Abstract
A cDNA clone from tomato fruit encodes a protein with strong homology with the rab11/YPT3 class of small GTPases that is thought to be involved in the control of protein trafficking within cells. The gene, LeRab11a, showed a pattern consistent with a single copy in DNA gel blots. The corresponding mRNA was developmentally regulated during fruit ripening, and its expression was inhibited in several ripening mutants. Its reduced expression in the Never-ripe mutant indicates that it may be induced by ethylene in fruit. The ripening-induced expression in tissues that are undergoing cell wall loosening immediately suggests a possible role in trafficking of cell wall–modifying enzymes. The message also was produced in leaves and flowers but not in roots. Antisense transformation was used to generate a “mutant phenotype.” Antisense fruit changed color as expected but failed to soften normally. This was accompanied by reduced levels of two cell wall hydrolases, pectinesterase and polygalacturonase. There were other phenotypic effects in the plants, including determinate growth, reduced apical dominance, branched inflorescences, abnormal floral structure, and ectopic shoots on the leaves. In some plants, ethylene production was reduced. These data suggest an alternative or additional role in exocytosis or endocytosis of homeotic proteins, hormone carriers, or receptors.
INTRODUCTION
The process by which proteins and other large molecules are trafficked through the cell has been fairly well characterized in yeast and mammalian systems, and many of the processes and components have been observed in plant cells (Sanderfoot and Raikhel, 1999). Secreted proteins that are made on the rough endoplasmic reticulum (ER) pass cotranslationally into the lumen of the ER. Coated vesicles carrying a protein cargo bud off from the ER and fuse with the Golgi apparatus. From the trans-Golgi network, other coated vesicles carry the cargo to the cell membrane. Cargos also may be transported to other membrane systems, such as the vacuole or prevacuolar compartment, or may be transported in the opposite direction through the Golgi stacks from trans to cis and from the Golgi apparatus to the ER. Endocytotic vesicles carry cargoes from the cell membrane; in animal systems, this pathway leads to a series of lytic compartments called endosomes. Each transport process is performed by a different type of coated vesicle with characteristic proteins on the surface that ensure the correct docking of the vesicle with its target. One class of proteins found on these vesicles is the Rab GTPases, of which there are ∼30 different families (Olkkonen and Stenmark, 1997). Each type of Rab is associated with a specific type of vesicle and probably plays a role in ensuring correct fusion (Takai et al., 2001).
Recently, we isolated a Rab-11–like GTPase from mango that was interesting because it was identified by its differential expression in ripe fruit but not in unripe fruit (Zainal et al., 1996). In accordance with a recent proposal (Bischoff et al., 1999), this clone, which was previously named mango RabX gene, is now renamed MiRab11a.
That a Rab GTPase should be expressed in a ripening-induced manner in fruit is intriguing. Ripening of tomato fruit involves a number of changes, including changes in color, flavor, and texture, that are accompanied by the expression of a characteristic set of genes (Gray et al., 1992). Change in texture is mediated by modifications to the cell wall, including the well-characterized solubilization of the pectin of the middle lamella, which is brought about by the secretion of a number of enzymes such as polygalacturonase and pectinesterase. These enzymes are synthesized on the rough endoplasmic reticulum, and the nascent polypeptides have typical signal peptides at the N terminus (Bird et al., 1988; Ray et al., 1988).
These cell wall–softening enzymes presumably are trafficked through the endomembrane system of the cell and secreted to the apoplast. It is a reasonable hypothesis, there-fore, that Rab GTPases might be essential for this process and that Rab11 might be one of the types involved. Homologs of several of these Rab11 GTPases have been found in plants (Bischoff et al., 1999), but little is known about their role in plant systems other than what can be inferred from animal systems. Even in mammalian systems, the role of Rab11 proteins is unclear. They have been shown to be associated with both the trans-Golgi network and the recycling endosomes, and they may play a role not only in vesicle docking but also in vesicle budding (Takai et al., 2001).
To elucidate the role of this class of GTPase, we identified an ortholog of the mango clone from a tomato fruit cDNA library that we have named LeRab11a. We investigated the pattern and control of RNA accumulation in fruit and other organs of the plant. We also blocked its expression by antisense transformation. This resulted in reduced softening of the fruit and interesting developmental abnormalities that mimic homeotic mutations.
RESULTS
cDNA Cloning and Sequence Analysis
An early ripening tomato fruit phage library (Picton et al., 1993) was screened using the mango rabX clone as a probe. One clone was isolated, and the resulting plasmid clone was named pNY650. The sequence has been deposited in the EMBL sequence database under accession number AJ245570.
The longest open reading frame begins at the first ATG and encodes a 218–amino acid protein of 24,192 kD with a pI of 5.82. The sequence of the predicted protein was compared with the SWISS-PROT database, and matches were found with several other GTPase proteins of the Rab11 class using the WU-blastp program on the EBI-EMBL server. The best match (94.5% identity) was with the Nicotiana plumbaginifolia Np-Ypt3 protein (Dallmann et al., 1992) (accession number Q01111). Others showing more than 80% identity were Lotus japonicus Rab11d (Q40194; 84.9% identity) and Rab11e (Q40195; 82.2%) (Borg et al., 1997), pea pra7 (Q08151; 84.4%) (Nagano et al., 1993), and rice Ric2 (P40393; 80.2%) (Kidou et al., 1993). For comparison, the mango probe used to screen the tomato library (Q43554) (Zainal et al., 1996) showed 70.5% identity, and the best nonplant match, mouse Rab11b (P46638) (Lai et al., 1994), showed 64.7% identity.
RNA and DNA Gel Blot Analyses of Wild-Type Plants
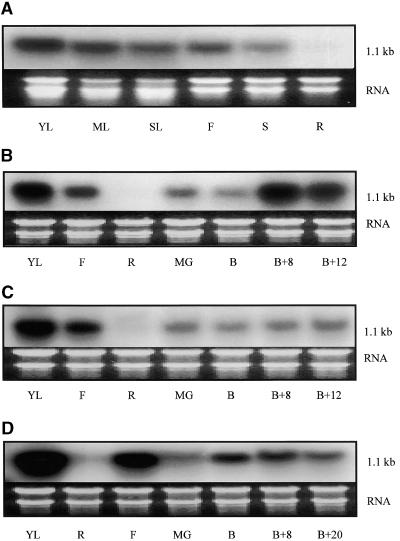
To determine the pattern of RNA accumulation, RNA samples from fruit at different stages of ripening, from leaves of different ages, and from stems, flowers, and roots were subjected to RNA gel blot analysis with the tomato Rab cDNA as probe. Figure 1 shows that the message, like its mango equivalent, accumulated much more strongly in ripe fruit than in mature green fruit. LeRab11a also showed strong expression in young leaves and flowers. Expression in older leaves and other organs was weaker, and no mRNA was detectable in roots.
Figure 1.
Accumulation of LeRab11a Transcripts in Different Organs and Developmental Stages of Wild-Type and Mutant Tomato Plants.
(A) Different organs of wild-type Ailsa Craig.
(B) Fruit ripening series of wild-type Ailsa Craig with root, leaf, and flower controls.
(C) Fruit ripening series of alc mutant with root, leaf, and flower controls.
(D) Fruit ripening series of Nr mutant with root, leaf, and flower controls.
