemical efficiency of photosystem II (PSII) was deduced from the characteristics of chlorophyll fluorescence (Oh et al., 1996) using a portable plant efficiency analyzer (Hansatech Instruments, Morfolk, England). The ratio of maximum variable fluorescence to maximum yield of fluorescence, which corresponds to the potential quantum yield of the photochemical reactions of PSII, was used as the measure of the photochemical efficiency of PSII (John et al., 1995; Raggi, 1995; Oh et al., 1997). Membrane ion leakage was determined by measuring electrolytes leaked from leaves. Two leaves were immersed in 3 mL of 400 mM mannitol at 22°C with gentle shaking for 3 hr, after which the initial conductivity was measured. Total conductivity was determined after boiling for 10 min. The conductivity was expressed as the percentage of the initial conductivity versus the total conductivity.
Isolation of Total RNA and RNA Gel Blot Analysis
Total cellular RNA was isolated with Tri-Reagent (Molecular Research Center, Cincinnati, OH) from control and hormone-treated leaves. Ten micrograms of total cellular RNA was size fractionated by electrophoresis through a 1.2% formaldehyde-agarose gel and transferred onto a nylon membrane (ICN Biomedicals, Costa Mesa, CA) according to standard methods (Sambrook et al., 1989). Radiolabeled probes were prepared using a random labeling kit according to the manufacturer's instructions (Amersham). After hybridization, the membranes were washed with 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at room temperature for 20 min and with 0.1 × SSC and 0.1% SDS at 55°C for 30 min, after which they were placed on x-ray film.
Genetic Mapping
The ore9-1 plants (Columbia [Col] background) were crossed to a wild-type Landsberg erecta (Ler) plant. From the F2 progeny, 984 plants showing the mutant phenotype were selected. Additional cleaved amplified polymorphic sequence (CAPS) markers were developed from Arabidopsis genome sequence data (http://www.arabidopsis.org). The new markers were MHK1011 (amplification with primers 5′-TTGACTCGCATTTGGTTAAACC-3′ and 5′-TCAACCTGTTTTGAGCAAGGAA-3′, yielding a 1.2-kb product with two and one BstXI sites in Col and Ler, respectively), F14N226 (amplification with primers 5′-CAATACTAGACGTCTTAAATGG-3′ and 5′-CAT-AGATAAGCTGTCGTTAATC-3′, yielding a 1.2-kb product with two and three DraI sites in Col and Ler, respectively), and F14N2213 (amplification with primers 5′-GATGATGATCGTCATTTTATGG-3′ and 5′-GATTTCTATTCGTGATCGAAAG-3′, yielding a 1.2-kb product with one and two HinfI sites in Col and Ler, respectively).
Complementation Test
The 4.5-kb DNA fragment containing 2.0 kb of the promoter region, the predicted open reading frame, and 0.4 kb of downstream sequence was amplified by polymerase chain reaction using two oligonucleotides, 5′-ATTGAGTTTGTACTCCGGATCCCTC-3′ and 5′-GACGGA-TCCTCTAAATAGTTTGAAACGG-3′. The fragment was cloned into the BamHI site of the pCAMBIA1300 vector (Molecular Research Center) and transformed into the ore9-1 plants. T2 seed from three independent T1 transgenic lines were plated on a medium supplemented with 20 μg/mL hygromycin. Hygromycin resistance was scored after 10 days. Phenotypes of T2 plants were scored for two senescence parameters, chlorophyll content and photochemical efficiency, during age-dependent senescence.
Yeast Two-Hybrid Experiments
The derivatives of the ORE9 gene were fused to the GAL4 binding domain of the pGBT9 vector. The ASK1 gene was fused to the GAL4 activation domain of the pGAD424 vector. The genes were cloned into the respective vectors after polymerase chain reaction with primers containing appropriate restriction sites. Yeast cells were grown on 1% yeast extract, 2% peptone, 2% dextrose, and 1.5% agar (for plates) or on synthetic minimal medium (0.67% yeast nitrogen base, the appropriate auxotrophic supplements, and 1.5% agar [for plates]) containing 2% dextrose. The yeast strain HF7c (MATa, ura3-52, his3-200, lys2-801, ade2-101, trp1-901, leu2-3,112, gal4-542, gal80-538, LYS2::GAL1UAS-GAL1TATA-HIS3, URA3::GAL417mer[×3]- CyC1TATA-lacZ) was used to assay for protein–protein interaction. Yeast was transformed with the appropriate plasmids by the lithium acetate method, and the transformants were selected on the appropriate synthetic minimal medium. For the β-galactosidase assay, yeast cells grown on synthetic minimal medium were transferred to a Whatman number 1 filter (Chung et al., 1997). The filter was placed in liquid nitrogen for 30 sec and then incubated in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 1 mM MgSO4) containing 0.82 mM 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. The filters were kept at 30°C and monitored for color change indicating β-galactosidase activity.