Each lane contained 20 μg of total RNA from tissues as follows: YL, young leaf; ML, mature leaf; SL, senescent leaf; F, whole flower; S, stem; R, root; MG, mature green fruit; B, breaker fruit; B+8, fruit 8 days after breaker; B+12, fruit 12 days after breaker; and B+20, fruit 20 days after breaker. Membranes were washed twice in 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at 65°C for 5 min and then twice in 0.1 × SSC and 0.1% SDS for 15 min at 65°C. All bands correspond to a transcript size of 1.1 kb.
To investigate further the mechanism of gene control in ripening fruit, RNA was extracted from different stages of fruit development in the ripening mutants alcobaca (alc) and Never-ripe (Nr). As shown in Figure 1, the accumulation of mRNA during the course of ripening was reduced greatly in these mutants. In contrast to the wild type (Figure 1B), the level of mRNA in the fruit of alc (Figure 1C) and Nr (Figure 1D) plants never approached that seen in the positive control (young leaf tissue).
DNA gel hybridization was performed with several different restriction endonucleases. Only one or two bands were seen in each case under stringent hybridization and washing conditions (data not shown).
Generation and Analysis of Transformants
A 960-bp (almost full length) cDNA fragment was cut from pNY650 with EcoRV and XbaI and ligated in a reverse orientation into pDH51 between the cauliflower mosaic virus (CaMV) 35S promoter and the CaMV 35S terminator to give pBDHR. This new gene construct was then cut out and ligated into pBIN19. Two very slightly different transformation plasmids were produced by excising the gene construct fragment from pDHR with either a partial digest of EcoRI, to give pBINDR, or a digest of PvuII and SmaI, to give pBINTR.
A half-length construct was produced by excising 540 bp of DNA from the 5′ end of the LeRab11 cDNA with EcoRV and BglII and ligating it into pGA643 (An et al., 1988) between the CaMV 35S promoter and terminator, again in the antisense orientation, to give pGATR.
A total of 53 primary transformants was produced, 13 with pBINDR, 14 with pBINTR, and 26 with pGATR. Thirty-two of these transformants that were grown into mature plants and characterized fully are described in Table 1. The presence of the transgene was confirmed by DNA gel blot and/or polymerase chain reaction (PCR) analysis. Three plants found not to contain the transgene were included as further controls, in addition to three control plants regenerated from callus that was not treated with Agrobacterium tumefaciens.
Table 1.
Summary of Phenotypes Associated with Primary Transformantsa
| Plant Lines | Construct | Transgene Present | Growth Habit | Ectopic Shoots | Branching Trusses | Other | Fruit Firmness |
|---|---|---|---|---|---|---|---|
| T6 | pGATR | + | D | ++ | +++ | − | ++ |
| T7 | pGATR | + | D | +++ | ++ | − | ++ |
| T13 | pGATR | + | D | ++ | +++ | − | ++ |
| T18 | pGATR | + | D | + | ++ | − | ++ |
| T20 | pGATR | + | D | +++ | ++ | − | ++ |
| T21 | pGATR | + | D | + | − | − | − |
| T32 | pGATR | + | D | + | ++ | − | ++ |
| T12 | pGATR | + | SD | +++ | ++ | − | + |
| T14 | pGATR | + | SD | + | − | − | ++ |
| T15 | pGATR | + | SD | + | ++ | − | + |
| T16 | pGATR | + | SD | +++ | ++ | − | +++ |
| T17 | pGATR | + | SD | ++ | ++ | − | ++ |
| T27 | pGATR | + | SD | ++ | ++ | − | + |
| T31 | pGATR | − | − | − | ++ | − | − |
| T3 | pBINDR | + | − | ++ | +++ | − | +++ |
| T5 | pBINDR | + | D | ++ | +++ | − | +++ |
| T25 | pBINDR | + | D | ++ | − | P | + |
| T26 | pBINDR | + | D | + | − | − | ++ |
| T29 | pBINDR | + | D | + | ++ | − | ++ |
| T30 | pBINDR | + | D | ++ | − | P, Y | + |
| T1 | pBINDR | + | SD | ++++ | ++ | − | ++ |
| T2 | pBINDR | + | − | − | ++ | − | ++ |
| T4 | pBINDR | − | − | − | − | − | − |
| T49 | pBINDR | + | − | − | − | − | − |
| T9 | pBINTR | + | D | + | − | P, Y | ++ |
| T10 | pBINTR | + | D | ++ | − | P, Y | ++ |
| T11 | pBINTR | + | D | ++ | ++ | − | ++ |
| T19 | pBINTR | + | D | + | ++ | − | ++ |
| T22 | pBINTR | + | D | − | ++ | − | +++ |
| T8 | pBINTR | + | SD | + | + | − | ++ |
| T24 | pBINTR | + | SD | − | + | − | − |
| T28 | pBINTR | − | − | − | − | − | − |
| WT1 | None | − | − | − | − | − | − |
| WT2 | None | NT | − | − | − | − | − |
| WT3 | None | NT | − | − | − | − | − |
a Data from some transformed lines were omitted to save space. D, determinate growth habit; SD, semideterminate, formed terminal flower on the main stem with two side shoots; P, abnormal petals; Y, yellow striped fruit; −, wild type ([WT]; transgene or mutant phenotype not detected); NT, not tested; +++, ++, +, very high, high, or moderate penetration, respectively.
Sense and Antisense Rab11 RNA in Transgenic Plants
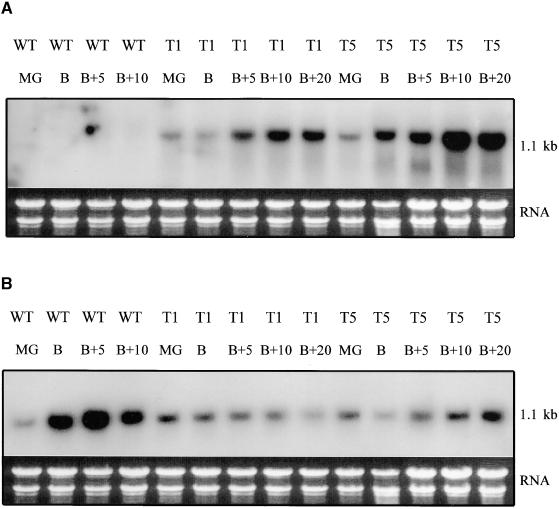
RNA gel blot analysis was used to detect the accumulation of transcripts from the transgene. The results, shown in Figure 2A, indicate that the antisense RNA was produced in fruit of transformed lines T1 and T5 but not in the Ailsa Craig wild-type parent. In contrast, the endogenous sense message, which increased considerably during ripening in the Ailsa Craig wild-type parent, either increased only slightly or actually decreased slightly as ripening progressed in the transformed lines (Figure 2B). The level of antisense RNA was reduced during the early stages of ripening compared with the later stages. Other transformed lines tested gave similar levels of sense and antisense RNA, as summarized in Table 2.
Figure 2.
RNA Gel Blot Analysis of Fruit RNA in Normal and Transgenic Lines.
Total RNA was extracted from fruit of two different transformed lines at different stages of ripening: mature green (MG), breaker (B), and five (B+5), ten (B+10), or twenty (B+20) days after breaker. It was then separated on a 1% agarose-formaldehyde gel and transferred onto a nylon membrane. The blot was hybridized with either a sense-specific or an antisense-specific 32P-labeled LeRab11a riboprobe. Membranes were washed twice in 2 × SSC and 0.1% SDS at 65°C for 5 min and then twice in 0.1 × SSC and 0.1% SDS for 15 min at 65°C. The stained rRNA bands are shown under each lane to show equivalence of loading.
(A) Transformed lines T1 (pBINDR) and T5 (pBINTR) and wild-type (WT) control tomato with an antisense-specific riboprobe.
(B) Transformed lines T1 and T5 and wild-type control tomato with a sense-specific riboprobe.
Table 2.
Summary of RNA Analysis of Primary Transformantsa
| Plant Lines |
Construct | Transgene Present |
Sense mRNA in Fruit |
Antisense mRNA in Fruit |
|---|---|---|---|---|
| T1 | pBINDR | + | + | ++ |
| T2 | pBINDR | + | ++ | ++ |
| T3 | pBINDR | + | + | +++ |
| T4 | pBINDR | − | ++++ | − |
| T5 | pBINDR | + | ++ | +++ |
| T6 | pGATR | + | ++ | +++ |
| T7 | pGATR | + | +++ | ++ |
| T8 | pBINTR | + | ++ | ++ |
| T9 | pBINTR | + | ++ | +++ |
| T10 | pBINTR | + | + | ++++ |
| T11 | pBINTR | + | ++ | NT |
| T12 | pGATR | + | ++ | NT |
| T13 | pGATR | + | ++ | NT |
| T14 | pGATR | + | ++ | NT |
| T15 | pGATR | + | ++ | NT |
| WT | None | − | ++++ | − |
a +, relative RNA band intensity; −, RNA not detectable; NT, not tested; WT, wild type.
Abnormal Phenotypes
A range of abnormal phenotypes was seen in the antisense transformed plants. Not all of the effects were strongly marked in every primary transformant, but most of the transformed lines showed several of the observed differences from the wild type (Table 1). These effects included reduced fruit softening and extended fruit life, reduced apical dominance and terminal inflorescences, highly branched compound inflorescences, and ectopic shoots arising from the rachis or rachillae of the leaves. These effects were seen with all three different constructs and in many independent transformants arising from transformations performed on different days. These effects were not seen in untransformed control plants taken through tissue culture and grown alongside the transgenic plants, nor were they seen in “transformants” found not to contain the transgene.
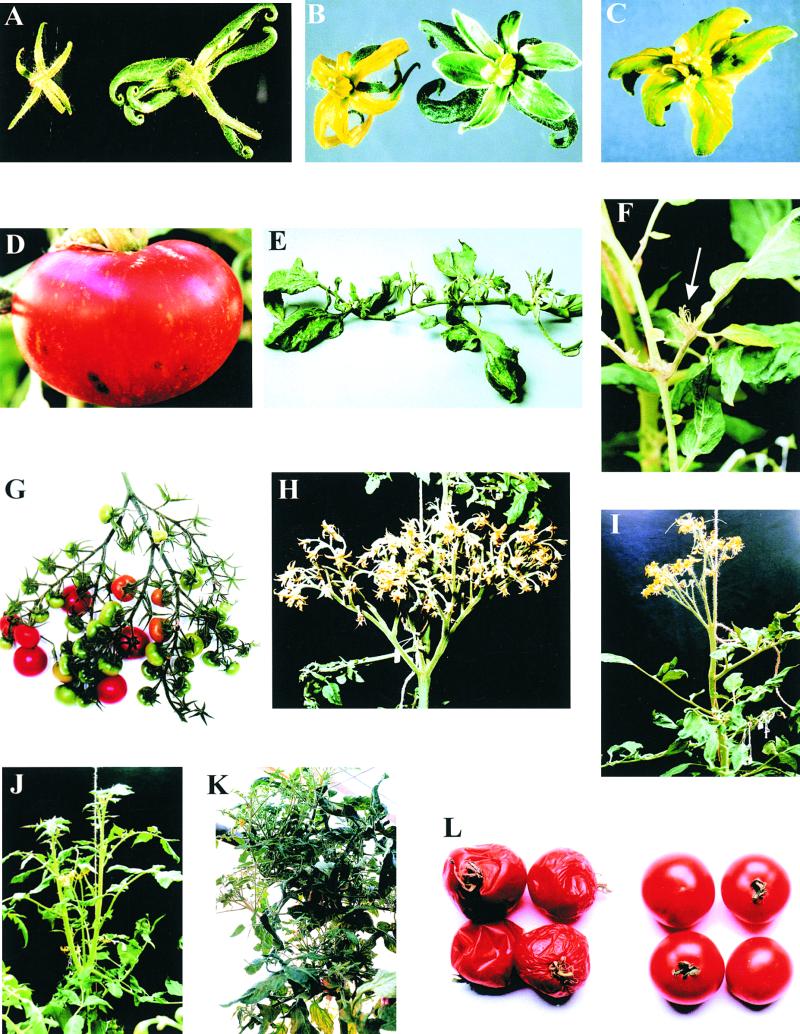
Some of the plants transformed with the full-length constructs pBINTR and pBINDR (T9, T10, T25, T30, T49, and T55) showed two additional effects. One was that the flowers were malformed in one of several ways, as shown in Figures 3A, 3B, and 3C. These included an unusually large calyx, a leaf-like corolla, and a flower structure in which the calyx and corolla were not readily distinguishable. These plants also gave rise to normal flowers. Some of the fruit from these plants showed an unusual yellow flecking (Figure 3D). These abnormal flower and flecked fruit phenotypes also were seen in the progeny of selfed primary transformants.
Figure 3.
Visual Phenotypes of Rab Antisense Transgenic Plants.
(A) Sepals of wild-type flower (left) and transformant flower (right).
(B) Flowers of wild-type (left) and transformant (right) plants.
(C) Petal of a transformant plant.
(D) Yellow stripes in fruit of transformed plant line T9.
(E) A mature leaf from a transformed plant showing ectopic shoots.
(F) The formation of new ectopic shoots on a leaf.
(G) and (H) Branching inflorescences of transformants.
(I) Typical determinate growth pattern shown by transformed line T3.
(J) Determinate growth pattern plus side shoots (line T1).
(K) A transformed plant (line T1).
(L) Fruit 30 days after picking from plants of transformed line T5 (right) and wild-type Ailsa Craig (left).
Leaf Morphology
The transformed plants often gave rise to small rosette-like structures on either the main axis (the rachis) or the side axes (rachillae) of the bipinnate leaves (Figure 3E). Although the development of these structures was slow initially, they usually grew into shoots that appeared normal apart from their ectopic origin. Sometimes, several would appear from each leaf (Figure 3F). These appeared more frequently in older plants. They were not seen in our untransformed controls growing alongside the transgenic plants, but such shoots are seen very rarely in Ailsa Craig.
Inflorescence Structure
Tomato plants normally bear an inflorescence that is a simple panicle. However, the transformed plants often bore inflorescences that were highly branched to give rise to patterns resembling either a compound panicle (Figure 3G) or a dichasium (Figure 3H). These patterns were not seen in untransformed controls.
Indeterminate Growth Pattern and Apical Dominance
Ailsa Craig is an indeterminate cultivar, but many of the transformed plants showed a determinate growth habit. Growth of these plants terminated in a compound inflorescence (Figure 3I), usually at a height of ∼1.5 m from the ground. Sometimes, growth would continue from leaf axils lower on the plant (Figure 3J).