In Vitro Binding Assay
pGEX or pGEX-ASK1 plasmids were transformed and expressed in Escherichia coli BL21 (DE3) pLys S strain. Glutathione S-transferase (GST) or GST-ASK1 fusion proteins were purified using glutathione–Sepharose 4B beads. Equal amounts of GST or GST-ASK1 fusion proteins were absorbed by the glutathione beads prewashed three times with 10 volumes of buffer B (20 mM sodium phosphate, pH 7.6, 150 mM NaCl, 10% glycerol, 0.5% Nonidet P-40, and 1 mM DTT) by incubation at 4°C for 1 hr on a rotating mixer. The beads were then washed three times with 1 mL of buffer B and stored at 4°C as a 50% slurry in buffer B. Radiolabeled ORE9 and the derivatives were generated using an in vitro transcription/translation system (Promega) and 35S-methionine (DuPont–New England Nuclear) as described (Chung et al., 2000). Equal amounts of 35S-labeled translation products were incubated with 60 μL of GST beads (50% slurry) in 1 mL of GB buffer (20 mM Tris-HCl, pH 7.5, 0.15% Nonidet P-40, 150 mM NaCl, and 1 mM EDTA). After 2 hr of incubation at 4°C on a rotating mixer, the beads were washed four times with 1 mL of GB buffer and boiled for 3 min in 30 μL of 2 × SDS sample buffer before analysis by SDS-PAGE.
GenBank Accession Numbers
The GenBank accession numbers are as follows: bacterial artificial chromosome clone F14N22, AC007087; 693-amino acid protein, AF305597.
Acknowledgments
We thank K.W. Kim, K.S. Lee, Y.O. Shin, K.H. Suh, and K.Y. Yu for excellent technical assistance. This work was supported by the National Research Laboratory Program of the Korean Ministry of Science and Technology.
References
- Abeles, F.B., Dunn, L.J., Morgens, P., Callaha, A., Dinterman, R.E., and Schmidt, J. (1988). Induction of 33-kD and 60-kD peroxidases during ethylene-induced senescence of cucumber cotyledons. Plant Physiol. 87, 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni, N. (1989). Interrelationship between ethylene and growth regulators in senescence of lettuce leaf disk. J. Plant Growth Regul. 8, 309–317. [Google Scholar]
- Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W., and Elledge, S.J. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274. [DOI] [PubMed] [Google Scholar]
- Bell, E., Creelman, R.A., and Mullet, J.E. (1995). A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 92, 8675–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston, V. (1994). Isolation of cDNA clones for genes that are expressed during leaf senescence in Brassica napus. Plant Physiol. 105, 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston, V. (1997). The molecular biology of leaf senescence. J. Exp. Bot. 48, 181–199. [Google Scholar]
- Callis, J., and Vierstra, R.D. (2000). Protein degradation in signaling. Curr. Opin. Plant Biol. 3, 381–386. [DOI] [PubMed] [Google Scholar]
- Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., and Ecker, J.R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144. [DOI] [PubMed] [Google Scholar]
- Chung, K.M., Song, O.K., and Jang, S.K. (1997). Hepatitis C virus nonstructural protein 5A contains potential transcriptional activator domains. Mol. Cell 7, 661–667. [PubMed] [Google Scholar]
- Chung, K.M., Lee, J., Kim, J.E., Song, O.K., Cho, S., Lim, J., Seedorf, M., Hahm, B., and Jang, S.K. (2000). Nonstructural protein 5A of hepatitis C virus inhibits the function of karyopherin β3. J. Virol. 74, 5233–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, K.L., and Tyers, M. (1999). The F-box: A new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog. Biophys. Mol. Biol. 72, 299–328. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A., Tierney, M.L., and Mullet, J.E. (1992). Jasmonic acid/methyl jasmonate accumulate in wounded soybean hypocotyls and modulate wound gene expression. Proc. Natl. Acad. Sci. USA 89, 4938–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, K.M., and Grierson, D. (1989). Identification of cDNA clones for tomato (Lycopersicon esculentum Mill.) mRNAs that accumulate during fruit ripening and leaf senescence in response to ethylene. Planta 179, 73–80. [DOI] [PubMed] [Google Scholar]