As part of good glasshouse practice, axillary shoots initially were removed from both transgenic and untransformed plants. However, additional shoots regenerated from the leaf axils of the transgenic plants more frequently than from the axils of untransformed plants. In wild-type Ailsa Craig, inflorescences will sometimes terminate in a vegetative shoot. However, this effect was much more marked in the transformed plants. The combination of this effect and the increased numbers of axillary shoots sometimes gave rise to plants with very bushy growth (Figure 3K).
Fruit Softening
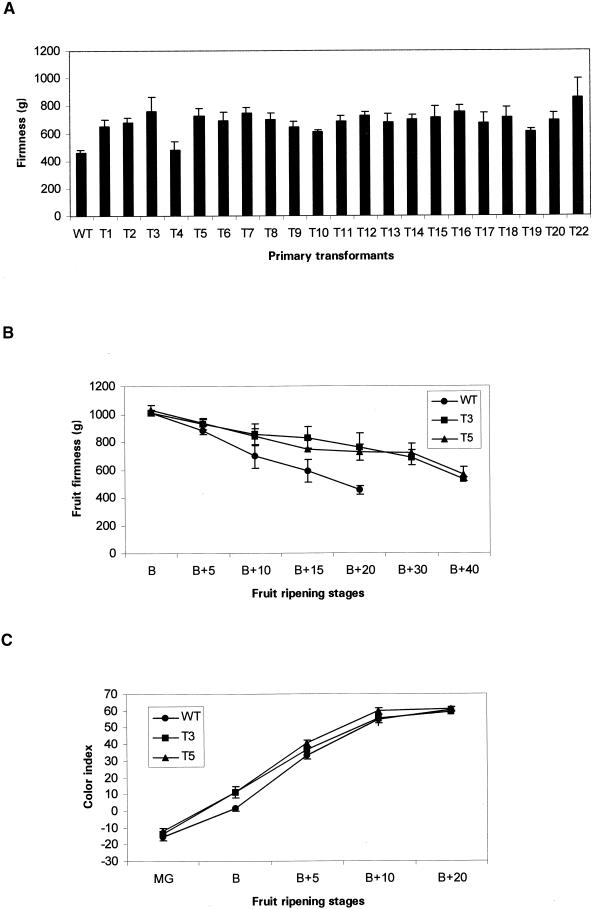
Because an effect on fruit softening had been anticipated, fruit firmness was measured 20 days after breaker stage (breaker stage being defined as the stage at which the onset of color change is first visible). The results for untransformed and 21 different transformant lines are shown in Figure 4A. All of the transformed lines had significantly firmer fruit than the untransformed control except for T4, which was found not to contain the transgene and therefore was used as an internal control. A time course for the softening of T3 (half- length construct) and T5 (full-length construct) fruit (Figure 4B) shows that the fruit were still firm enough to measure after 40 days, whereas the untransformed controls were too soft to test after 20 days. However, color development in the transformed and untransformed fruit was not significantly different (Figure 4C). The appearance of representative fruit at different ripening stages is shown in Figure 3L.
Figure 4.
Fruit Firmness and Color Index of Antisense Transformants and Wild-Type Tomato during Ripening.
(A) Firmness of ripe fruit (20 days after breaker stage [B+20]) from different transformants T1 to T20, T22, and wild-type (WT) tomato.
(B) Firmness of transformed lines T3 and T5 and wild-type control tomato during different ripening stages. Ripening stages shown here and in panel (C) are: mature green (MG), breaker (B), and five (B+5), ten (B+10), fifteen (B+15), twenty (B+20), thirty (B+30) or forty (B+40) days after breaker.
(C) Fruit color index of transformants T3 and T5 and wild-type tomato during ripening.
Error bars indicate ±se.
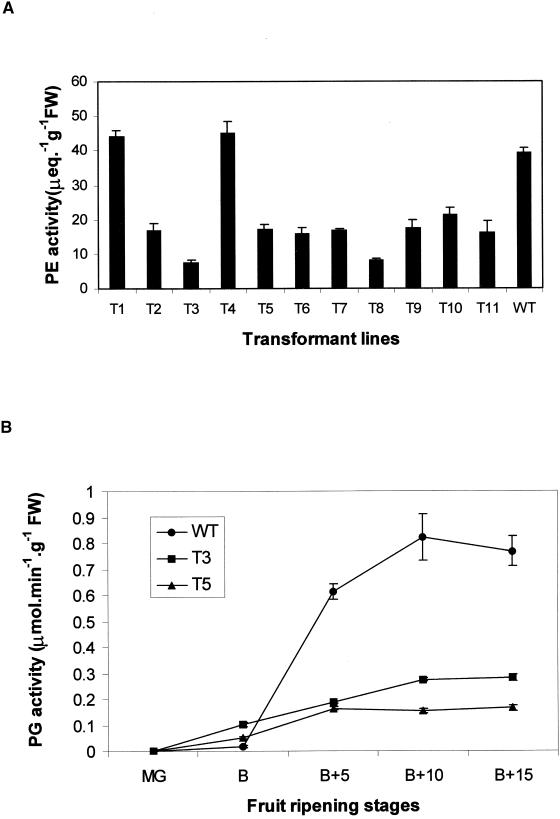
To investigate whether the reduced softening was attributable to an effect on the trafficking of cell wall hydrolases, the levels of polygalacturonase (PG) and pectinesterase (PE) were assayed. The results, shown in Figure 5, indicate that the levels of both PG and PE were reduced in a wide range of the transgenic fruit but not in the wild-type controls or in the untransformed T4 line.
Figure 5.
Cell Wall Hydrolase Activity.
(A) PE activity extracted from wild type (WT) and transformed lines T1 to T11 at breaker +10. μeq, microequivalents of H+ ions.
(B) PG activity extracted from fruit of wild type and transformed lines T3 and T5 at different ripening stages: mature green (MG), breaker (B), and five (B+5), ten (B+10), or fifteen (B+15) days after breaker.
FW, fresh weight.
Error bars indicate ±se.
Ethylene Production
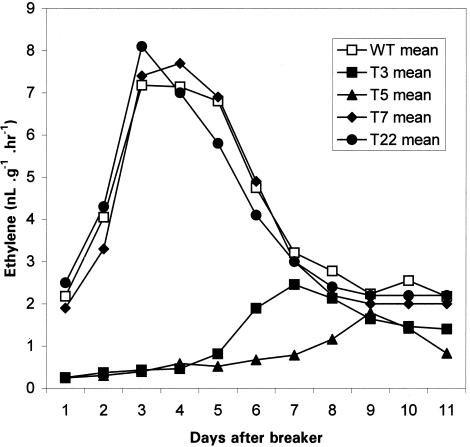
Ethylene levels in the fruit were measured during ripening. The peak of ethylene production was much smaller and much later in some of the transgenic fruit than in the wild type. However, the levels of ethylene were normal in other transgenic fruit and did not correlate with the reduction in fruit softening. Figure 6 shows data from some representative lines of each type.
Figure 6.
Ethylene Production in Transformants and Wild-Type (WT) Tomato Fruit during Ripening.
Daily measurements were taken from untransformed control wild-type fruit and transformed lines T3, T5, T7, and T22 commencing at the start of color change. Data are expressed as means of values obtained from four fruit.
Inheritance of the Mutant Phenotype
The inheritance of growth habit, ectopic shoots, branching inflorescences, and fruit firmness in the progeny (R1 plants) of the primary transformants (R0 plants) also was analyzed. The inheritance of the transgene was tested by PCR amplification of the transgene and by estimating the percentage of R2 seedlings that grew on kanamycin agar. This latter test allowed plants to be designated tentatively as azygous, hemizygous, or homozygous. The results of the analysis of the T1 and T5 primary transformants are listed in Table 3. The four abnormal phenotypes all cosegregated with the T-DNA construct, and in general, the severity of the symptoms was greater in the homozygous progeny than in the hemizygous progeny. The level of ethylene production was reduced in the hemizygous line T3R1-9 and the homozygous line T3R1-4 but similar to that of the wild-type untransformed control in the azygous line T3R1-6 (data not shown).
Table 3.
Properties of R1 Transgenic Plants and Wild-Type Controlsa
| R1 Plant | Growth Habit | Ectopic Shoots | Branching Truss | Fruit Firmness | PCR for Transgene | Inheritance Assay |
|---|---|---|---|---|---|---|
| T1R1-1 | D | + | + | + | + | Hemizygous |
| T1R1-2 | SD | +++ | ++ | ++ | + | Homozygous |
| T1R1-3 | D | +++ | ++ | +++ | + | Homozygous |
| T1R1-4 | − | − | − | − | − | Azygous |
| T1R1-5 | SD | +++ | +++ | ++ | + | Homozygous |
| T1R1-6 | − | − | − | − | − | Azygous |
| T1R1-7 | D | ++ | + | + | + | Hemizygous |
| T1R1-8 | SD | + | + | + | + | Hemizygous |
| T1R1-9 | D | ++ | + | ++ | + | Hemizygous |
| T1R1-10 | − | − | + | − | − | Azygous |
| WT1 | − | − | − | − | − | Azygous |
| T3R1-1 | SD | ++ | ++ | ++ | + | Hemizygous |
| T3R1-2 | SD | ++ | + | ++ | + | Hemizygous |
| T3R1-3 | − | − | − | + | − | Azygous |
| T3R1-4 | D | ++ | +++ | +++ | + | Homozygous |
| T3R1-5 | D | − | + | ++ | + | Hemizygous |
| T3R1-6 | − | − | + | − | − | Azygous |
| T3R1-7 | SD | − | ++ | ++ | + | Hemizygous |
| T3R1-8 | − | − | − | − | − | Azygous |
| T3R1-9 | SD | ++ | ++ | ++ | + | Hemizygous |
| T3R1-10 | D | + | ++ | +++ | + | Homozygous |
| WT2 | − | − | − | − | − | Azygous |
a D, determinate growth habit; SD, semideterminate, formed terminal flower on the main stem with two side shoots; −, wild type ([WT]; transgene or mutant phenotype not detected); +++, ++, +, very high, high, or moderate penetration, respectively.
DISCUSSION
The similarity of the ripening-related expression of LeRab11 to that of MiRab11a (formerly called mango RabX; Zainal et al., 1996) suggests that the two clones not only have sequence homology but probably are functionally equivalent. The reduced expression in the ripening mutant alc further confirms that LeRab11 is controlled as part of a ripening response, because the fruit of alc plants fail to express many ripening-related proteins and also have a much reduced ethylene climacteric response (Mutschler, 1984; Mutschler et al., 1988). Many genes expressed in a ripening-related manner are regulated by ethylene in fruit. The Nr tomato mutant is unable to perceive ethylene because of a mutation in an ethylene receptor ortholog (Wilkinson et al., 1995), and the lack of expression of LeRab11a in Nr fruit suggests that the LeRab11a gene is regulated in an ethylene-dependent manner in fruit. However, the lower level of expression in senescent leaves compared with young leaves is not consistent with ethylene inducibility in tissues other than fruit. It is interesting that the highest levels of LeRab11 RNA were seen in fruit, leaves, and flowers, because these were the main organs that showed abnormalities in the antisense plants.
The different phenotypic effects that were seen in the antisense plants occurred in many independent transformants. They also were seen to cosegregate with the transgene construct. The observed effects therefore must be attributable to reduced LeRab11 activity in the plants. However, the phenotypic effects were limited in that there was no overall effect on the height or rate of growth of the plants, so the trafficking of cell wall components in expanding cells could not have been affected, nor could that of other gross cellular components. This finding suggests that a limited number of proteins or other cellular components were affected. It is entirely possible that two or more closely related Rab11 GTPase gene family members were suppressed in our plants. The extra range of phenotypes seen in plants transformed with the full-length constructs is consistent with the suppression of gene family members with different physiological and cellular roles. This also would explain why two different cellular processes, exocytosis and endocytosis, may have been affected in the antisense plants (see below).
A search of the tomato expressed sequence tag database at The Institute for Genomic Research (Rockville, MD) revealed four other Rab11 GTPase genes in tomato with 73 to 79% nucleotide identity and several others with lesser identity. Unfortunately, the level of nucleotide identity is not the only factor, or even the most important factor, in whether transgene-induced gene silencing will occur in a given plant. The presence of other gene family members that were not suppressed also may explain why the level of sense message was reduced only partly in antisense fruit. Even if two or three closely related gene family members were suppressed in any given plant, there could be several other highly expressed members that were unaffected by the antisense transgene. This is consistent with the pattern seen in Lotus japonicus, in which 10 different types of Rab11 cDNA clones were detected, some of which were much more abundant than others (Borg et al., 1997).
The reduced softening of the fruit was very marked. This finding supports the hypothesis (Zainal et al., 1996) that ripening-induced Rab11 GTPases are involved in trafficking of cell wall–modifying enzymes to the apoplast. The reduction in ethylene levels in some transgenic lines also could have contributed to the reduced softening. However, several transgenic lines with essentially normal levels of ethylene production also exhibited substantially reduced fruit softening. Therefore, although the reduced ethylene levels may have had an additive effect on fruit softening in some lines, it could not have been the primary cause of the reduced softening.
The observation that the levels of PG and PE were reduced in the transgenic fruit compared with those in wild-type fruit is consistent with the disruption of trafficking of cell wall hydrolases in transgenic plants. Because PE is known to be synthesized by the fruit before the ethylene climacteric (Hobson, 1963), this result provides further evidence that the reduction in fruit softening was not mediated by ethylene. Presumably, any proenzyme synthesized in the cell may have been targeted incorrectly for degradation or accumulated in cellular compartments in which post-translational processing to the active enzyme did not occur. These findings are consistent with the observation that the Rab11 protein is essential for the transport of proteins from the trans-Golgi network to the plasma membrane in animal cells (Chen et al., 1998) and suggest fundamental similarities in Rab11 GTPase function between the two kingdoms.