- Fan, L., Zheng, S., and Wang, X. (1997). Antisense suppression of phospholipase Dα retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 9, 2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik, P., Durr, A., and Fleck, J. (1994). Differential expression of several E2-type ubiquitin carrier protein genes at different developmental stages in Arabidopsis thaliana and Nicotiana sylvestris. Mol. Gen. Genet. 244, 548–556. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseuw, E. Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić, V., and Bleecker, A.B. (1995). Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8, 595–602. [Google Scholar]
- Gut, H., Ruts, C., Matile, P., and Thomas, H. (1987). Leaf senescence in a non-yellowing mutant of Festuca pratensis: Degradation of carotenoids. Physiol. Plant. 70, 659–663. [Google Scholar]
- Hensel, L.L., Grbić, V., Baumgarten, D.A., and Bleeker, A.B. (1993). Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, I., Drake, R., Farrell, A., Cooper, W., Lee, P., Horton, P., and Grierson, D. (1995). Delayed leaf senescence in ethylene-deficient ACC-oxidase antisense tomato plants: Molecular and physiological analysis. Plant J. 7, 483–490. [Google Scholar]
- Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 18, 350–382. [Google Scholar]
- Lohman, K.N., Gan, S., John, M.C., and Amasino, R.M. (1994). Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plant. 92, 322–328. [Google Scholar]
- Nam, H.G. (1997). Molecular genetic analysis of leaf senescence. Curr. Opin. Biotechnol. 8, 200–207. [DOI] [PubMed] [Google Scholar]
- Nelson, D.C., Lasswell, J., Rogg, L.E., Cohen, M.A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331–340. [DOI] [PubMed] [Google Scholar]
- Noodén, L.D. (1988). The phenomenon of senescence and aging. In Senescence and Aging in Plants, L.D. Noodén and A.C. Leopold, eds (San Diego, CA: Academic Press) pp. 2–50.
- Oh, S.A., Lee, S.Y., Chung, I.K., Lee, C.H., and Nam, H.G. (1996). A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 30, 739–754. [DOI] [PubMed] [Google Scholar]
- Oh, S.A., Park, J.-H., Lee, G.I., Paek, K.H., Park, S.K., and Nam, H.G. (1997). Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. 12, 527–535. [DOI] [PubMed] [Google Scholar]
- Park, J.-H., Oh, S.A., Kim, Y.H., Woo, H.R., and Nam, H.G. (1998). Differential expression of senescence-associated mRNAs during leaf senescence induced by different senescence-inducing factors in Arabidopsis. Plant Mol. Biol. 37, 445–454. [DOI] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. (1998). Combinational control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet. 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Raggi, V. (1995). CO2 assimilation, respiration and chlorophyll fluorescence in peach leaves infected by Taphrina deformans. Physiol. Plant. 93, 540–544. [Google Scholar]
- Reymond, P., and Farmer, E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Klenz, J.E., Kohalmi, S.E., Risseuw, E., Haughn, G.W., and Crosby, W.L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20, 433–445. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Smart, C.M. (1994). Gene expression during leaf senescence. New Phytol. 126, 419–448. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89, 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, H. (1987). Sid: A Mendelian locus controlling thylakoid membrane disassembly in senescing leaves of Festuca pratensis. Theor. Appl. Genet. 73, 551–555. [DOI] [PubMed] [Google Scholar]
- Thomas, H., and Stoddart, J.L. (1980). Leaf senescence. Annu. Rev. Plant Physiol. 31, 83–111. [Google Scholar]
- Thomas, J.H. (1993). Thinking about genetic redundancy. Trends Genet. 9, 395–399. [DOI] [PubMed] [Google Scholar]
- Vierstra, R.D. (1993). Protein degradation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 385–410. [Google Scholar]
- Weaver, L.M., Gan, S., Quirino, B., and Amasino, R.M. (1998). A comparison of the expression patterns of several senescence associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37, 455–469. [DOI] [PubMed] [Google Scholar]
- Weidhase, R.A., Lehmann, J., Kramell, H., Sembdner, G., and Parthier, B. (1987). Degradation of ribulose-1,5-bisphosphate carboxylase and chlorophyll in senescing barley leaf segments triggered by jasmonic acid methyl ester and counteraction by cytokinin. Physiol. Plant. 69, 161–166. [Google Scholar]
- Xin, D.-X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Zacarias, L., and Reid, M.S. (1990). Role of growth regulators in the senescence of Arabidopsis thaliana leaves. Physiol. Plant. 80, 549–554. [Google Scholar]
- Zeevaart, J.A.D., and Creelman, R.A. (1988). Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 439–473. [Google Scholar]