Several of the effects seen in our plants are quite different from any seen in mutants or transformants affected in fruit ripening or ethylene production. However, some tomato mutants are known to have defects similar to those seen in our plants. For example, the blind mutant terminates in a single flower or inflorescence and has ectopic shoots on the leaves. leafy, macrocalyx, and compound inflorescence (s) mutants also show some of the symptoms exhibited by our plants (Rick and Butler, 1956), but none of these mutants exhibits the full syndrome of effects. Some of the effects that were seen in the LeRab11 antisense plants mimic those of disrupting the expression of homeobox genes. Mutation or antisense silencing of the tomato SELF-PRUNING gene gives rise to determinate plants with a terminal inflorescence (Pneuli et al., 1998), and a petunia MADS box mutant, gp, has a calyx-like corolla (van der Krol et al., 1993). Overexpression of the maize KNOTTED-1 gene in tobacco gives rumpled leaves with ectopic shoots (Sinha et al., 1993), and overexpression of the homologous KNAT-1 gene in Arabidopsis also gives rise to deformed leaves and ectopic shoots (Lincoln et al., 1994; Chuck et al., 1996). Expression of the maize KNOTTED-1 gene in tomato only produces more highly compound leaves (Hareven et al., 1996), but overexpression of the tomato homolog LeT6 also produces floral meristems on the leaf rachis (Janssen et al., 1998). It is possible, therefore, that the LeRab11 protein might be involved in the trafficking of these proteins and that disruption of the trafficking results in abnormally low levels in some tissues or even abnormally high levels in cells that normally export them. Interestingly, the KNOTTED-1 protein is trafficked through the plasmodesmata (Lucas et al., 1995). This raises the intriguing possibility that the tomato protein might be connected with trafficking through the plasmodesmata.
However, it is possible that the effect is less direct. The rumpled leaves and abnormal flower morphology seen in tobacco plants expressing a NTH15 homeobox transgene was accompanied by reduced GA1 levels and increased levels of cytokinin (Tamaoki et al., 1997), and plants with increased cytokinin levels caused by transformation with the Agrobacterium ipt gene form ectopic buds on leaves (Estruch et al., 1991). Increased levels of cytokinin and abnormal flower development were both seen in tobacco plants expressing sense or antisense copies of a rice Rab gene (Kamada et al., 1992; Sano et al., 1994). We have confirmed that ethylene levels were perturbed in our plants, suggesting that phytohormone levels may be involved in producing at least some of the phenotypic effects in plants expressing Rab antisense transgenes as well as in those expressing homeobox genes.
It is also possible that LeRab11 and proteins like it are involved in the trafficking of hormone transporters or receptors to the cell membrane. However, disruption of trafficking to the cell membrane would be expected to lead to reduced levels of homeotic proteins, hormones, and hormone receptors, whereas an increase in the levels of these would most readily explain the phenotype that we observed. A second, more likely possibility is that endocytosis of integral membrane proteins was disrupted in our antisense plants. The role of Rab11-type GTPases in plants has been inferred so far from their known role in mammalian systems, in which the role of rab11b proteins is uncertain and several different roles have been reported (Olkkonen and Stenmark, 1997). Rab11a proteins seem to be associated with recycling endosomes and probably are involved in endocytotic processes as well as secretion. Certain classes of G protein–coupled receptors are desensitized by means of endocytosis and either destruction of the whole receptor-ligand complex in lysosomes or recycling of the receptor through the endosome (Bünemann et al., 1999). Recently, it was shown that this type of endocytotic recycling of β(2)-adrenergic receptors to lysosomes involves the rab11-positive perinuclear recycling compartment (Moore et al., 1999). If endocytotic receptor down-regulatory mechanisms exist in plants, then disruption of them may give rise to a failure to completely switch off hormone-mediated developmental programs or to increased sensitivity to low levels of signaling molecules and could explain most of the phenotypic effects seen in our plants.
METHODS
Plant Material and Growth Conditions
Plants of wild-type tomato (Lycopersicon esculentum cv Ailsa Craig) and mutants alcobaca (Kopeliovitch et al., 1980) and Never-ripe (Rick and Butler, 1956) were grown under glass with minimum day temperatures of 22°C and minimum night temperatures of 18°C. Light was supplemented to give a 16-hr day.
For plant transformation, the seed were surface-sterilized by soaking in 50% sodium hypochlorite solution for 10 min and rinsing three to four times in sterile distilled water. Seed were sown in MSR3 medium (4.4 g/L Murashige and Skoog [1962] salts, 30 g/L sucrose, 0.1% [v/v] R3 vitamins [1 g/L thiamine, 0.5 g/L nicotinic acid, 0.5 g/L pyridoxine], pH 5.9, and 8 g/L bacteriological agar) and were germinated in a growth room in 16 hr of light at 26°C.
Plants regenerated from tissue culture that had developed roots were transferred to Levington F2 compost (Levington Horticulture, Ipswich, UK) for the first week. The plants were grown in a tissue culture room (16 hr of light, 8 hr of dark, 24°C) and then transferred to the glasshouse (day, 25°C; night, 18°C).
Screening of the cDNA library
Screening was performed by plaque hybridization using a single stranded mango RabX cDNA as a probe. Filters were prehybridized for 5 hr at 65°C in 5 × SSPE (1 × SSPE is 0.115 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA, pH 7.4), 5 × Denhardt's solution (1 × Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), 0.1% SDS, and 100 μg/mL denatured salmon sperm DNA. Hybridization was performed in the same solution supplemented with 2.5 μL of 32P-dCTP–labeled (10 μCi/μL) probe for 18 hr at 65°C. Filters were washed twice with 2 × SSPE and 0.1% SDS at 65°C for 10 min and once with 1 × SSPE and 0.1% SDS at 65°C for 15 min. Finally, the membranes were washed at high stringency with 0.1 × SSPE and 0.1% SDS at 65°C for 10 min. The membranes were dried and autoradiographed. Positive λZAPII clones were isolated and processed to pBluescript SK− phagemids.
DNA and RNA Preparation
Total RNA was extracted from fruit, leaves, stems, flowers, or roots as described by Jones et al. (1998) with two modifications. First, the RNA extraction buffer contained phenol supplemented with 14% (v/v) m-cresol and 0.1% (w/v) 8-hydroxyquinoline instead of phenol alone. Second, lithium chloride precipitation was performed between the first ethanol precipitation and the second phenol extraction as follows. The dried pellet was dissolved in sterile deionized water. An equal volume of 8 M lithium chloride was added, and the solution was placed at −20°C for 2 hr. The precipitate was collected by centrifugation at 12,000g for 30 min at 0 to 5°C, and the pellet was washed with chilled 70% ethanol.
RNA Gel Blot Analysis
Total RNA was fractionated by gel electrophoresis on a denaturing 1.5% agarose gel containing 8% formaldehyde and blotted onto a GeneScreen membrane (DuPont–New England Nuclear) using 20 mM sodium phosphate as transfer buffer. The RNA was fixed to the membrane using a UV cross-linker (Stratalinker; Stratagene, Cambridge, UK) for 30 sec at high power. The probes were amplified from plasmid DNA by 35 cycles of polymerase chain reaction (PCR) and labeled using Rediprime (Amersham International) and 32P-dCTP (110 TBq/mmol; Amersham International) according to the manufacturer's instructions. Membranes were prehybridized in Rapid-hyb buffer (Amersham International) at 65°C for 4 hr and hybridized with the probe at 65°C for 16 hr according to the manufacturer's instructions. Membranes were washed twice in 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at 65°C for 5 min, then twice in 0.1 × SSC and 0.1% SDS for 15 min at 65°C. Filters were then exposed to Kodak X-Omat AR film using intensifying screens for 24 hr at −80°C.
DNA Gel Blot Analysis
Samples of tomato genomic DNA (25 μg) were digested with 25 units of EcoRI, EcoRV, HindIII, BglII, or XhoI. The reactions were incubated at 37°C for 16 hr, and then the fragments were separated by gel electrophoresis in a 1% agarose gel (in 40 mM Tris-acetate, 1 mM EDTA, and 0.5 μg/mL ethidium bromide) at 40 V for 4 hr. The digested tomato genomic DNA was transferred on a nylon membrane and hybridized to radiolabeled LeRab11a cDNA. Hybridization was performed in 20 mL of a solution containing 5 × SSPE, 5 × Denhardt's solution, 1% SDS, and 0.2 mg/mL denatured salmon sperm DNA at 65°C for 16 hr. The DNA gel blot was washed at high stringency (twice for 5 min each in 2 × SSC and 0.1% SDS and twice at 65°C for 15 min each in 0.1 × SSC and 0.1% SDS) and exposed to x-ray film (Kodak X-Omat AR).
Plant Transformation
A 960-bp (almost full length) cDNA fragment was cut from pNY650 with EcoRV and XbaI, gel purified, and ligated in a reverse orientation into the SmaI and XbaI sites of pDH51 (between the cauliflower mosaic virus [CaMV] 35S promoter and the CaMV 35S terminator) to give pDHR. This new gene construct was then cut out with PvuII and ligated into the SmaI site of pBIN19 (Bevan, 1984) to produce pBINTR. pBINDR was similar but was produced by cutting the gene construct out of pDHR by a partial EcoRI digest and ligating it into the EcoRI site of pBIN19.
A half-length construct was produced by excising 540 bp of DNA from the 5′ end of the LeRab11 cDNA with EcoRV and BglII and ligating it into pGA643 (An et al., 1988) between the CaMV 35S promoter and terminator, again in the antisense orientation, to give pGATR. The antisense constructs then were transferred from Escherichia coli JM109 to Agrobacterium tumefaciens LBA 4404 by direct transformation. This procedure and the transformation of plants were performed as described by Jones et al. (1998).
PCR
PCR was performed in 50 μL of amplification mixture containing 5 μL of 10 × Taq DNA polymerase buffer (Promega), 15 to 30 ng of template DNA, deoxynucleotide triphosphates at 0.2 mM each, and two primers, each at 1 nM. Reactions were started at 95°C by the addition of 1 unit of Taq polymerase (Promega) followed by 35 incubation cycles of 95°C for 35 sec, 50°C for 1 min, and 72°C for 2 min. The PCR products were analyzed on a 1.2% agarose gel. To prepare hybridization probes, T3 and T7 primers were used. To confirm the presence of the transgene in Agrobacterium and in transformed plants, the primers used were 5′-GGACCCCCACCCACGAGGA-3′ (35S promoter) and 5′-GGGATACTGCTGGTCAAG-3′ (LeRab11a coding sequence).
Analysis of Tomato Fruit Firmness
All assays were performed on freshly harvested fruit. The fruit were tagged at the breaker stage, and fruit were harvested at the required period after the breaker stage (0, 5, 10, 15, 20, and 40 days after the breaker stage). Firmness was measured with a Stevens-LFRA texture analyzer (C. Stevens and Son Ltd., St. Albans, UK) fitted with a circular probe of 13 mm in diameter. A speed of 2 mm/sec was used to compress fruit by 4 mm. Six fruit were used for each treatment, and the experiment was repeated four or five times.
Ethylene Measurements
Fruit were placed in a sealed container at 25°C for 1 hr. One-milliliter samples were withdrawn for ethylene measurement by gas chromatography. The measurements were performed as described by Picton et al. (1993).
Extraction and Assay of PG and PE
Pectinesterase (PE) was extracted as follows. Ten grams of frozen pericarp was ground to a fine powder as described for the extraction of polygalacturonase (PG). Forty milliliters of acetone (−20°C) was added to each sample and stirred using a compressor stirrer (Buddeburg, Mannheim, Germany) for 10 min. The mixture was filtered, using a vacuum, through Whatman glass microfiber No. 1 filter paper. The precipitate was washed with 40 mL of −20°C acetone. The pellet was placed in a desiccator with silica gel and left overnight at room temperature under vacuum. The powder was weighed and resuspended in 20 mL of 1 M NaCl and 50 mM sodium acetate buffer, pH 6.0, and stirred overnight at 4°C. The stirred extract was centrifuged at 10,000g for 30 min. The supernatant was placed in dialysis tubing and then dialyzed overnight against 0.15 M NaCl and 50 mM sodium acetate, pH 6.0, at 4°C. Finally, the supernatant was filtered through Whatman No. 1 filter paper. The volume of filtrate was measured, and the filtrate was stored at 4°C as crude extract. PE activity was assayed titermetrically as described by Tucker et al. (1982). PG was extracted and assayed as reported by Tucker et al. (1980).
Kanamycin Assay of Seedlings
Seed were surface sterilized as described above for plant materials and sown on MSR3 medium supplemented with 75 μg/mL kanamycin. Each pot contained 20 seed. The seed were germinated in a culture room under 16 hr of light at 25°C. After 2 weeks, the seedlings were scored for their ability to germinate and establish roots.
Acknowledgments
We are grateful to Don Grierson for the use of the tomato fruit cDNA library. We acknowledge the support of C.L. in the form of an Overseas Research Studentship award from the Committee of Vice-Chancellors and Principals and other awards from The Henry Lester Trust, The Great Britain-China Educational Trust, and the Gilchrist Educational Trust. Z.Z. was supported by studentship funding from Universiti Kebangsaan Malaysia.
References
- An, G., Ebert, P.R., Mitra, A., and Ha, S.B. (1988). Binary vectors. In Plant Molecular Biology Manual, S.B. Gelvin, R.A. Schilperoort, and D.P.S. Verma, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. A3 1–19.
- Bevan, M.W. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, C.R., Smith, C.J.S., Ray, J.A., Moureau, P., Bevan, M.W., Bird, A.S., Hughes, S., Morris, P.C., Grierson, D., and Schuch, W. (1988). The tomato polygalacturonase gene and ripening-specific expression in transgenic plants. Plant Mol. Biol. 11, 651–662. [DOI] [PubMed] [Google Scholar]
- Bischoff, F., Molendijk, A., Rajendrakumar, C.S.V., and Palme, K. (1999). GTP-binding proteins in plants. Cell. Mol. Life Sci. 55, 233–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg, S., Brandstrup, B., Jensen, T.J., and Poulsen, C. (1997). Identification of new protein species among 33 different small GTP-binding proteins encoded by cDNAs from Lotus japonicus, and expression of corresponding mRNAs in developing root nodules. Plant J. 11, 237–250. [DOI] [PubMed] [Google Scholar]
- Bünemann, M., Lee, K.B., Pals-Rylaarsdam, R., Roseberry, A.G., and Hosey, M.M. (1999). Desensitisation of G-protein-coupled receptors in the cardiovascular system. Annu. Rev. Physiol. 61, 169–192. [DOI] [PubMed] [Google Scholar]
- Chen, W., Feng, Y., Chen, D., and Wandinger-Ness, A. (1998). Rab11 is required for trans-Golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol. Biol. Cell 9, 3241–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8, 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann, G., Sticher, L., Marshallsay, C., and Nagy, F. (1992). Molecular characterisation of tobacco cDNAs encoding two small GTP-binding proteins. Plant Mol. Biol. 19, 847–857. [DOI] [PubMed] [Google Scholar]
- Estruch, J.J., Prinsen, E., Van Onckelen, H., Schell, J., and Spena, A. (1991). Viviparous leaves produced by somatic activation of an inactive cytokinin-synthesising gene. Science 254, 1364–1367. [DOI] [PubMed] [Google Scholar]
- Gray, J., Picton, S., Shabbeer, J., Schuch, W., and Grierson, D. (1992). Molecular biology of fruit ripening and its manipulation with antisense genes. Plant Mol. Biol. 19, 69–87. [DOI] [PubMed] [Google Scholar]
- Hareven, D., Gutfinger, T., Parnis, A., Eshed, Y., and Lifschitz, E. (1996). The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell 84, 735–744. [DOI] [PubMed] [Google Scholar]
- Hobson G.E.. (1963). Pectinesterase in normal and abnormal tomato fruit. Biochem. J. 86, 358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, B.-J., Lund, L., and Sinha, N. (1998). Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 117, 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C.G., Scothern, G.P., Lycett, G.W., and Tucker, G.A. (1998). Co-ordinated gene silencing by a single chimeric gene construct. Planta 204, 499–505. [Google Scholar]
- Kamada, L., Yamauchi, S., Youssefian, S., and Sano, H. (1992). Transgenic tobacco plants expressing rgp1, a gene encoding a ras-related GTP-binding protein from rice, show distinct morphological characteristics. Plant J. 2, 799–807. [Google Scholar]
- Kidou, S., Anai, T., Umeda, M., Aotsuka, S., Tsuge, T., Kato, A., and Uchimiya, H. (1993). Molecular structure of ras-related small GTP-binding protein genes of rice plants and GTPase activities of gene products in Escherichia coli. FEBS Lett. 332, 282–286. [DOI] [PubMed] [Google Scholar]
- Kopeliovitch, E., Mizrahi, Y., Rabinowitch, H.D., and Kedar, N. (1980). Physiology of the tomato mutant alcobaca. Physiol. Plant. 48, 307–311. [Google Scholar]
- Lai, F., Stubbs, L., and Artzt, K. (1994). Molecular analysis of mouse Rab11b: A new type of mammalian YPT/Rab protein. Genomics 22, 610–616. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is ex-pressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6, 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, W.J., Bouché-Pillon, S., Jackson, D.P., Nguyen, L., Baker, L., Ding, B., and Hake, S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270, 1980–1983. [DOI] [PubMed] [Google Scholar]
- Moore, R.H., Tuffaha, A., Millman, E.E., Dai, W., Hall, H.S., Dickey, B.F., and Knoll, B.J. (1999). Agonist-induced sorting of human β2-adrenergic receptors to lysosomes during downregulation. J. Cell Sci. 112, 329–338. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Mutschler, M.A. (1984). Ripening and storage characteristics of the alcobaca ripening mutant in tomato. J. Am. Soc. Hortic. Sci. 109, 504–507. [Google Scholar]
- Mutschler, M.A., Gutteri, M., Kinzer, S., Grierson, D., and Tucker, G.A. (1988). Changes in ripening-related processes in tomato conditioned by the alc mutant. Theor. Appl. Genet. 76, 285–292. [DOI] [PubMed] [Google Scholar]
- Nagano, Y., Murai, N., Matsuno, R., and Sasaki, Y. (1993). Isolation and characterisation of cDNAs that encode eleven small GTP-binding proteins from Pisum sativum. Plant Cell Physiol. 34, 447–455. [PubMed] [Google Scholar]
- Olkkonen, V.M., and Stenmark, H. (1997). Role of rab GTPases in membrane traffic. Int. Rev. Cytol. 176, 1–85. [DOI] [PubMed] [Google Scholar]
- Picton, S., Gray, J., Barton, S. AbuBakar, U., Lowe, A., and Grierson, D. (1993). cDNA cloning and characterisation of novel ripening related mRNAs with altered patterns of accumulation in the ripening inhibitor (rin) tomato ripening mutant. Plant Mol. Biol. 23, 193–207. [DOI] [PubMed] [Google Scholar]
- Pneuli, L., Carmel-Goren, L., Hareven, D., Gutfinger, T., Alvarez, J., Ganal, M., Zamir, D., and Lifschitz, E. (1998). The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125, 1979–1989. [DOI] [PubMed] [Google Scholar]
- Ray, J., Knapp, J., Grierson, D., Bird, C., and Schuch, W. (1988). Identification and sequence determination of a cDNA clone for tomato pectin esterase. Eur. J. Biochem. 174, 119–124. [DOI] [PubMed] [Google Scholar]
- Rick, C.M., and Butler, L. (1956). Cytogenetics of the tomato. Adv. Genet. 8, 267–382. [Google Scholar]
- Sanderfoot, A.A., and Raikhel, N.V. (1999). The specificity of vesicle trafficking: Coat proteins and SNAREs. Plant Cell 11, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, H., Seo, S., Orudgev, E., Youssefian, S., Ishizuka, K., and Ohashi, Y. (1994). Expression of the gene for a small GTP binding protein in transgenic tobacco elevates endogenous cytokinin levels, abnormally induces salicylic-acid in response to wounding, and increases resistance to tobacco mosaic-virus infection. Proc. Natl. Acad. Sci. USA 91, 10556–10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, N.R., Williams, R.E., and Hake, S. (1993). Overexpression of the maize homeobox gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 7, 787–795. [DOI] [PubMed] [Google Scholar]
- Takai, Y., Sasaki, T., and Matozaki, T. (2001). Small GTP-binding proteins. Physiol. Rev. 81, 153–208. [DOI] [PubMed] [Google Scholar]
- Tamaoki, M., Kusaba, S., Kano-Murakami, Y., and Matsuoka, M. (1997). Ectopic expression of a tobacco homeobox gene, NTH15, dramatically alters leaf morphology and hormone levels in transgenic tobacco. Plant Cell Physiol. 38, 917–927. [DOI] [PubMed] [Google Scholar]
- Tucker, G.A., Robertson, N.G., and Grierson, D. (1980). Changes in polygalacturonase isoenzymes during the ‘ripening’ of normal and mutant tomato fruit. Eur. J. Biochem. 112, 119–124. [DOI] [PubMed] [Google Scholar]
- Tucker, G.A., Robertson, N.G., and Grierson, D. (1982). Purification and changes in activities of tomato pectinesterase isoenzymes. J. Sci. Food Agric. 33, 396–400. [Google Scholar]
- van der Krol, A.R., Brunelle, A., Tsuchimoto, S., and Chua, N.-H. (1993). Functional analysis of petunia floral homeotic MADS box gene pMADS1. Genes Dev. 7, 1214–1228. [DOI] [PubMed] [Google Scholar]
- Wilkinson, J.Q., Lanahan, M.B., Yen, H.-C., Giovannoni, J.J., and Klee, H.J. (1995). An ethylene-inducible component of signal transduction encoded by Never-ripe. Science 270, 1807–1809. [DOI] [PubMed] [Google Scholar]
- Zainal, Z., Tucker, G.A., and Lycett, G.W. (1996). A rab11-like gene is developmentally regulated in ripening mango (Mangifera indica L.) fruit. Biochim. Biophys. Acta 1314, 187–190. [DOI] [PubMed] [Google Scholar]